Abstract
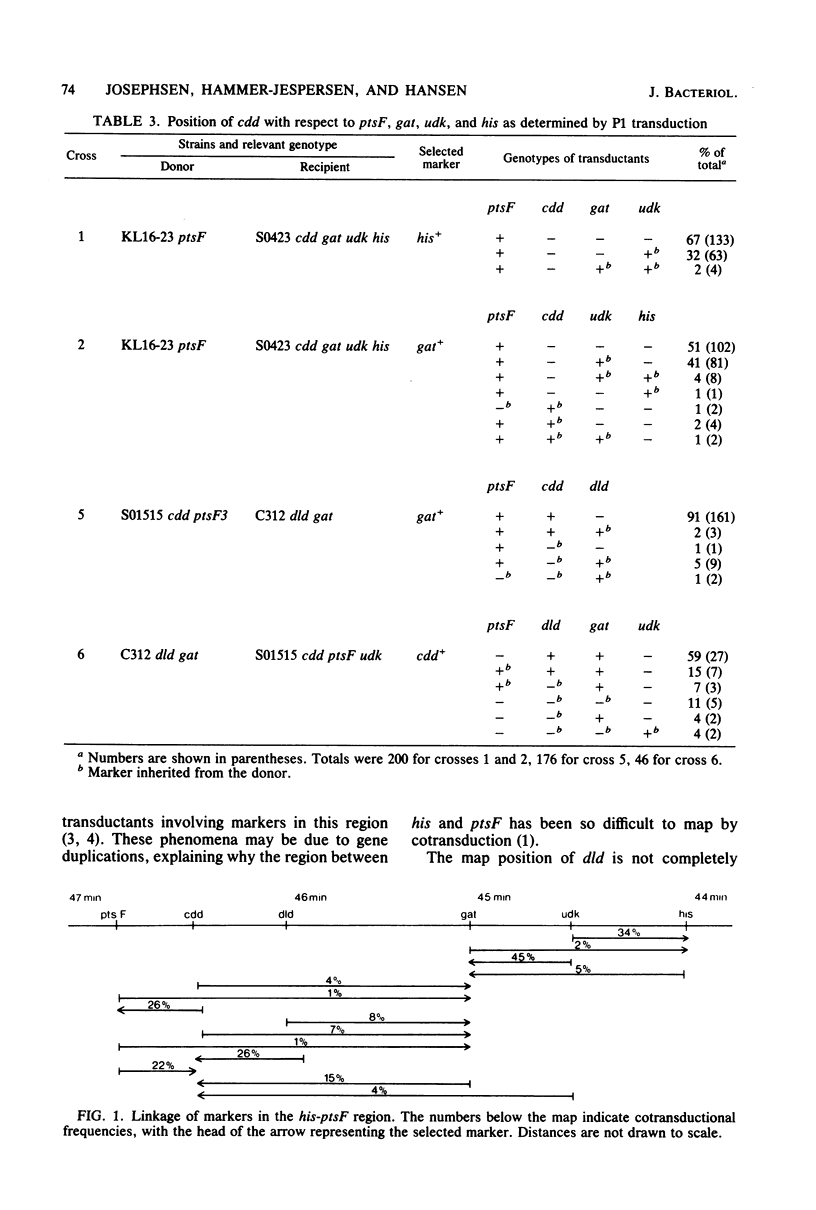
The structural gene encoding cytidine deaminase (cdd) has been mapped in Escherichia coli K-12. It is located counterclockwise to ptsF between 46 and 47 min. The gene order in this region of the E. coli chromosome was found to be his-udk-gat-dld-cdd-ptsF.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C. F., Ingraham J. L., Neuhard J., Thomassen E. Metabolism of pyrimidines and pyrimidine nucleosides by Salmonella typhimurium. J Bacteriol. 1972 Apr;110(1):219–228. doi: 10.1128/jb.110.1.219-228.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T. P1 transduction: formation of heterogenotes upon cotransduction of bacterial genes with a P2 prophage. Virology. 1972 Jan;47(1):76–93. doi: 10.1016/0042-6822(72)90241-3. [DOI] [PubMed] [Google Scholar]
- Boos W., Steinacher I., Engelhardt-Altendorf D. Mapping of mglB, the structural gene of the galactose-binding protein of Escherichia coli. Mol Gen Genet. 1981;184(3):508–518. doi: 10.1007/BF00352531. [DOI] [PubMed] [Google Scholar]
- Fuchs J. A., Karlström H. O. Mapping of nrdA and nrdB in Escherichia coli K-12. J Bacteriol. 1976 Dec;128(3):810–814. doi: 10.1128/jb.128.3.810-814.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer-Jespersen K., Munch-Petersen A. Mutants of Escherichia coli unable to metabolize cytidine: isolation and characterization. Mol Gen Genet. 1973 Nov 2;126(2):177–186. doi: 10.1007/BF00330992. [DOI] [PubMed] [Google Scholar]
- Hammer-Jespersen K., Munch-Ptersen A. Multiple regulation of nucleoside catabolizing enzymes: regulation of the deo operon by the cytR and deoR gene products. Mol Gen Genet. 1975;137(4):327–335. doi: 10.1007/BF00703258. [DOI] [PubMed] [Google Scholar]
- Josephsen J., Hammer-Jespersen K. Fusion of the lac genes to the promotor for the cytidine deaminase gene of Escherichia coli K-12. Mol Gen Genet. 1981;182(1):154–158. doi: 10.1007/BF00422783. [DOI] [PubMed] [Google Scholar]
- Lengeler J. Analysis of mutations affecting the dissmilation of galactitol (dulcitol) in Escherichia coli K 12. Mol Gen Genet. 1977 Mar 28;152(1):83–91. doi: 10.1007/BF00264944. [DOI] [PubMed] [Google Scholar]
