Abstract
A subunit of the preprotein translocon of the outer envelope of chloroplasts (Toc complex) of 64 kD is described, Toc64. Toc64 copurifies on sucrose density gradients with the isolated Toc complex. Furthermore, it can be cross-linked in intact chloroplasts to a high molecular weight complex containing both Toc and Tic subunits and a precursor protein. The 0 Å cross-linker CuCl2 yields the reversible formation of disulfide bridge(s) between Toc64 and the established Toc complex subunits in purified outer envelope membranes. Toc64 contains three tetratricopeptide repeat motifs that are exposed at the chloroplast cytosol interface. We propose that Toc64 functions early in preprotein translocation, maybe as a docking protein for cytosolic cofactors of the protein import into chloroplasts.
Keywords: protein import, Toc complex, chloroplasts, outer envelope, tetratricopeptide repeat motif
Introduction
Most chloroplastic proteins are nuclear encoded, synthesized in the cytosol, and posttranslationally imported into the organelle. Sorting in plant cells is complex, as mitochondria and chloroplasts or plastids coexist in every cell. Mitochondrial and chloroplast proteins are therefore synthesized in the cytosol as preproteins with an NH2-terminal targeting sequence, which is both necessary and sufficient for directing the preproteins to the correct organelle. Chloroplast targeting signals are loosely characterized by three distinct domains (von Heijne et al. 1989). The N-proximal portion is devoid of positively charged residues as well as the amino acids glycine and proline. The central region is rich in the hydroxylated amino acids serine and threonine and overall is positively charged. The COOH-terminal part is predicted to form an amphiphilic β-strand. The precursor forms of prominent chloroplast proteins like the small subunit of stromal Rubisco (SSU), the thylakoid a/b binding protein (LHCP), or the thylakoid lumen localized subunits of the oxygen evolving complex of 23 kD and 33 kD (OE23, OE33) contain a phosphorylation site within the chloroplast targeting signal (Waegemann and Soll 1996). A phosphorylated precursor protein can still bind to the chloroplast import machinery, but must become dephosphorylated before translocating through the membranes. The protein kinase is found in the cytosol of pea mesophyll cells or in wheat germ extract. When preSSU or preOE33 are synthesized in a wheat germ lysate, they form an oligomeric guidance complex with other proteins, two of which are hsp70 and 14-3-3 (May and Soll 2000). Formation of the guidance complex requires an intact phosphorylation site in the chloroplastic precursor protein. PreSSU present in this complex imports with a fourfold higher rate into chloroplasts than the nonbound free precursor form.
Recognition and translocation of different precursor proteins is initiated by subunits of the so-called translocon at the outer envelope of chloroplasts (Toc complex). Established subunits of the Toc complex are: Toc160, a large nucleotide binding protein that could function as a receptor component (Hirsch et al. 1994; Perry and Keegstra 1994; Bölter et al. 1998; Bauer et al. 2000); Toc75, which forms the aqueous preprotein translocation channel (Perry and Keegstra 1994; Hinnah et al. 1997); and Toc34, a second nucleotide binding protein that might regulate precursor protein binding and translocation (Kessler et al. 1994; Seedorf et al. 1995, Jarvis et al. 1998). Although it is likely that more polypeptides are involved in building the Toc complex, none have been identified so far.
The Toc complex acts jointly with the translocon of the inner envelope of chloroplasts (Tic complex) to fully import a precursor protein into the stroma, where processing to the mature form occurs. Several subunits of the Tic complex have been identified. Tic110 might be involved in the formation of joint translocation sites and in recruiting Hsp93 as well as chaperonin 60 to the stromal site of the translocon (Kessler and Blobel 1996; Lübeck et al. 1996; Akita et al. 1997; Nielsen et al. 1997). Tic55 contains a Rieske-type iron sulfur center and a mononuclear binding site and might act as a redox regulator for the functional state of the translocon (Caliebe et al. 1997). Tic40 is a recently described subunit of the inner envelope import machinery (Stahl et al. 1999), with homologies to the previously described Cim/Com44 (Ko et al. 1995). Tic22 is a peripheral protein at the lumenal site of the membrane but of unknown function. Tic20 is proposed to form part of the translocation channel (Kouranov et al. 1998).
To our knowledge all precursor proteins that can be posttranslationally imported into chloroplasts use this general import pathway via the Toc and Tic complexes. Interaction of the precursor proteins with the Toc complex requires ATP as well as GTP (Olsen et al. 1989; Kouranov and Schnell 1997; Young et al. 1999). Both nucleotides might be required at multiple sites, since Toc160 and Toc34 are both GTP binding proteins (Kessler et al. 1994; Seedorf et al. 1995), and Toc160 as well as the Toc-associated hsp70 most likely requires ATP for their action. ATP <50 μM results in precursor protein binding that engages both the Toc and the Tic complex (Waegemann and Soll 1991). Complete translocation is efficient only at ATP concentrations >100 μM in the stroma, which probably fuels the action of stromal chaperones in pulling the preproteins across the membranes (Flügge and Hinz 1986; Schindler and Soll, 1986; Olsen et al. 1989; Theg et al. 1989).
Membrane protein complexes containing Toc as well as Tic subunits can be enriched from intact chloroplasts by detergent solubilization either in the presence or absence of precursor protein (Soll and Waegemann 1992; Schnell et al. 1994). These findings suggest that at least some import sites are rather stationary and do not form exclusively upon translocating of a precursor protein. A partially purified Toc complex can be enriched from isolated chloroplast outer envelope membranes (Waegemann and Soll 1991). This complex contained the Toc subunits Toc160 (respectively its proteolytic fragment of 86 kD), Toc75, Toc34, and hsp70, as well as some Tic110 and the large and small Rubisco subunits as contaminants. A few unidentified proteins were also detectable in the isolated active Toc complex. One of these proteins had an apparent molecular mass of 64 kD. Here we describe this chloroplast outer envelope protein of 64 kD as a new subunit of the complex and propose to name it Toc64. Toc64 exposes a large polypeptide domain, including three tetratricopeptide repeats (TPR) on the cytosolic surface of the organelle, which might interact with cytosolic proteins involved in translocation.
Materials and Methods
Isolation of a cDNA Clone of Toc64
Purified outer envelope membrane proteins were separated by SDS-PAGE. After staining with Coomassie brilliant blue, the putative Toc64-containing protein band was excised from the gel and treated with cyanogen bromide (Sigma Chemical Co.) or endoproteinase Glu-C (Roche Diagnostics) to obtain peptide fragments. The incubation mixture was separated on a second SDS-PAGE, polypeptides transferred to polyvinylidene difluoride membrane, and the amino acid sequence determined by standard procedures. The NH2-terminal sequence of Toc64 was determined directly without further treatment. The NH2-terminal and internal sequences received exhibited homologies to the Arabidopsis thaliana cDNA expressed sequence tag (EST) clone 180A6T7 (sequence data available from EMBL/GenBank/DDBJ under accession no. H36863). A pea cDNA expression library (UniZAPXR; Stratagene) was screened using digoxigenin-labeled PCR product obtained from the first 150 coding nucleotides of the Arabidopsis thaliana EST clone. A full-length pea Toc64 cDNA clone was isolated and both DNA strands were sequenced (Sanger et al. 1977).
Cloning and Overexpression
To engineer pea Toc64 with a six-histidine tag at its COOH terminus, flanking NdeI and XhoI restriction sites were introduced via PCR and used for cloning into the expression vector pET21b (Novagen). The plasmid was transferred into Escherichia coli BL21 (DE3) cells (Novagen). The resulting overexpressed Toc64his was purified by chromatography on Ni-NTA agarose according to the manufacturer's recommendation (Qiagen). The same procedure was used to overexpress the amino acids 1–476, which lacked the TPR motif–containing domain (Toc64hisΔTPR) and the COOH-terminal amino acids 477–593 consisting of the three tetratricopeptide repeats (Toc64TPRhis).
