Abstract
Molecular biology databases are an integral part of biological research. To date, many databases were established with varied options to access associated biological data. Depending on the data being annotated, some are architecturally similar while others are specialized. In order to provide a partial solution to data integration, we report Database of Databases (DoD2007), constructed using html and javascript. The database has a web-based user interface with simple global search, specific database search, keyword help as well as links to abstracts, full-text and database home pages. Majority of data were derived form Nucleic Acids Research database issue and other published resources. The current release includes 15 categories with updated descriptions and links to 1082 databases, of which, 209 are new entries. New databases included in this issue are represented with ‘+’ sign before the name and a ‘*’ symbol provided for those that remained silent.
Availability
The database is freely available at http://www.progenebio.in/DoD/index.htm.
Keywords: molecular biology, database, javascript, data integration
Background
Molecular Biology databases have become an integral part of scientific research. They are widely used to understand the underlying mechanism of genomes, expression patterns, bimolecular interactions, metabolism, understanding evolutionary relationships etc. as well as providing knowledge that helps to examine specific state of a disease or condition and assist in drug discovery and development. This sort of biological knowledge is disseminated to a variety of scientific researchers through specialized databases made possible through internet technologies, software's and tools. As more and more genomes are being sequenced and annotated, huge amount of data are accumulating. [1] Biological databases designed would cater to meet the needs of the scientific community. But depending on the data being annotated, some are architecturally similar while the others are specialized. The challenges to develop an integrated system are due to several factors such as variety and amount of data available and data heterogeneity in different sources. [2] Therefore, data integration has proved problematic and to provide a partial solution, we report an update on collection of molecular biology databases with a search interface, Database of Databases (DoD2007). The latest edition, DoD2007 was compiled with similar capabilities [3,4] but with additional features such as a global search for all databases and links to database entries from category list, added patents database as a new category and one sub-category each in nucleotide, Genomics database and two in Protein Sequence database, respectively.
Methodology
DoD2007 was updated from the reported database issue of Nucleic Acids Research [5] and various journal sources. The 14 categories reported in our previous update [4] remain unchanged except a new category; ‘patents db’ has been incorporated in DoD2007 update, keeping in view the importance of patent rights in scientific discipline. Currently, this category includes two databases, patome [6] and DNA patents database. [7] DoD2007 was constructed using html and javascript. The modes of access of databases listed in DoD2007 vary depending on the content, format and access methods. Majority of the databases are provided with direct access to search data from DoD2007. Some databases require multiple search items while some others require sequence or annotation to be pasted in the search box and therefore they are provided with a direct link to the database home page. Over time, DoD2007 has come to enhance the listed molecular biology databases as its use by researchers, educators and students in diverse disciplines has expanded. The recent release DoD2007 contains three distinct newly developed functionalities, which are outlined below.
First, we have developed a user interface to search databases globally. In other words, a global search can be utilized to search the categories reported in DoD2007, database names and other pages located in the database. Keywords used to search the database can be general or specific and depends on the terms indexed for search.
Second, an organized category list was created in a separate page. The availability of these databases as a list is intended to promote better understanding of database segregation used to generate categories and to quickly recognize the new ones with those that are inaccessible. These databases have links to the database entries because a link back to the main page makes it easy for both new and proficient users to conduct a variety of searches. However, some users may not realize the keywords that need to be used to search a database and even some users may show interest in the published resource. Hence, a keyword help and links to abstract or full-text articles were created in respective main pages for intuitive and efficient presentation. Finally, a number of sub-categories are created such as ‘Operons’ and ‘Comparative genomics’ in ‘nucleotide db’ and ‘Genomics db’ respectively along with two sub-categories viz. ‘Protein Interactions’ and ‘Amino Acid Repeats’ in ‘Protein Sequence db’.
Features of DoD2007
DoD2007 is a unique, categorized and easy-to-use interface for all molecular biology databases. New databases in various categories were appended and the number of databases in DOD2007 reached up to 1082 that are 209 databases ahead from the previous issue. Major additions, 49 new entries, were reported in Genomics database. However, the list is not complete. A brief description of databases under all categories was given as a pop-up window with a provision to view full-text of the published article and a direct link to the database home page. The search option under ‘link to database’ leads to the respective home pages of the database. Boolean search items are not allowed in the DoD2007 but spaces between search strings enables the search. During the search, results are displayed in a separate window as this helps in further database scan without losing the home page.