- Lengeler J. Mutations affecting transport of the hexitols D-mannitol, D-glucitol, and galactitol in Escherichia coli K-12: isolation and mapping. J Bacteriol. 1975 Oct;124(1):26–38. doi: 10.1128/jb.124.1.26-38.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch-Petersen A., Mygind B., Nicolaisen A., Pihl N. J. Nucleoside transport in cells and membrane vesicles from Escherichia coli K12. J Biol Chem. 1979 May 25;254(10):3730–3737. [PubMed] [Google Scholar]
- Munch-Petersen A., Mygind B. Nucleoside transport systems in Escherichia coli K12: specificity and regulation. J Cell Physiol. 1976 Dec;89(4):551–559. doi: 10.1002/jcp.1040890410. [DOI] [PubMed] [Google Scholar]
- Munch-Petersen A., Nygaard P., Hammer-Jespersen K., Fiil N. Mutants constitutive for nucleoside-catabolizing enzymes in Escherichia coli K12. Isolation, charactrization and mapping. Eur J Biochem. 1972 May 23;27(2):208–215. doi: 10.1111/j.1432-1033.1972.tb01828.x. [DOI] [PubMed] [Google Scholar]
- Neuhard J., Thomassen E. Altered deoxyribonucleotide pools in P2 eductants of Escherichia coli K-12 due to deletion of the dcd gene. J Bacteriol. 1976 May;126(2):999–1001. doi: 10.1128/jb.126.2.999-1001.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A. M. Xylitol and D-arabitol toxicities due to derepressed fructose, galactitol, and sorbitol phosphotransferases of Escherichia coli. J Bacteriol. 1977 Oct;132(1):166–173. doi: 10.1128/jb.132.1.166-173.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner J. L. Formation, induction, and curing of bacteriophage P1 lysogens. Virology. 1972 Jun;48(3):679–689. doi: 10.1016/0042-6822(72)90152-3. [DOI] [PubMed] [Google Scholar]
- Schnier J., Isono K. The gene for ribosomal protein L25 (rplY) maps at 47.3 min near nalA in Escherichia coli K-12. Mol Gen Genet. 1979 Nov;176(3):313–318. doi: 10.1007/BF00333093. [DOI] [PubMed] [Google Scholar]
- Sunshine M. G., Kelly B. Extent of host deletions associated with bacteriophage P2-mediated eduction. J Bacteriol. 1971 Nov;108(2):695–704. doi: 10.1128/jb.108.2.695-704.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor H., Hafner E. W., Tabor C. W. Construction of an Escherichia coli strain unable to synthesize putrescine, spermidine, or cadaverine: characterization of two genes controlling lysine decarboxylase. J Bacteriol. 1980 Dec;144(3):952–956. doi: 10.1128/jb.144.3.952-956.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]


