Abstract
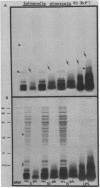
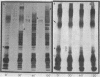
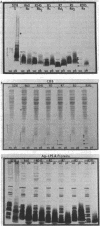
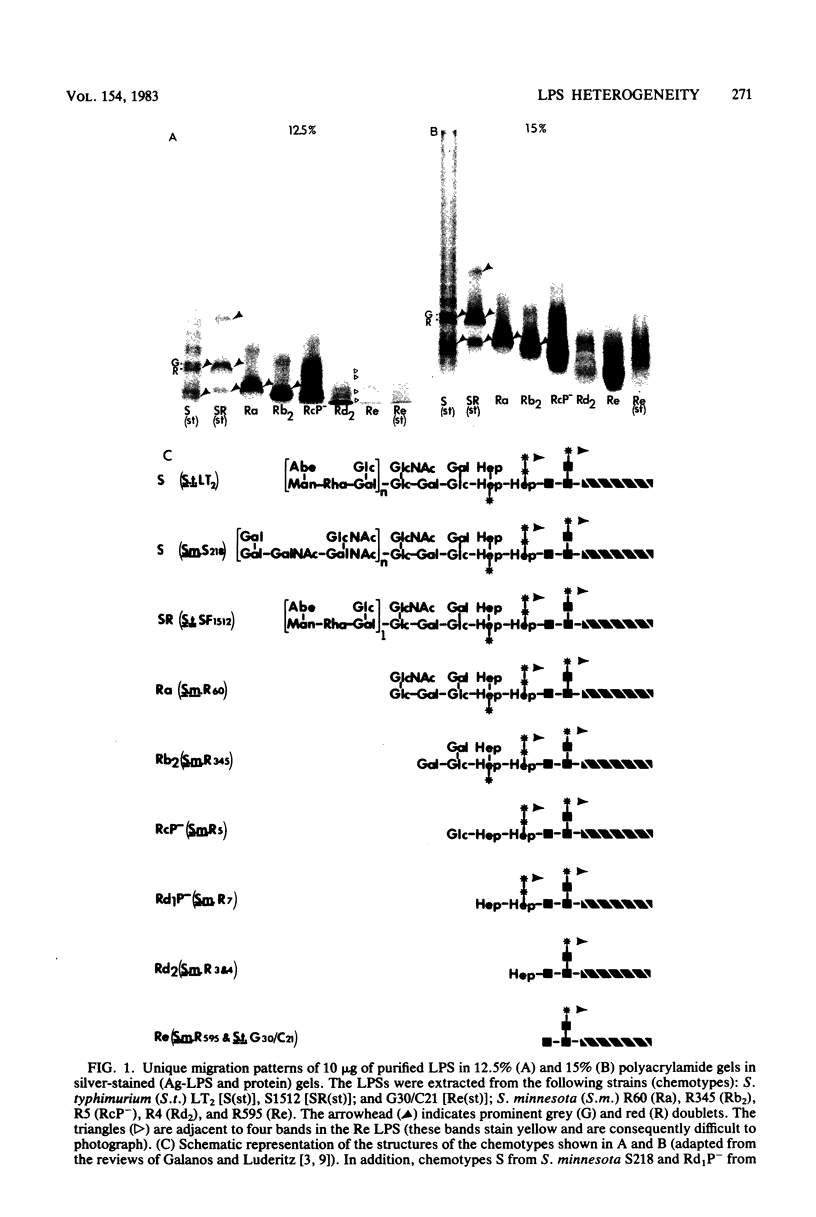
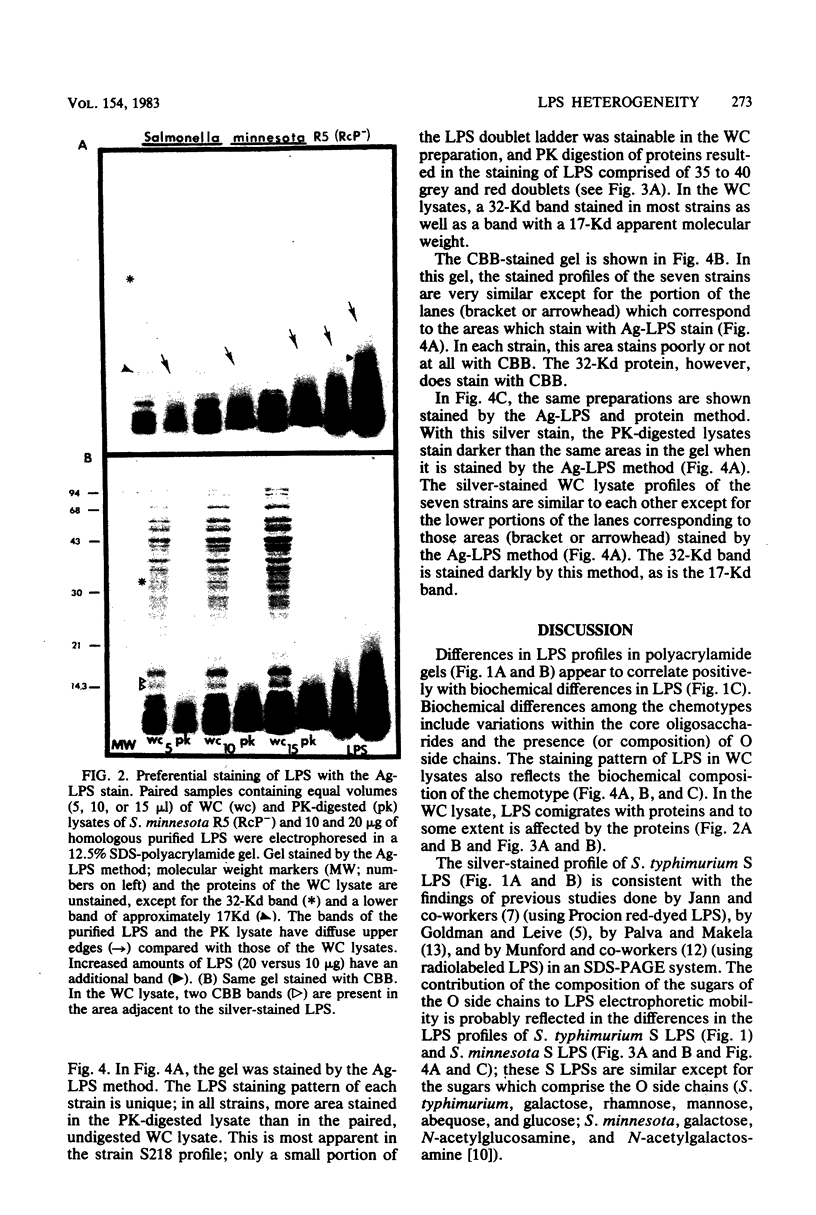
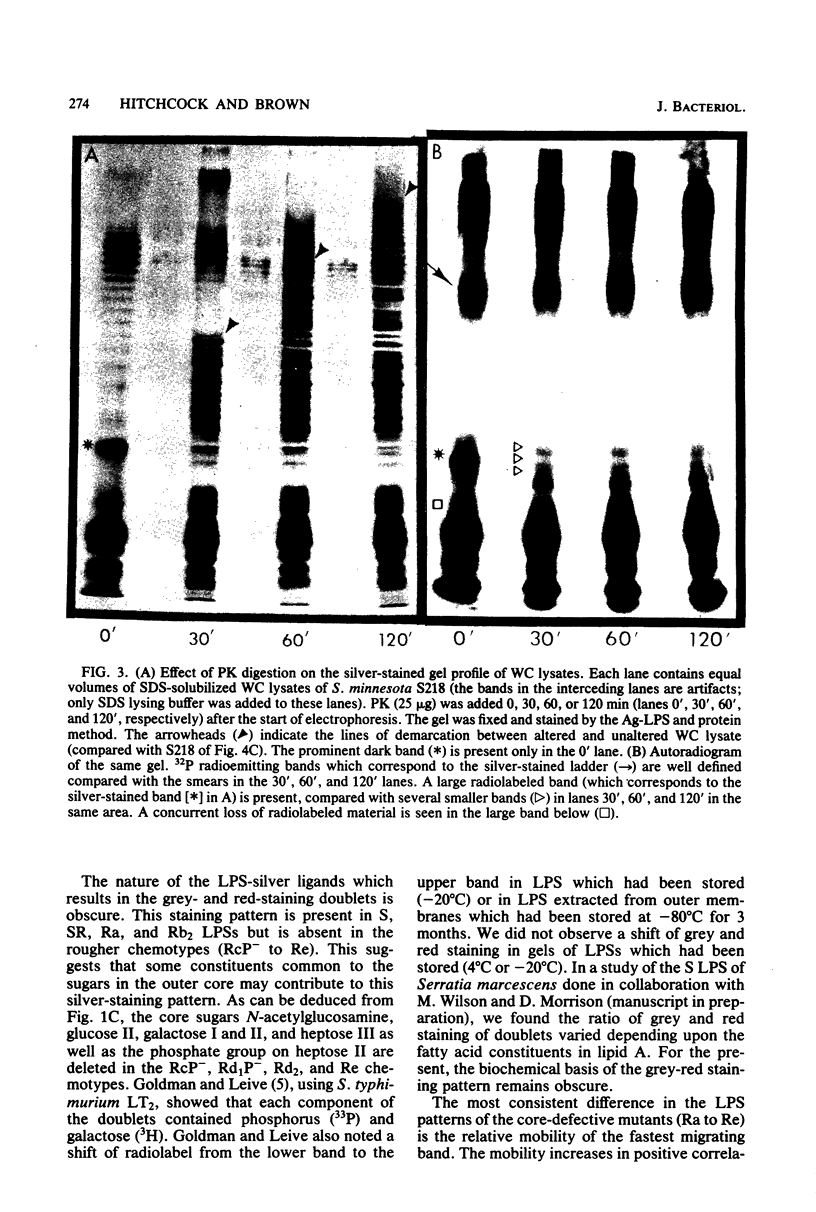
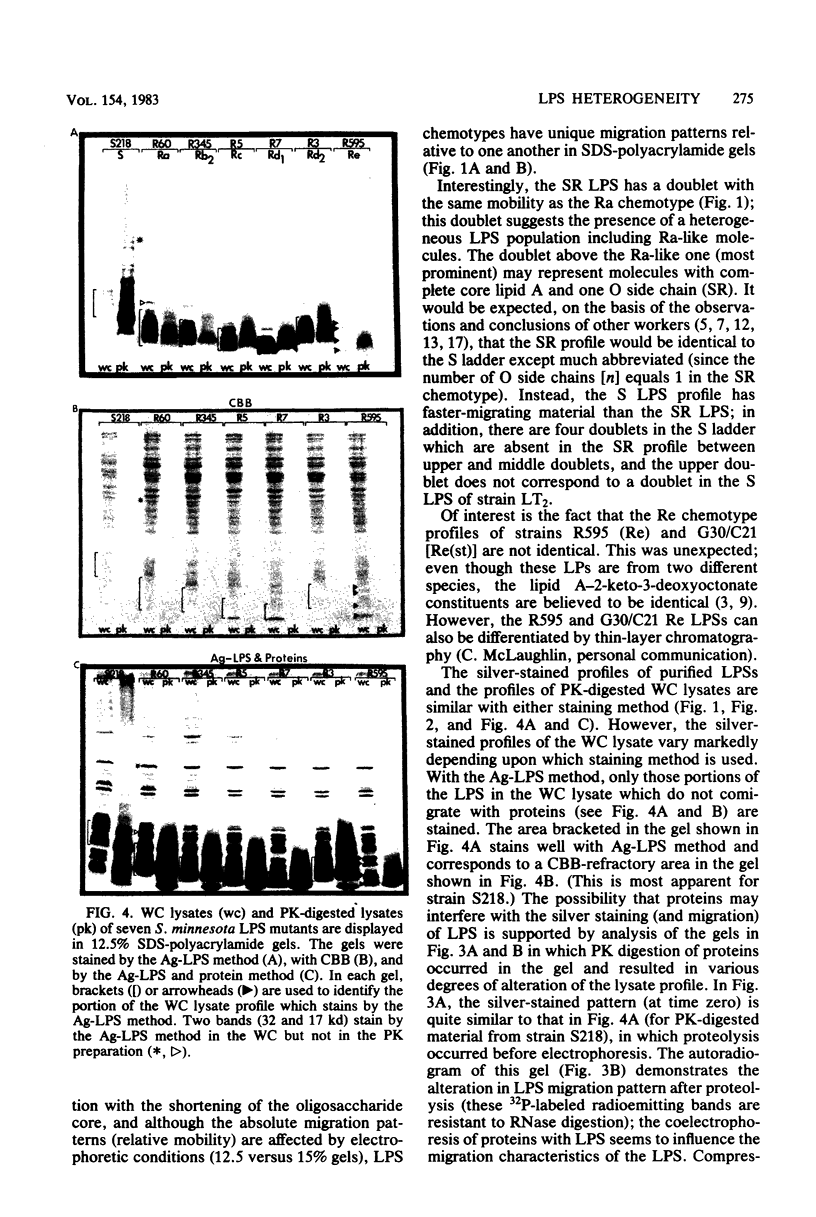
The morphological heterogeneity of lipopolysaccharides (LPSs) among salmonella mutants with different LPS chemotypes was analyzed in silver-stained polyacrylamide gels. The biochemical differences in the LPS chemotypes were reflected in the unique profiles of the purified LPSs. The LPS profiles in the whole-cell lysates were also unique for each chemotype. (Whole-cell lysates were assessed by a method which preferentially silver stains LPS and by a proteinase K digest of whole-cell lysates. The silver-stained LPS profiles of proteinase K-digested lysates were similar to the homologous