Isolation of Chloroplasts and Chloroplast Membrane Vesicles
Intact chloroplasts were isolated from 10–12-d-old pea seedlings (Pisum sativum Linnee, variety Golf) as described before (Waegemann and Soll 1991). Outer and inner envelope vesicles were purified from pea chloroplasts (Keegstra and Yousif 1986). Thylakoid membranes were washed at least twice (50 mM NaPi buffer, pH 7.4, and 10 mM NaCl) before use. Mixed envelope vesicles were enriched after hypotonic lysis of chloroplasts in Tris buffer (5 mM Tris/HCl, pH 7.5, and 1 mM EDTA) for 30 min on ice, followed by a thylakoid sedimentation step through a 1-M sucrose cushion in Tris buffer for 1 h at 330,000 g. The enriched envelope vesicles were recovered from the sucrose-buffer interphase, diluted, and washed twice with Tris buffer.
Precursor Binding and Chemical Cross-linking
Wheat germ lysate was isolated as described (Anderson et al. 1983) and used in vitro for protein translation in the presence of [35S]methionine and [35S]cysteine (1.175 Ci/mmol) for 75 min at 25°C. Binding experiments contained 40 μl translation product and intact chloroplasts equivalent to 750 μg chlorophyll in 1.5 ml binding buffer (330 mM sorbitol, 50 mM Hepes/KOH, pH 7.6, 3 mM MgCl2, 100 μM ATP, 20 mM potassium gluconate, 10 mM methionine, 10 mM cysteine, 10 mM NaHCO3, and 2% [wt/vol] BSA) for 10 min on ice. The chloroplasts were recovered through a 40% (vol/vol) Percoll cushion (Schindler et al. 1987) and washed once in wash medium (330 mM sorbitol, 50 mM Hepes/KOH, pH 7.6, and 3 mM MgCl2) before cross-linking was performed with 1 mM CuCl2 in 1.5 ml wash medium on ice for 15 min. The reaction was stopped with 5 mM EDTA, and the chloroplasts were reisolated in wash medium containing 1 mM EDTA by centrifugation.
Outer envelope membrane vesicles equivalent to 40 μg protein were sedimented for 10 min at 160,000 g, resuspended in Hepes buffer (25 mM Hepes/KOH, pH 7.6, and 3 mM MgCl2), and cross-linked for 15 min on ice with CuCl2 or dithiobis[succinimidyl propionate] (DSP; Pierce), respectively, in a total volume of 60 μl (Akita et al. 1997). The proteins were separated on a nonreducing 4 to 10% polyacrylamide gradient SDS gel with a 60:1 acrylamide/bisacrylamide ratio. Cross-linking products were visualized with Coomassie brilliant blue, excised from the gel, and cleaved with 20 mM DTT for 30 min. After a second reducing 10 to 12.5% gradient SDS-PAGE, the proteins were analyzed by silverstaining or by blotting and immunodecoration, respectively.
Protease Treatment
Chloroplasts were treated with 1 μg thermolysin per 2 μg chlorophyll for 30 min on ice in wash medium containing 0.5 mM CaCl2. After addition of 10 mM EDTA, intact chloroplasts were reisolated through a 40% (vol/vol) Percoll cushion with 10 mM EDTA and washed twice in wash medium. Afterwards, mixed envelope vesicles were purified for further analysis. Thermolysin treatment of envelope membrane vesicles was carried out for 2 min at 25°C in 25 mM Hepes/KOH, pH 7.6, 5 mM MgCl2, and 0.5 mM CaCl2, with a ratio of 1 μg thermolysin per 2 μg protein. The reaction was stopped with 10 mM EDTA and membranes recovered by centrifugation at 160,000 g for 10 min. The pellet was resuspended in 25 mM Hepes/KOH, pH 7.6, and 10 mM EDTA.
Raising of Antiserum and Immunoprecipitation
An antiserum was raised in a rabbit against heterologously expressed Toc64his using Freund's complete adjuvant as immune stimulans. An aliquot of the αToc64 antiserum was incubated with recombinant Toc64ΔTPRhis or Toc64TPRhis, respectively, which had been covalently coupled to cyanogen bromide-activated Sepharose 4B (Amersham Pharmacia Biotech). The flow through was used as a peptide-specific serum, i.e., αToc64ΔTPR and αToc64TPR. The binding specificity was controlled in immunoblotting assays using Toc64his, Toc64ΔTPRhis, and Toc64-TPRhis. For immunoprecipitation experiments, αToc64 or preimmune sera were used (Mason et al. 1988).
Miscellaneous Methods
Extraction of peripheral proteins from membrane vesicles was accomplished by 1 mM NaCl, pH 11.5 (0.5 M Na2CO3), or 4 M urea, respectively, for 10 min on ice, followed by a separation into soluble and insoluble proteins by centrifugation for 10 min at 160,000 g. Outer envelope membrane vesicles were solubilized by digitonin and fractionated on a 5 to 20% (wt/vol) sucrose density gradient as detailed before (Waegemann and Soll 1991). Protein concentrations were determined as described (Lowry et al. 1951). Proteins were separated by 12.5% (acrylamide/bisacrylamide ratio of 30:1) SDS-PAGE (Laemmli 1970), if not indicated otherwise. Chlorophyll concentrations were measured by the method of Arnon 1949.
Results
To identify further components of the chloroplast protein import apparatus, outer envelope membranes were solubilized with digitonin and fractionated on a sucrose density gradient as described before (Waegemann and Soll 1991). Fractions obtained from the gradient were analyzed by SDS-PAGE and polypeptides visualized by silverstaining. A protein with an apparent molecular mass of 64 kD, now named Toc64, cofractionated with the established Toc subunits, namely Toc34, Toc75, and the 86-kD fragment of Toc160 (Fig. 1, Toc). The small and large subunits of Rubisco (SSU and LSU), the most abundant stromal protein, were also present as contaminants in this fraction at ∼14 kD and 54 kD, respectively (Waegemann and Soll 1991). Toc64 is also detectable in total outer envelope proteins (Fig. 1, OE). Toc64 was purified by SDS-PAGE from outer envelope membrane vesicles, and an NH2-terminal as well as internal amino acid sequences were obtained. A dbest search using the peptide sequence information led to an EST cDNA from Arabidopsis thaliana (sequence data available from EMBL/GenBank/DDBJ under accession no. H36863). Heterologous screening of a pea cDNA expression library with this cDNA resulted in the isolation of a cDNA clone (sequence data available from EMBL/GenBank/DDBJ under accession no. AF179282) from pea coding for Toc64. The isolated full-length clone had an open reading frame coding for 593 amino acids with a calculated molecular mass of 64.5 kD (Fig. 2 a). All nine peptide sequences were found in the deduced open reading frame, verifying that the isolated pea cDNA clone encodes the 64 kD protein.
Figure 1.
