Abstract
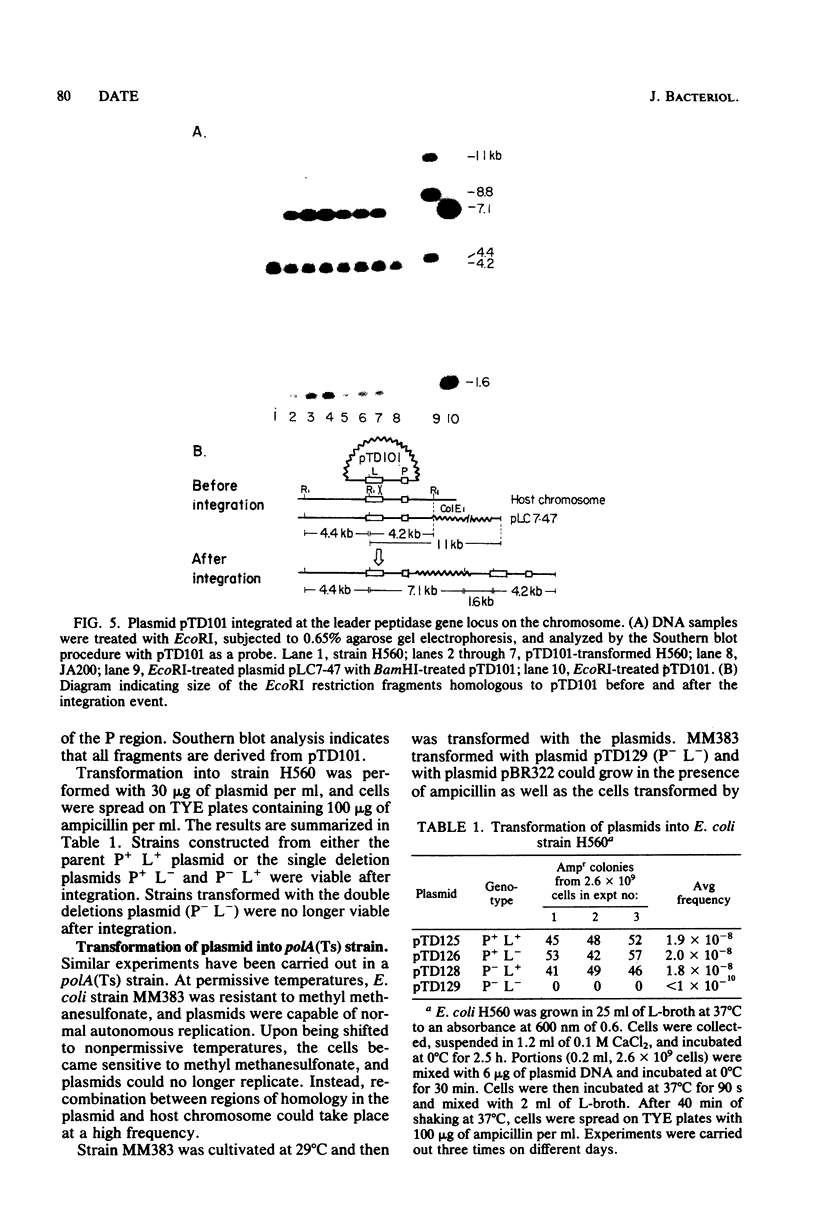
It was previously shown that two separate regions of DNA are required for expression of the cloned leader peptidase gene on plasmid pTD101 (T. Date and W. Wickner, Proc. Natl. Acad. Sci. U.S.A. 78:6106-6110, 1981). Both loci have been mapped in detail, and their roles have been established. A 1.3-kilobase region, termed the L region, encodes the 37,000-dalton leader peptidase protein. Another region, termed the P region, is about 1.5 kilobases away from the L region and is less than 350 base pairs long. The P region acts in cis to the L region, suggesting that it plays a role as a promoter. A technique for inactivation of the leader peptidase gene on the Escherichia coli chromosome has been developed to examine whether the leader peptidase which we had cloned is essential for cell growth. A specific plasmid (P− L−) which deletes both the P region and a substantial portion of the L region was constructed and transformed into a polA mutant strain. The plasmid cannot replicate in this strain; thus, the plasmid-borne ampicillin resistance is lost unless the plasmid DNA recombines into the chromosome. Integration of the P− L− plasmid did not yield any viable ampicillin-resistant cells, whereas the three control plasmids, P+ L+, P+ L−, and P− L+, did. When the P− L− plasmid was transformed into the polA(Ts) strain, the strain could only grow in the presence of ampicillin at a permissive temperature, suggesting that integration of the plasmid into the host chromosome leads to inactivation of the chromosomal leader peptidase gene. Southern hybridization analysis demonstrated that the integration of plasmids into the chromosome occurred at the homologous site. This study demonstrates that expression of the leader peptidase gene is critical for cell growth.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boeke J. D., Russel M., Model P. Processing of filamentous phage pre-coat protein. Effect of sequence variations near the signal peptidase cleavage site. J Mol Biol. 1980 Dec 5;144(2):103–116. doi: 10.1016/0022-2836(80)90027-3. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Collins C. J., Jackson D. A., DeVries A. J. Biochemical construction of specific chimeric plasmids from ColE1 DNA and unfractionated Escherichia coli DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3838–3842. doi: 10.1073/pnas.73.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Ingram J. M., Cheng K. J. Structure and function of the cell envelope of gram-negative bacteria. Bacteriol Rev. 1974 Mar;38(1):87–110. doi: 10.1128/br.38.1.87-110.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date T., Wickner W. Isolation of the Escherichia coli leader peptidase gene and effects of leader peptidase overproduction in vivo. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6106–6110. doi: 10.1073/pnas.78.10.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M., Ichihara S., Mizushima S. Accumulation of glyceride-containing precursor of the outer membrane lipoprotein in the cytoplasmic membrane of Escherichia coli treated with globomycin. J Biol Chem. 1980 Apr 25;255(8):3707–3712. [PubMed] [Google Scholar]
