Abstract
Myxococcus xanthus cells exhibit directed motility up phosphatidylethanolamine (PE) gradients, and we suggest that PE behaves as a chemoattractant. Computer-assisted stop-motion digital microscopy was used to record cell movements in slide culture. PE decreased cellular reversal frequency with molecular specificity that was correlated with the fatty acid composition. Synthetic dilauroyl (di C12:0) PE and dioleoyl (di C18:1 ω9c) PE suppressed direction reversals and stimulated movement up the gradient. Sensory adaptation occurred about 1 hr after the onset of stimulation. Null mutants in a methylated chemotaxis protein homolog (FrzCD) and a CheA/CheY homolog (FrzE) moved up a PE gradient at a reduced rate. The mutants displayed normal excitation but were defective in adaptation. A dominant, hyper-reversal mutant in the M. xanthus methyl accepting chemotaxis protein homolog, frzCD224, failed to respond to PE stimulation, which argued that PE was a transduced stimulus. Neither dilauroyl PE nor dioleoyl PE is present at high enough concentrations in vegetative or developmental PE to account for all of the chemotactic activity. It appears then that there are additional, as yet unknown, PE species that serve as autoattractants. We report on a discrete phospholipid chemoattractant in a gliding bacterium
Gliding, a form of motility requiring a surface, is as widely distributed in the bacterial world as swimming though the mechanism of translocation remains unknown (1). Although no chemoattractant has yet been identified, the discovery of directed movement might provide insight into the mechanism of gliding just as chemotaxis has enlightened bacterial swimming behavior. We chose to investigate chemotaxis in the gliding bacterium Myxococcus xanthus because its developmental program depends on directed motility for fruiting body morphogenesis.
M. xanthus is a Gram-type negative soil bacterium that uses amino acids as a carbon, energy, and nitrogen source. Amino acid depletion initiates a social developmental cycle in which tens of thousands of cells aggregate, construct a fruiting body, and sporulate (for review see ref. 2). Fruiting body morphogenesis is temporally and spatially synchronized by at least five intercellular signals (3, 4). The mechanism by which the cells direct their movement into the fruiting body remains unknown. When M. xanthus cells were separated from preformed fruiting bodies by a thin layer of agar, the nascent fruiting bodies formed over the preformed fruiting bodies, suggesting that the preformed structures secreted a morphogenic determinant with chemotactic properties (5). However, an exhaustive survey of biological compounds failed to identify any chemoattractants. Furthermore, chemotaxis was deemed unlikely for M. xanthus because the time required for a small soluble molecule to diffuse the length of the bacterium is short compared with the time required for M. xanthus to glide the same distance (6). Evidence for (7) and against (8) biased movement up steep gradients of rich mixtures like Casitone and yeast extract has been presented, but no single chemical attractant has been discovered.
We demonstrate that M. xanthus cells direct their movement up gradients of the phospholipid, phosphatidylethanolamine (PE). The mechanism of directed movement shares several features with chemotactic behavior of enteric bacteria.
METHODS
Strains and Growth Conditions.
M. xanthus DK1622, DZ4169 (frzCD), DZ4148 (frzE), and DZ4024 (frzCD224) were grown at 32°C in CYE [10 g/liter of Difco Casitone, 5 g/liter of Difco yeast extract, 10 mM 3-[N-morpholino]propanesulfonic acid (Mops), pH 7.6, and 4 mM MgSO4] broth with vigorous shaking.
Phospholipid Purification.