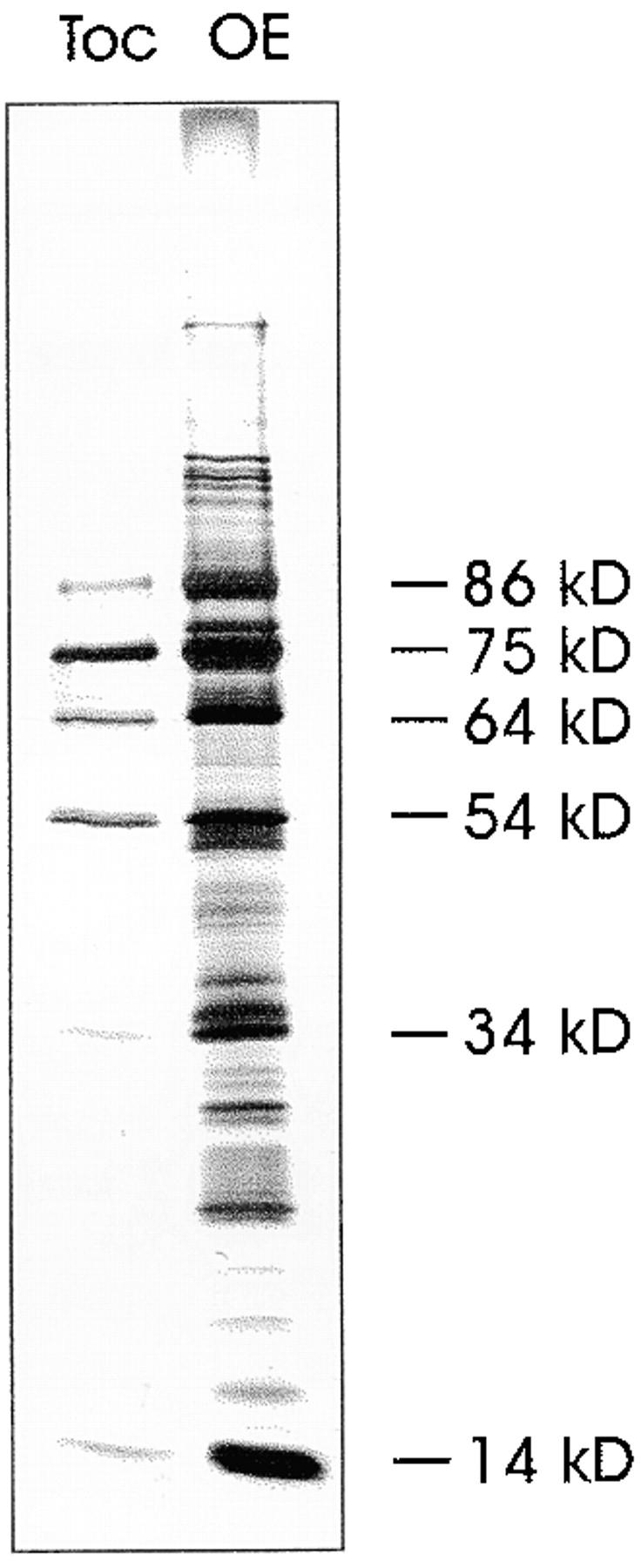
Polypeptide composition of isolated pea chloroplast outer envelope (OE) membranes and the purified Toc complex. A silverstained SDS-PAGE is shown.
Figure 2.
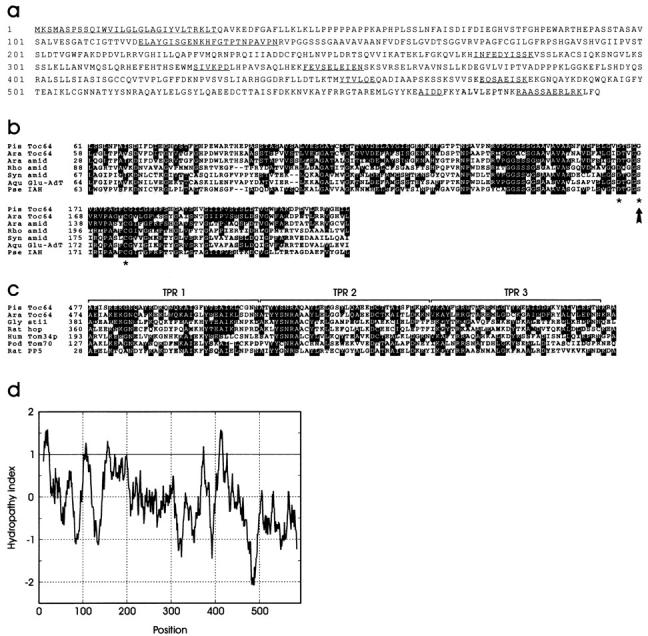
Sequence analysis of Toc64. (a) Protein sequence of pea Toc64 as deduced from the cDNA clone. The determined amino acid sequences are underlined. These sequence data are available from GenBank/EMBL/DDBJ under accession no. AF179282. (b) Sequence comparison of pea (Pis) Toc64 with a Toc64 homologue from Arabidopsis thaliana (Ara; sequence data available from EMBL/GenBank/DDBJ under accession no. H36863), amidases (amid) from Arabidopsis thaliana (sequence data available from EMBL/GenBank/DDBJ under accession no. AC000106), Rhodococcus rhodochrous (Rho; sequence data available from EMBL/GenBank/DDBJ under accession no. D16207), and Synechocystis sp. (Syn; sequence data available from EMBL/GenBank/DDBJ under accession no. D90907), Aquifex aeolicus glutamyl-tRNAGln amidotransferase subunit A (Aqu Glu-AdT; sequence data available from EMBL/GenBank/DDBJ under accession no. AE000680), and Pseudomonas syringae indoleacetamide hydrolase (Pse IAH; sequence data available from EMBL/GenBank/DDBJ under accession no. U04358) using the ClustalW 1.7 and BOXSHADE programs. The active residues of the amidases are indicated by asterisks. The arrow shows the amino acid exchange of Toc64. (c) Sequence alignment of TPR motifs of different polypeptides: pea Toc64, its homologue from Arabidopsis thaliana, hop from Rattus norvegicus (Rat; sequence data available from EMBL/GenBank/DDBJ under accession no. CAA75351) and its plant homologue Sti1 from Glycine max (Gly; sequence data available from EMBL/GenBank/DDBJ under accession no. S56658), human hTom34p (Hum; sequence data available from EMBL/GenBank/DDBJ under accession no. U58970), Podospora anserina (Pod) Tom70 (sequence data available from EMBL/GenBank/DDBJ under accession no. Y14750), and serine/threonine protein phosphatase 5 (PP5) from Rattus norvegicus (sequence data available from EMBL/GenBank/DDBJ under accession no. P53042) using the ClustalW 1.7 and BOXSHADE programs. (d) Hydropathy analysis of Toc64 according to Kyte and Doolittle 1982 using a window size of 19 amino acids.
Comparison of the coding sequence of Toc64 with known proteins in the database revealed distinct homologies to two different polypeptide classes. One is the relation to amidases and indole acetamide hydrolases (Fig. 2 b). However, one of three essential active site residues (Kobayashi et al. 1997; Fig. 2 b, indicated by asterisks) has been changed in Toc64 from a serine to a glycine (Fig. 2 b, indicated by an arrow). Enzymatic assays of recombinant Toc64 to test for amidase or amidotransferase activity were not successful (data not shown). Furthermore, amidases and related proteins are generally soluble proteins. However, Toc64 reveals several areas of high hydrophobicity (Fig. 2 d) that could anchor the protein into a membrane. The most prominent hydrophobic motifs at the extreme NH2-terminus and between amino acids 410–425 have especially no resemblance to sequences of amidases. The Arabidopsis sequence data base reveals two EST sequences, one (sequence data available from EMBL/GenBank/DDBJ under accession no. H36863) that has higher homology to pea Toc64 and contains the same amino acid exchange, and a second with similar homology that contains the conservative serine at the active site (sequence data available from EMBL/GenBank/DDBJ under accession no. AC000106) (Fig. 2 b), thus indicating that Toc64 might no longer serve as an enzyme with amidase or related function. The second module that we detected at the COOH-terminal part of Toc64 is a threefold repeat of a TPR motif (Fig. 2 c). The TPR motifs have been implied in mediating protein–protein interaction (Goebl and Yanagida 1991; Lamb et al. 1995). They are found in a wide range of proteins, including protein import receptors of mitochondria and peroxisomes, i.e., translocon at the outer membrane of mitochondria (Tom) 70 (Hase et al. 1983), hTom34 (Nuttall et al. 1997), and Pex5 (Brocard et al. 1994), respectively.