- Hussain M., Ichihara S., Mizushima S. Mechanism of signal peptide cleavage in the biosynthesis of the major lipoprotein of the Escherichia coli outer membrane. J Biol Chem. 1982 May 10;257(9):5177–5182. [PubMed] [Google Scholar]
- Inouye H., Barnes W., Beckwith J. Signal sequence of alkaline phosphatase of Escherichia coli. J Bacteriol. 1982 Feb;149(2):434–439. doi: 10.1128/jb.149.2.434-439.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury D. T., Helinski D. R. DNA polymerase as a requirement for the maintenance of the bacterial plasmid colicinogenic factor E1. Biochem Biophys Res Commun. 1970 Dec 24;41(6):1538–1544. doi: 10.1016/0006-291x(70)90562-0. [DOI] [PubMed] [Google Scholar]
- Monk M., Kinross J. Conditional lethality of recA and recB derivatives of a strain of Escherichia coli K-12 with a temperature-sensitive deoxyribonucleic acid polymerase I. J Bacteriol. 1972 Mar;109(3):971–978. doi: 10.1128/jb.109.3.971-978.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn M. J., Gander J. E., Parisi E., Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J Biol Chem. 1972 Jun 25;247(12):3962–3972. [PubMed] [Google Scholar]
- Osborn M. J., Wu H. C. Proteins of the outer membrane of gram-negative bacteria. Annu Rev Microbiol. 1980;34:369–422. doi: 10.1146/annurev.mi.34.100180.002101. [DOI] [PubMed] [Google Scholar]
- Pratt D., Tzagoloff H., Beaudoin J. Conditional lethal mutants of the small filamentous coliphage M13. II. Two genes for coat proteins. Virology. 1969 Sep;39(1):42–53. doi: 10.1016/0042-6822(69)90346-8. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Vosberg H. P., Hoffmann-Berling H. DNA synthesis in nucleotide-permeable Escherichia coli cells. I. Preparation and properties of ether-treated cells. J Mol Biol. 1971 Jun 28;58(3):739–753. doi: 10.1016/0022-2836(71)90037-4. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W. Assembly of proteins into membranes. Science. 1980 Nov 21;210(4472):861–868. doi: 10.1126/science.7001628. [DOI] [PubMed] [Google Scholar]
- Wickner W., Killick T. Membrane-associated assembly of M13 phage in extracts of virus-infected Escherichia coli. Proc Natl Acad Sci U S A. 1977 Feb;74(2):505–509. doi: 10.1073/pnas.74.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe P. B., Silver P., Wickner W. The isolation of homogeneous leader peptidase from a strain of Escherichia coli which overproduces the enzyme. J Biol Chem. 1982 Jul 10;257(13):7898–7902. [PubMed] [Google Scholar]
- Yamaguchi K., Tomizawa J. Establishment of Escherichia coli cells with an integrated high copy number plasmid. Mol Gen Genet. 1980;178(3):525–533. doi: 10.1007/BF00337857. [DOI] [PubMed] [Google Scholar]
- Zwizinski C., Date T., Wickner W. Leader peptidase is found in both the inner and outer membranes of Escherichia coli. J Biol Chem. 1981 Apr 10;256(7):3593–3597. [PubMed] [Google Scholar]
- Zwizinski C., Wickner W. Purification and characterization of leader (signal) peptidase from Escherichia coli. J Biol Chem. 1980 Aug 25;255(16):7973–7977. [PubMed] [Google Scholar]
- de Vries F. A., Collins C. J., Jackson D. A. Joining of simian virus 40 DNA molecules at endonuclease R Eco Ri sites by polynucleotide ligase and analysis of the products by agarose gel electrophoresis. Biochim Biophys Acta. 1976 Jul 2;435(3):213–227. doi: 10.1016/0005-2787(76)90103-9. [DOI] [PubMed] [Google Scholar]