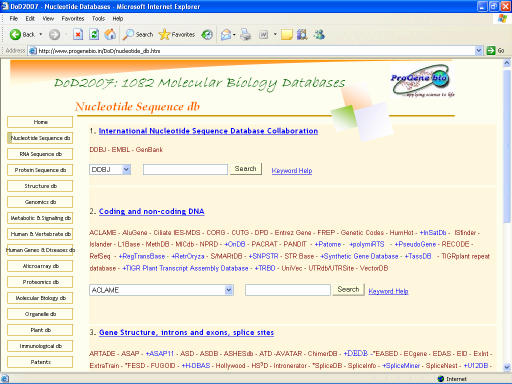
DoD2007 facilitates integrated database search to scan a number of relevant databases from respective categories. A few databases incorporated in DoD2007 require user log-in or restricted access and a few are inaccessible or discontinued. To distinguish such databases, new entries in DoD2007 are provided with a ‘+’ sign and inaccessible ones are recognized by a ‘*’ sign before the name. The current list of database categories and the number of entries in each category are given in table 1 (supplementary material). An image of the database is given in figure 1. Since its first appearance in the year 2005, DoD constituted 719 molecular biology databases and the year 2005 resulted in an addition of 154 entries making up to 873 databases (DoD2006) and the year 2006 has 209 new entries being appended in DoD2007. On the other hand, nearly 4 percent of databases are inaccessible such as HLA Ligand/Motif database [8] of Immunological database or discontinued while few others remained silent such as European rRNA Database [9] and Small RNA Database [10] of RNA sequence database etc.
Figure 1.
Screen-shot image of DoD2007 showing sub-categories of nucleotide sequence databases
On an average, there is a continuous increase in the number of databases which specifies the importance of such databases to biological community. Therefore, in order to account for the rise in number of databases, a graph as shown in figure 2 was plotted for each category since year 2004. The displayed graph showed a substantial increase in Nucleotide, Protein and Genomics databases respectively.
Figure 2.
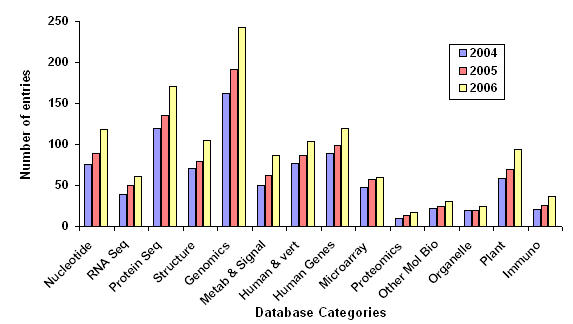
Year wise (2004-2006) number of database entries in 14 categories of database of databases
Utility
DoD2007 provides the updated descriptions and links to existing and new databases that serves as an interface and user-friendly access to molecular biology scientific community. Number of inaccessible databases reported in the years 2005 and 2006 remain unchanged in DoD2007 so as to enable the users to know the type of database that once existed.
Conclusion
DoD2007, a freely available web-based database resource enabling ease of access, serves as a general reference source both for community of researchers working in molecular biology and educators who deal with particular databases of their interest. Brief description about the databases and keyword help makes the users familiar with the contents of the database, respective keywords and database links. The database shall be updated on a yearly basis.
Supplementary material
References
- 1.Stein LD. Nat Rev Genet. 2003;4:337. doi: 10.1038/nrg1065. [DOI] [PubMed] [Google Scholar]
- 2.Hernandez T, Kambhampati S. ACM SIGMOD Record. 1992;33:51. [Google Scholar]
- 3.Babu PA, et al. In Silico Biol. 2005;5:605. [PubMed] [Google Scholar]
- 4.Babu PA, et al. Bioinformation. 2006;1:228. [PMC free article] [PubMed] [Google Scholar]
- 5.Galperin MY, et al. Nucleic Acids Res. 2007;35:D3. doi: 10.1093/nar/gkl1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee B, et al. Nucleic Acids Res. 2007;35:D47. doi: 10.1093/nar/gkl807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. http://dnapatents.georgetown.edu/aboutdpd.htm.
- 8. http://hlaligand.ouhsc.edu/
- 9. http://bioinformatics.psb.ugent.be/webtools/rRNA/
- 10. http://mbcr.bcm.tmc.edu/smallRNA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.