We wanted to obtain experimental evidence for the outer envelope localization of Toc64. This was done in two ways. An antiserum was raised against the heterologously expressed protein containing a hexa-histidine tag, Toc64his, after purification over a metal chelating matrix. The antiserum recognized polypeptides in the stroma as well as in outer envelope vesicles (Fig. 3 a). Neither thylakoids nor inner envelope membranes contained any immunoreactive proteins. A polypeptide at 64 kD was recognized by the antiserum in the purified outer envelope fraction (Fig. 3 a, lane 4). To exclude a contamination of the outer envelope membrane by the stromal proteins, the membranes were extracted with 1 M NaCl, 0.5 M Na2CO3, or 4 M urea. In every case Toc64 was not extractable and behaved like an integral membrane protein (Fig. 3 b, P and S). A soluble stromal contaminant migrated as a doublet between 60–64 kD on SDS-PAGE (Fig. 3 a, lane 1), which could be the α and β subunit of chaperonin 60 (Musgrove et al. 1987). To test if the soluble protein recognized by αToc64 is indeed identical with chaperonin 60, the soluble stroma fraction was subjected to centrifugation (150,000 g, 12 h) in order to pellet the native chaperonin complex that has a molecular mass of 840 kD. Centrifugation should result in less immunoreactive material in the stroma. This was found to be the case when both soluble protein fractions were analyzed with Toc64 antiserum (Fig. 3 a, lanes 5 and 6). Furthermore, an antiserum raised against chloroplastic chaperonin 60 clearly recognized a polypeptide doublet in the stromal fraction with identical running behavior as αToc64. Again, the immunoreactive proteins were largely removed by centrifugation (Fig. 3 a, lanes 7 and 8). We thus conclude that the stromal proteins recognized by αToc64 are the α and β subunits of chaperonin 60.
Figure 3.
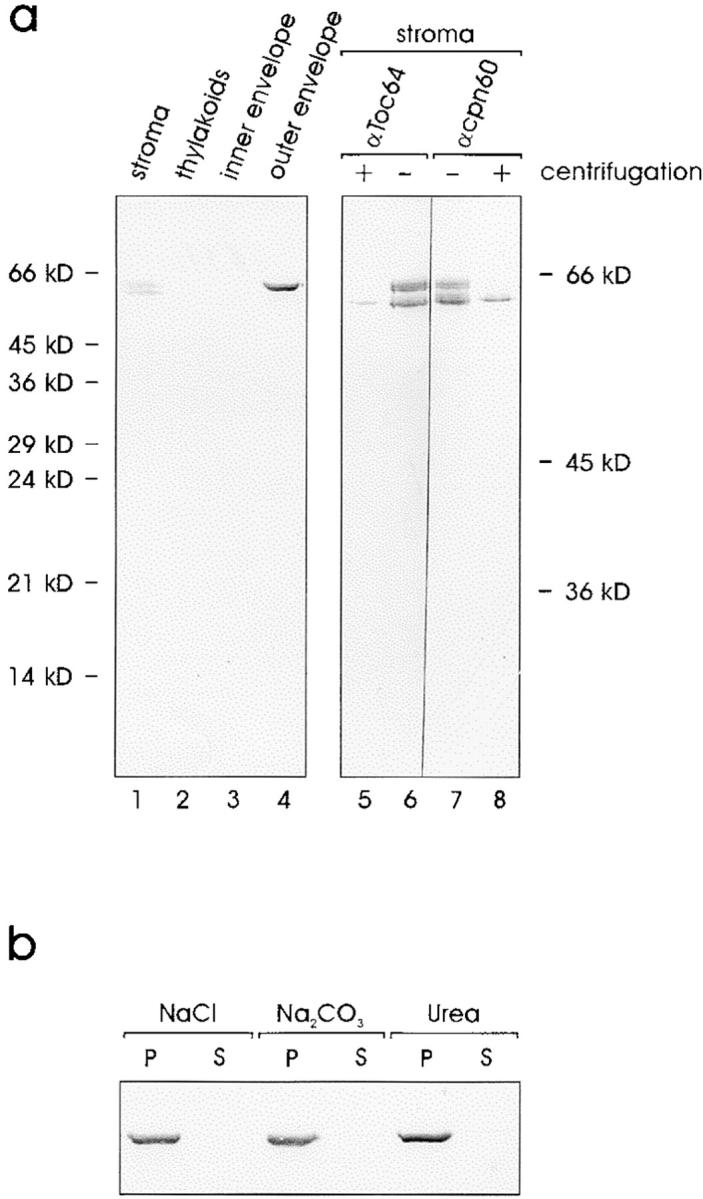
Toc64 is an integral protein of the outer envelope of pea chloroplasts. (a) Immunoblot analysis of the different subcompartments using αToc64 antiserum. Proteins were separated on a 12.5% (wt/vol) acrylamide containing running gel (lanes 1–4). Total soluble chloroplast proteins were immunodecorated with αToc64 (lanes 5 and 6) or with antiserum against chaperonin 60 (lanes 7 and 8) before or after centrifugation. Proteins were separated on a 15% (wt/vol) acrylamide containing running gel at 35 mA for 5 h. (b) Purified outer envelope membranes were treated with 1 mM NaCl, 0.5 M Na2CO3, or 4 M urea as indicated and separated into soluble (S) and insoluble (P) protein fractions. An immunoblot is shown.
Is the 64-kD protein detected in the purified Toc complex (Fig. 1) identical to the outer envelope polypeptide for which we obtained the cDNA (Fig. 2 a)? To address this question the protein composition of the purified Toc complex was analyzed by immunoblotting (Fig. 4). The αToc64 antiserum recognized a polypeptide of the expected size in the enriched Toc complex fraction, which also contained Toc160 and its proteolytic fragments (Waegemann et al. 1992; Bauer et al. 2000), Toc75 and Toc34, but not the outer envelope protein OEP16, a protein not associated with the general import pathway. We conclude that the 64-kD protein seen in the isolated Toc complex is identical to the protein encoded by the cDNA described in Fig. 2. To obtain further evidence of the presence of Toc64 in the import machinery, isolated outer envelope membranes were treated with the thiol oxidant CuCl2 and the NH2-group selective thiol cleavable cross-linker DSP. SDS-PAGE revealed that several high molecular weight cross-linking products had been formed (Fig. 5 a, lanes Cu2+ and DSP). CuCl2 and DSP formed cross-linked products of similar size, i.e., ∼600 kD and ∼300 kD, respectively (Fig. 5 a). The complexes were formed as a function of the CuCl2 cross-linker concentration, indicating the selectivity of cross-linking (Fig. 5 b). The disulfide bond formed by oxidation in the presence of Cu2+ has a length of ∼2 Å, indicating that the constituents of such complexes are in close physical proximity. Gel slices containing prominent high molecular complexes (Fig. 5, a and b, indicated by arrows) were incubated under reducing conditions to cleave the cross-links, and run on a second SDS-PAGE. One half of the sample was stained with silver (Fig. 5 c, left panel) and the other half was used for immunoblotting (Fig. 5 c, right panel). Silverstaining revealed that complexes 1–4 contained Toc160 or its proteolytic fragment of 86 kD (Toc160*), Toc75, Toc34, and Toc64 as the only prominent polypeptides. Minor contaminations can be seen around 50–55 kD. The nature of these proteins remains to be determined. The identities of the Toc subunits was confirmed by immunoblotting (Fig. 5 c, right panel). Nonproteolysed Toc160 was mainly present in the high molecular weight complexes 1 and 2. We do not believe that this is responsible for the size difference observed between complexes 1 and 2 and complexes 3 and 4, respectively. Complexes 1 and 2 might represent a Toc dimer, whereas 3 and 4 are probably Toc monomers.
Figure 4.