purified LPS and could be used to preliminarily characterize the LPS chemotype before purification.) In summary, biochemical variation in LPS composition can be detected in silver-stained polyacrylamide gels.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- DULBECCO R., VOGT M. Plaque formation and isolation of pure lines with poliomyelitis viruses. J Exp Med. 1954 Feb;99(2):167–182. doi: 10.1084/jem.99.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanos C., Lüderitz O., Westphal O. A new method for the extraction of R lipopolysaccharides. Eur J Biochem. 1969 Jun;9(2):245–249. doi: 10.1111/j.1432-1033.1969.tb00601.x. [DOI] [PubMed] [Google Scholar]
- Goldman R. C., Leive L. Heterogeneity of antigenic-side-chain length in lipopolysaccharide from Escherichia coli 0111 and Salmonella typhimurium LT2. Eur J Biochem. 1980;107(1):145–153. doi: 10.1111/j.1432-1033.1980.tb04635.x. [DOI] [PubMed] [Google Scholar]
- James J. F., Swanson J. Studies on gonococcus infection. XIII. Occurrence of color/opacity colonial variants in clinical cultures. Infect Immun. 1978 Jan;19(1):332–340. doi: 10.1128/iai.19.1.332-340.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jann B., Reske K., Jann K. Heterogeneity of lipopolysaccharides. Analysis of polysaccharide chain lengths by sodium dodecylsulfate-polyacrylamide gel electrophoresis. Eur J Biochem. 1975 Dec 1;60(1):239–246. doi: 10.1111/j.1432-1033.1975.tb20996.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lüderitz O., Staub A. M., Westphal O. Immunochemistry of O and R antigens of Salmonella and related Enterobacteriaceae. Bacteriol Rev. 1966 Mar;30(1):192–255. doi: 10.1128/br.30.1.192-255.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer L. W., Holmes K. K., Falkow S. Characterization of plasmid deoxyribonucleic acid from Neisseria gonorrhoeae. Infect Immun. 1974 Oct;10(4):712–717. doi: 10.1128/iai.10.4.712-717.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munford R. S., Hall C. L., Rick P. D. Size heterogeneity of Salmonella typhimurium lipopolysaccharides in outer membranes and culture supernatant membrane fragments. J Bacteriol. 1980 Nov;144(2):630–640. doi: 10.1128/jb.144.2.630-640.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palva E. T., Mäkelä P. H. Lipopolysaccharide heterogeneity in Salmonella typhimurium analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Eur J Biochem. 1980;107(1):137–143. doi: 10.1111/j.1432-1033.1980.tb04634.x. [DOI] [PubMed] [Google Scholar]
- Tsai C. M., Frasch C. E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982 Jan 1;119(1):115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]