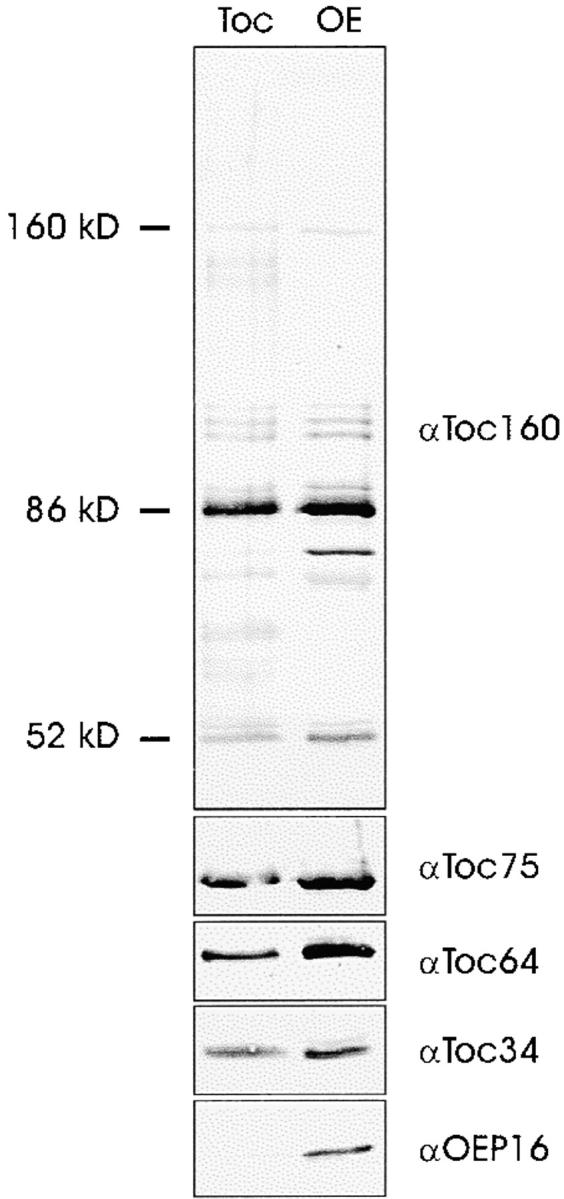
Immunoblot analysis of the purified Toc complex or isolated outer envelope membranes using the antisera indicated. The position of Toc160 and its typical proteolytical fragments is indicated. Identical samples were separated in distinct SDS-PAGE lanes and analyzed with antisera against Toc160, or Toc75, Toc64, Toc34, and OEP16, respectively.
Figure 5.
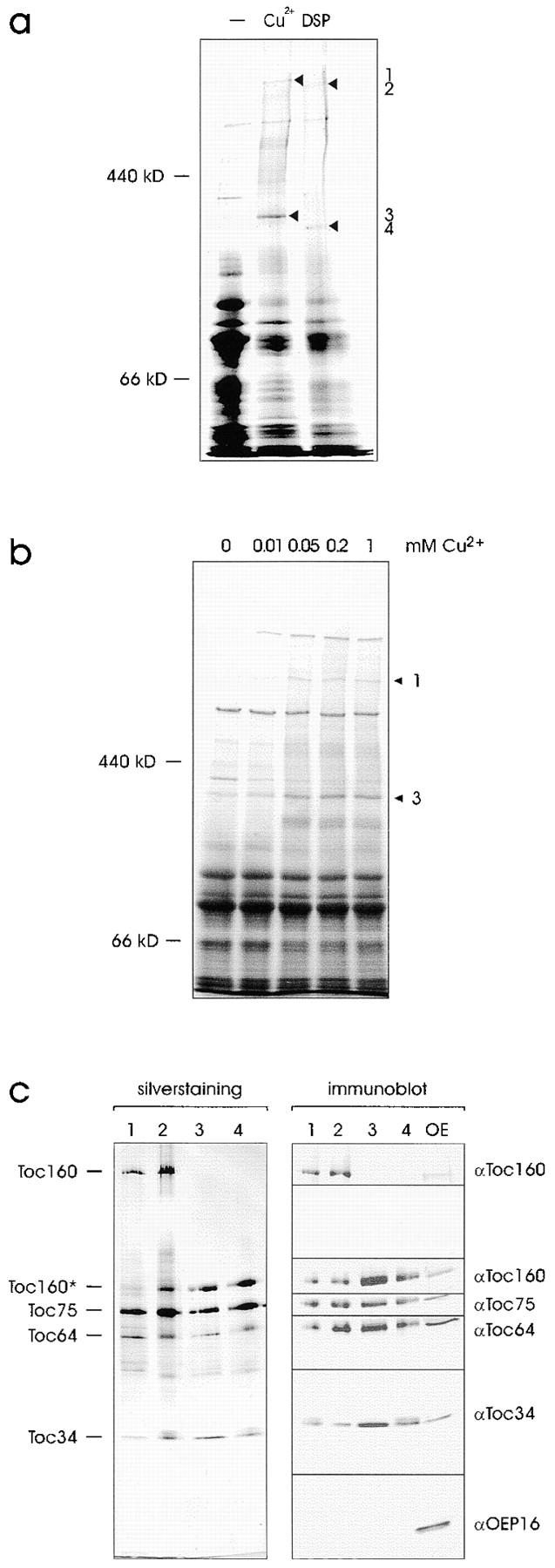
Toc64 can be cross-linked to other Toc complex subunits. (a) Outer envelope membranes (40 μg protein) were either not treated or treated with 1 mM CuCl2 or 1 mM DSP. (b) High molecular weight cross-links form in a concentration-dependent manner in the presence of CuCl2. Cross-linking was performed for 15 min using the concentrations indicated. Cross-linked products were separated by SDS-PAGE and visualized by staining with Coomassie brilliant blue. Numbers 1 and 3 label identical bands as in a. (c) Prominent cross-linked products (bands 1–4 in a) were cut out of the gel, divided into equal aliquots, and separated by a second SDS-PAGE under reducing conditions. Proteins were analyzed either by silverstaining (left panel) or immunoblotting using the antisera indicated (right panel).
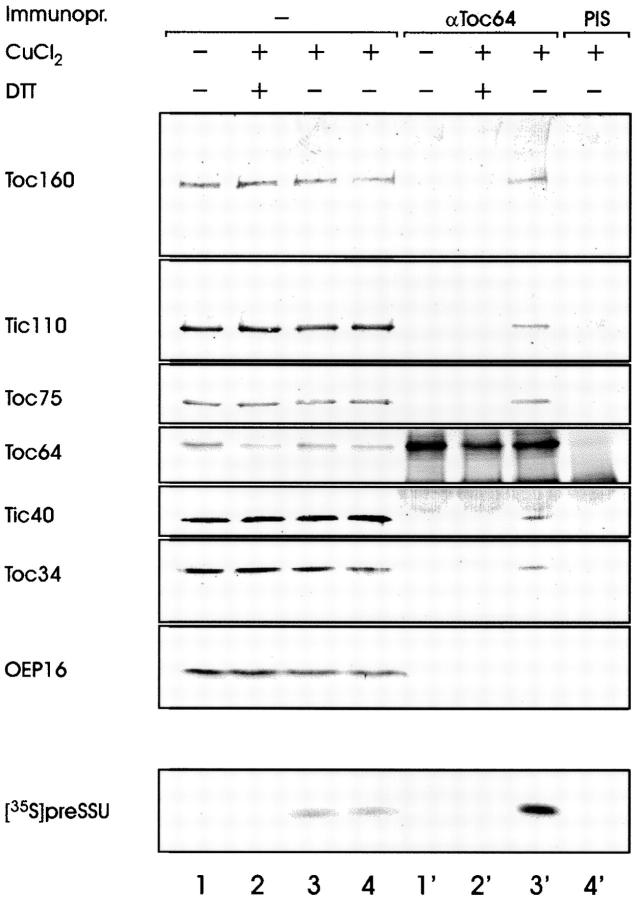
To obtain further evidence for the involvement of Toc64 in protein translocation, intact pea chloroplasts were incubated with preSSU in the presence of 100 μM ATP at 4°C. Under these conditions the precursor engages both the Toc and the Tic complex but it cannot be fully translocated due to limitations in the ATP level. Chloroplasts were purified from the binding experiment, washed, and incubated with 1 mM CuCl2 for 15 min. After addition of 15 mM EDTA, organelles were lysed in hypotonic buffer and a total membrane fraction recovered by centrifugation. Membranes were extracted with 4 M urea and centrifuged through a 1-M sucrose cushion. Envelope membranes were recovered from the 1-M sucrose interface, dissolved in SDS, and subjected to immunoprecipitation using preimmune or αToc64 antiserum. The Toc64 antiserum was able to coimmunoprecipitate [35S]preSSU only in the presence of cross-linking reagent (compare Fig. 6, lanes 1′ and 3′). Together with preSSU, the translocon subunit Toc160, Tic110, Tic40, and Toc34 could be detected in the precipitate. The outer envelope protein OEP16 was absent. When the cross-linked membranes were treated with DTT before the immunoprecipitation to convert the disulfide bridges back to thiol groups, only Toc64 was precipitated by αToc64 (Fig. 6, lane 2′). These results indicated that the complex was formed only by disulfide bridges. Taken together the data presented in Fig. 5 and Fig. 6 provide strong evidence that Toc64 is a bona fide subunit of the chloroplast protein import machinery.
Figure 6.
Toc64 is recovered in a precursor protein containing membrane complex. Intact pea chloroplasts were incubated with 35S-labeled preSSU at 4°C and 100 μM ATP for 5 min. Chloroplasts were recovered from the incubation mixture, washed, and subjected to cross-linking in the absence or presence of 1 mM CuCl2. Organelles were lysed, the membrane fractions extracted with 4 M urea, and envelope membranes enriched by centrifugation on a sucrose cushion. Envelope membranes were incubated with or without 20 mM DTT, washed, solubilized in SDS, and used for coimmunoprecipitation by αToc64 or preimmune serum as indicated. Lanes 1–4 contain 3% of the reaction mixture that was used in the immunoprecipitations presented in lanes 1′–4′. The immunoprecipitates were further analyzed by SDS-PAGE followed by immunoblotting using the antisera indicated or for the presence of [35S]preSSU by autoradiography.
Proteins destinated for the stroma import via the general import pathway, whereas outer envelope proteins insert independently of a cleavable transit sequence in general but by internal targeting information. After incubation of intact chloroplasts with radiolabeled Toc64 translation product, the protein associated with the organelles without an obvious shift in molecular size (Fig. 7 a, lanes 1 and 2). This indicates that Toc64 does not contain a cleavable targeting signal that directs it into the stroma. Toc64 also inserted into purified outer envelope vesicles (Fig. 7 a, lane 4). Protease treatment of chloroplast or envelope-associated radiolabeled Toc64 resulted in similar fragmentation patterns (Fig. 7 a, lanes 3 and 5), indicating a similar insertion pathway into the organelle and into isolated membranes. Chloroplast-associated radiolabeled Toc64 was resistant to extraction at pH 11 (data not shown). These results further support that the cDNA clone described in Fig. 2 is full-length and that Toc64 is a genuine outer envelope protein.
Figure 7.
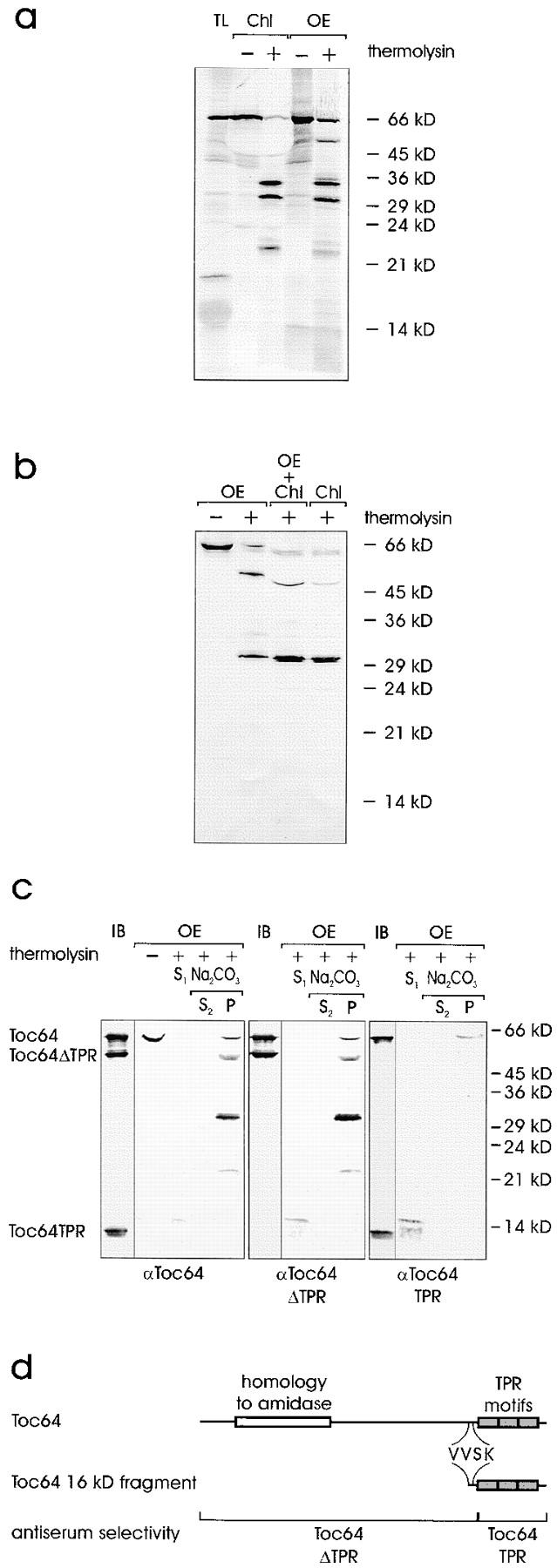
Topological arrangement of Toc64 in the outer envelope. (a) Insertion of 35S-labeled Toc64 translation product (TL) either into pea chloroplasts (Chl) or isolated outer envelope membranes (OE). After insertion, organelles or membranes were either not treated or treated with the protease thermolysin as indicated. TL, 10% of translation product added to an insertion assay. A fluorogram is shown. (b) Intact chloroplasts or purified outer envelope were either not treated or treated with the protease thermolysin as indicated. Chloroplasts were treated at pH 11 as described for Fig. 6. Proteins were separated by SDS-PAGE and analyzed by immunoblotting using αToc64. Lane 3 contains a mixture of chloroplasts and envelope, which were combined after completion of proteolysis. (c) The topology of Toc64 was analyzed in right-side-out outer envelope membrane vesicles before or after treatment with thermolysin as indicated. Lanes marked IB contain 3 μg of recombinant Toc64, Toc64ΔTPR, and Toc64TPR protein. Thermolysin-treated membranes were separated into a supernatant (S) and a membrane fraction. The membranes were further treated with 0.5 M Na2CO3 and fractionated into a soluble (S) and insoluble (P) protein fractions as indicated. The immunoblots were incubated with total Toc64 serum (left panel) or serum that recognized the NH2-terminal portion of Toc64 (αToc64ΔTPR, middle panel) or the COOH-terminal part of Toc64 (αToc64TPR, right panel). (d) Schematic representation of Toc64. The localization of distinct protein domains is indicated in the upper lane. The middle lane shows the beginning of the soluble 16-kD fragment as determined by sequencing. The lower lane indicates the position and size of truncated Toc64 proteins, which were heterologously expressed and used to affinity-isolate the different antibody populations.
TPR peptide domains are thought to mediate protein–protein interaction (for review see Lamb et al. 1995). To understand Toc64 function, it is important to determine the side of the outer envelope membrane to which the TPR motif is exposed. We verified that the isolated envelope vesicles had a right-side-out orientation as proposed (Waegemann et al. 1992). Intact chloroplasts and purified outer envelope vesicles were treated with the nonpenetrating protease thermolysin. Subsequently, mixed envelope vesicles were isolated from the chloroplasts and subjected to an extraction with 0.5 M Na2CO3. The proteolytic fragmentation pattern of Toc64 was then analyzed by immunoblotting (Fig. 7 b). Identical fragments were determined, indicating that the outer envelope vesicles had a right-side-out orientation. This is further supported by the results in Fig. 7 a, which had demonstrated that radiolabeled inserted Toc64 also yielded almost identical breakdown patterns as found in situ. The slight shift of the immunostained bands at 50 kD in Fig. 7 b is due to the highly abundant large subunit of Rubisco present in the chloroplast membrane fraction even after extraction at pH 11.
Toc64ΔTPR, which represented amino acids 1–476 of wild-type Toc64 but did not contain the TPR motif, and Toc64TPR, which contained amino acids 477–593, were heterologously expressed and purified (Fig. 7 d, lower lane). Both polypeptides were coupled to CNBr-activated Sepharose and αToc64 antiserum was passed over the matrices, resulting in antibody populations that either recognized only the NH2-terminal portion of Toc64 (αToc64ΔTPR; Fig. 7 c, middle panel, IB) but not the TPR motif–containing domains, or an antiserum that recognized the TPR motif–containing domains (αToc64TPR) but not the NH2-terminal moiety of Toc64 (Fig. 7 c, right panel, IB). Outer envelope vesicles were treated with the protease thermolysin, separated into a supernatant and a membrane fraction, and the membranes were further treated at pH 11 to distinguish between membrane-bound and peripherally-associated Toc64 fragments. The supernatant after protease treatment contained a 16-kD fragment that was 3–4 kD larger than the TPR motif–containing domain itself. It was recognized by all antisera populations used (Fig. 7 c, lanes S1). The other Toc64 proteolytic fragments were recovered in the high pH insoluble fraction and recognized only by αToc64 and αToc64ΔTPR (Fig. 7 c, lanes S2 and P). αToc64TPR recognized only the undigested Toc64 polypeptide. NH2-terminal sequencing of the 16-kD fragment yielded the amino acid sequence VVSK (Fig. 7 d). This result not only explains why the αToc64ΔTPR antiserum recognizes the 16-kD fragment (Fig. 7 d), but it clearly demonstrates that the TPR motif–containing domain is exposed to the cytosol. Upon proteolytic cleavage it can be recovered as a soluble fragment.
Discussion
The outer envelope protein Toc64 is proposed to be a new subunit of the preprotein translocon of pea chloroplasts. Different lines of experimental evidence support this conclusion: (a) Toc64 is a prominent constituent of the Toc complex purified from outer envelope membranes; (b) Toc64 can be cross-linked to other Toc components by the reversible formation of disulfide bridges. The disulfide bond has a length of ∼2 Å, indicating a very close physical proximity between Toc64 and another Toc component; and (c) Toc64 is present in a joint Toc–Tic complex that contains a trapped precursor protein.
What could be the functional role of Toc64? Toc64 seems to be built from two independent modules. One module exhibits homologies to prokaryotic and eukaryotic amidases. The enzymatic function as an amidase seems to be inactivated due to a point mutation (Ser170 → Gly170) in its active site and the addition of hydrophobic sequences that convert it to a membrane polypeptide. We failed to measure any amidase activity either from isolated envelope membranes or the overexpressed protein. Although these data do not exclude an amidase function, we feel that it is unlikely. The second module is a threefold repeated TPR motif. TPR motifs occur in polypeptides of different function and localization. They are generally proposed to mediate protein–protein interactions (for review see Lamb et al. 1995). To elucidate the possible role of Toc64 and the TPR motifs we first studied the topology of the polypeptide. Protease treatment of intact chloroplasts and isolated outer envelope membrane vesicles demonstrated that Toc64 exposes protease-sensitive peptide domains on the organellar surface. The identical proteolytic peptide pattern obtained in both systems also corroborated earlier findings that outer envelope vesicles are isolated primarily in right-side-out orientation. Proteolysis of Toc64 yielded three prominent membrane-embedded peptides. One further fragment of ∼16 kD apparent molecular mass appeared as a soluble peptide in the incubation medium after proteolysis, indicating that it was tightly folded and protected from further proteolysis. The soluble 16-kD fragment was recognized by affinity-purified antisera, one which specifically recognized the TPR domain only, and another that recognized TPR-less polypeptide, respectively. The data suggested that the soluble fragment contained the TPR motifs as well as a short amino acid region N-proximal of it. Sequencing of this fragment verified that the 16-kD peptide originated from the proposed region of Toc64 containing the TPR motifs. We conclude that the TPR motifs are exposed on the chloroplast surface.
TPR motifs have been described for several subunits of different protein translocation machineries. The mitochondrial protein import receptor Tom70 contains seven repeats of the TPR motif. Tom70 was proposed to interact preferentially with precursor proteins that requires the mitochondrial import stimulating factor (Komiya et al. 1997). The peroxisomal protein import related protein Pex5 also contains seven repeats of the TPR motif (Brocard et al. 1994). Hence, it is tempting to speculate that in protein import into chloroplasts the TPR domain of Toc64 fulfills a similar receptor function. This could take place by direct interaction with the targeting signal, as in Pex5 (Terlecky et al. 1995), or it could happen by interaction with a soluble cytosolic complex as demonstrated for Tom70. The latter hypothesis is supported by our recent finding that chloroplastic precursor proteins exist in an oligomeric guidance complex together with 14-3-3 proteins and hsp70. Preliminary evidence indicates that this precursor containing guidance complex is specifically recognized by Toc64 (Sohrt, K., and J. Soll, unpublished observations). We propose that Toc64 functions early in the binding event of preproteins to chloroplasts and maybe even before the major import receptor Toc160 (Bauer et al. 2000).
Acknowledgments
We thank Dr. P. Viitanen for the antiserum against chaperonin 60.
This investigation was supported by grants from the Deutsche Forschungsgemeinschaft, Fonds der Chemischen Industrie, and Human Frontier Science Program.
Footnotes
Abbreviations used in this paper: DSP, dithiobis[succinimidyl propionate]; EST, expressed sequence tag; OE, oxygen evolving; SSU, Rubisco small subunit; Tic, translocon at the inner membrane of chloroplasts; Toc, translocon at the outer membrane of chloroplasts; Tom, translocon at the outer membrane of mitochondria; TPR, tetratricopeptide repeat.
References
- Akita M., Nielsen E., Keegstra K. Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 1997;136:983–994. doi: 10.1083/jcb.136.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C.W., Straus J.W., Dudock B.S. Preparation of a cell-free protein-synthesizing system from wheat germ. Methods Enzymol. 1983;101:635–644. doi: 10.1016/0076-6879(83)01044-7. [DOI] [PubMed] [Google Scholar]
- Arnon D.J. Copper enzymes in isolated chloroplasts. Polyphenyloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J., Chen K., Hiltbunner A., Wehrli E., Eugster M., Schnell D., Kessler F. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature. 2000;403:203–207. doi: 10.1038/35003214. [DOI] [PubMed] [Google Scholar]
- Bölter B., May T., Soll J. A protein import receptor in pea chloroplasts, Toc86, is only a proteolytical fragment of a larger polypeptide. FEBS Lett. 1998;441:59–62. doi: 10.1016/s0014-5793(98)01525-7. [DOI] [PubMed] [Google Scholar]
- Brocard C., Kragler F., Simon M.M., Schuster T., Hartig A. The tetratricopeptide repeat-domain of the PAS10 protein of Saccharomyces cerevisiae is essential for binding the peroxisomal targeting signal-SKL. Biochem. Biophys. Res. Commun. 1994;204:1016–1022. doi: 10.1006/bbrc.1994.2564. [DOI] [PubMed] [Google Scholar]
- Caliebe A., Grimm R., Kaiser G., Lübeck J., Soll J., Heins L. The chloroplastic protein import machinery contains a Rieske-type iron-sulfur cluster and a mononuclear iron-binding protein. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:7342–7350. doi: 10.1093/emboj/16.24.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge U.I., Hinz G. Energy dependence of protein translocation into chloroplasts. Eur. J. Biochem. 1986;160:563–570. doi: 10.1111/j.1432-1033.1986.tb10075.x. [DOI] [PubMed] [Google Scholar]
- Goebl M., Yanagida M. The TPR snap helixa novel protein repeat motif from mitosis to transcription. Trends Biochem. Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- Hase T., Riezman H., Suda K., Schatz G. Import of proteins into mitochondrianucleotide sequence of the gene for a 70-kd protein of the yeast mitochondrial outer membrane. EMBO (Eur. Mol. Biol. Organ.) J. 1983;2:2169–2172. doi: 10.1002/j.1460-2075.1983.tb01718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnah S., Hill K., Wagner R., Schlicher T., Soll J. Reconstitution of a chloroplast protein import channel. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:7351–7360. doi: 10.1093/emboj/16.24.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Muckel E., Heemeyer F., von Heijne G., Soll J. A receptor component of the chloroplast protein translocation machinery. Science. 1994;266:1989–1992. doi: 10.1126/science.7801125. [DOI] [PubMed] [Google Scholar]
- Jarvis P., Chen L.J., Li H., Peto C.A., Fankhauser C., Chory J. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science. 1998;282:100–103. doi: 10.1126/science.282.5386.100. [DOI] [PubMed] [Google Scholar]
- Keegstra K., Yousif A.E. Isolation and characterization of chloroplast envelope membranes. Methods Enzymol. 1986;118:316–325. [Google Scholar]
- Kessler F., Blobel G. Interaction of the protein import and folding machineries of the chloroplast. Proc. Natl. Acad. Sci. USA. 1996;93:7684–7689. doi: 10.1073/pnas.93.15.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F., Blobel G., Patel H.A., Schnell D.J. Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science. 1994;266:1035–1039. doi: 10.1126/science.7973656. [DOI] [PubMed] [Google Scholar]
- Ko K., Budd D., Seibert F., Kourtz L., Ko Z.W. Isolation and characterization of a cDNA clone encoding a member of the Cim/Com44 envelope components of the chloroplast protein import apparatus. J. Biol. Chem. 1995;270:28601–28608. doi: 10.1074/jbc.270.48.28601. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Fujiwara Y., Goda M., Komeda H., Shimizu S. Identification of active sites in amidaseevolutionary relationship between amide bond- and peptide bond-cleaving enzymes. Proc. Natl. Acad. Sci. USA. 1997;94:11986–11991. doi: 10.1073/pnas.94.22.11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya T., Rospert S., Schatz G., Mihara K. Binding of mitochondrial precursor proteins to the cytoplasmic domains of the import receptors Tom70 and Tom20 is determined by cytoplasmic chaperones. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:4267–4275. doi: 10.1093/emboj/16.14.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A., Schnell D.J. Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J. Cell Biol. 1997;139:1677–1685. doi: 10.1083/jcb.139.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A., Chen X., Fuks B., Schnell D.J. Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 1998;143:991–1002. doi: 10.1083/jcb.143.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–142. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the heads of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamb J.R., Tugendreich S., Hieter P. Tetratrico peptide repeat interactionsto TPR or not to TPR? Trends Biochem. Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Lübeck J., Soll J., Akita M., Nielsen E., Keegstra K. Topology of IEP110, a component of the chloroplastic protein import machinery present in the inner envelope membrane. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:4230–4238. [PMC free article] [PubMed] [Google Scholar]
- Mason H.S., Guerro D., Boyer J.S., Mullet J.E. Proteins homologous to leaf glycoproteins are abundant in stems of darkgrown soybean seedlings. Analysis of proteins and cDNAs. Plant Mol. Biol. 1988;11:845–856. doi: 10.1007/BF00019524. [DOI] [PubMed] [Google Scholar]
- May T., Soll J. 14-3-3 Proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell. 2000;12:53–64. doi: 10.1105/tpc.12.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove J.E., Johnson R.A., Ellis R.J. Dissociation of the ribulosebisphosphate-carboxylase large-subunit binding protein into dissimilar subunits. Eur. J. Biochem. 1987;163:529–534. doi: 10.1111/j.1432-1033.1987.tb10900.x. [DOI] [PubMed] [Google Scholar]
- Nielsen E., Akita M., Davila-Aponte J., Keegstra K. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal HSP100 molecular chaperone. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:935–946. doi: 10.1093/emboj/16.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttall S.D., Hanson B.J., Mori M., Hoogenraad N.J. hTom34a novel translocase for the import of proteins into human mitochondria. DNA Cell Biol. 1997;16:1067–1074. doi: 10.1089/dna.1997.16.1067. [DOI] [PubMed] [Google Scholar]
- Olsen L.J., Theg S.M., Selman B.R., Keegstra K. ATP is required for the binding of precursor proteins to chloroplasts. J. Biol. Chem. 1989;264:6724–6729. [PubMed] [Google Scholar]
- Perry S.E., Keegstra K. Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell. 1994;6:93–105. doi: 10.1105/tpc.6.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Soulsen A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C., Hracky R., Soll J. Protein transport in chloroplastsATP is prerequisite. Z. Nuturforsch. 1987;42:103–108. [Google Scholar]
- Schnell D.J., Kessler F., Blobel G. Isolation of components of the chloroplast protein import machinery. Science. 1994;266:1007–1012. doi: 10.1126/science.7973649. [DOI] [PubMed] [Google Scholar]
- Seedorf M., Waegemann K., Soll J. A constituent of the chloroplast import complex represents a new type of GTP-binding protein. Plant J. 1995;7:401–411. doi: 10.1046/j.1365-313x.1995.7030401.x. [DOI] [PubMed] [Google Scholar]
- Soll J., Waegemann K. A functionally active protein import complex from chloroplasts. Plant J. 1992;2:253–256. [Google Scholar]
- Stahl T., Glockmann C., Soll J., Heins L. Tic40, a new “old” subunit of the chloroplast protein import translocation. J. Biol. Chem. 1999;274:37467–37472. doi: 10.1074/jbc.274.52.37467. [DOI] [PubMed] [Google Scholar]
- Terlecky S.R., Nuttley W.M., McCollum D., Sock E., Subramani S. The Pichia pastoris peroxisomal protein PAS8p is the receptor for the C-terminal tripeptide peroxisomal targeting signal. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:3627–3634. doi: 10.1002/j.1460-2075.1995.tb00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theg S.M., Bauerle C., Olsen L.J., Selman B.R., Keegstra K. Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J. Biol. Chem. 1989;264:6730–6736. [PubMed] [Google Scholar]
- von Heijne G., Steppuhn J., Herrmann R.G. Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Waegemann K., Soll J. Characterization of the protein import apparatus in isolated outer envelopes of chloroplasts. Plant J. 1991;1:149–158. [Google Scholar]
- Waegemann K., Soll J. Phosphorylation of the transit sequence of chloroplast precursor proteins. J. Biol. Chem. 1996;271:6545–6554. doi: 10.1074/jbc.271.11.6545. [DOI] [PubMed] [Google Scholar]
- Waegemann K., Eichacker L., Soll J. Outer envelope membranes from chloroplasts are isolated as right-site-out vesicles. Planta. 1992;187:89–94. doi: 10.1007/BF00201628. [DOI] [PubMed] [Google Scholar]
- Young M.E., Keegstra K., Froehlich J.E. GTP promotes the formation of early-import intermediates but is not required during the translocation step of protein import into chloroplasts. Plant Physiol. 1999;121:237–244. doi: 10.1104/pp.121.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]