To 0.4 g (wet cell weight) of M. xanthus cells, 3.75 ml of methanol/chloroform (2:1) was added and vortexed for 1 hr. The suspension was centrifuged at 10,000 × g for 5 min, and the supernatant was saved. The pellet was re-extracted with 4.75 ml of methanol/chloroform/water (2:1:0.8), centrifuged again at 10,000 × g, and the supernatants were combined. Chloroform (2.5 ml) and 2.5 ml of water were added to the combined supernatants, vortexed, and centrifuged to separate the chloroform and aqueous layers. The chloroform layer was collected and dried under nitrogen. An adsorption column was prepared with 2 g of Supelclean LC-SI (Supelco 5–7200) in a 12-ml syringe barrel. The column was washed with 15 ml of hexane/methyl tert butyl ether (MTBE)/acetic acid (100:2:0.2). The extraction residue was resuspended in 1 ml of hexane/MTBE/acetic acid (100:2:0.2) and applied to the column. The column was washed with 15 ml of MTBE/acetic acid (100:0.2), and then the active fraction was eluted with 15 ml of MTBE/methanol/ammonium acetate (10:4:1), pH 7.6 and dried under nitrogen. The residue was resuspended to 10 mg/ml in chloroform and separated by silica gel G TLC (Fisher 05–719-800) for 2 hr in 200 ml of chloroform/acetone/methanol/acetic acid/water (10:4:2:2:1), sections of the plate were excised, extracted in methanol, and dried under nitrogen. PE was detected colorimetrically after saturation with 0.5% ninhydrin solution (3% acetic acid in 1-butanol) and incubation for 3 min at 100°C.
Fluorescent Phospholipid Gradients.
One microgram of N-(5-dimethylaminonaphthalene-1-sulfonyl)-sn-glycero-3-phosphoethanolamine triethylammonium salt (Dansyl-labeled) from chicken egg PE (Molecular Probes) in 10 μl of chloroform was dried on top of 1.5% agar (Difco) containing TPM buffer [10 mM Tris (hydroxymethyl) aminomethane HCl (pH 8.0), 8 mM MgSO4, and 1 mM KHPO4-KH2PO4] and incubated at 32°C for 18 hr. Sections of agar (5 mm × 3 mm) were cut along a radius beginning at the application point. Chloroform (1.5 ml) was added to each agar section and incubated with shaking at 32°C for 1 hr. The fluorescent lipid in the chloroform extracts was quantified by using a AMINCO⋅Bowman Series 2 luminescence spectrometer (excitation λ: 334 nm; detection λ: 520 nm).
Swarm Expansion Assay.
Ten microliters of chloroform (containing PE when appropriate) was dried over 0.8 cm2 of TPM agar and incubated at 32°C for 18 hr. M. xanthus DK1622 cells were resuspended to 5 × 109 cells/ml in India ink/Mops [10 mM 3-[N-morpholino]propanesulfonic acid), pH 7.6, 8 mM MgSO4, 10% India ink] buffer, and 2 μl was spotted within 3 mm of the solvent application spot. The plate was incubated at 32°C for 6 hr. Images were digitally captured at ×15 magnification by using a Wild Heerbrugg dissecting microscope, a Sony Power HAD 3CCD color video camera, and Power Macintosh 9500 with Adobe Premier software.
Reversal Period Assay.
Phospholipids and phospholipid derivatives were purchased from Sigma. Vegetative PE, developmental PE, and dioleoyl PE were dissolved in chloroform, and dilauroyl PE, dilauroyl glycerol, and dioleoyl glycerol were dissolved in chloroform/methanol (1:1). Four microliters of test compound was applied to an area of about 0.4 cm2 and dried for 10 min on a thin layer of 0.7% Difco agar containing 10 mM Mops, 4 mM MgSO4, pH 7.6 mounted on a microscope slide. Five microliters of M. xanthus cells diluted to 5 × 107 cells/ml in Mops buffer was dried on top of the test compound and incubated at room temperature for 10 min to allow the liquid grown cells to adjust to the surface. Slide cultures were observed at room temperature with a Leitz light microscope (Laborlux D) for 45 min at ×640 magnification. Stop-motion digital movies were produced by using the same equipment as in the swarm expansion assay and Adobe Premier movie-making software with a frame capture rate of 12 frames/min. To determine cellular reversal frequency, the paths of 20 isolated cells were followed, and reversals were manually enumerated.
Adaptation rates were measured as for the reversal period assay except that the test compound and cells were placed on 1.5% agar containing TPM buffer. The higher agar concentration provided a surface that was more resistant to desiccation during the extended recording period. Plate cultures were observed at room temperature with a Leitz light microscope (Laborlux D) for 120 min at ×640 magnification. Stop-motion digital movies were produced with a frame capture rate of 12 frames/min. Each film was divided into 15- or 20-min segments, and reversal frequencies were calculated for each interval.
RESULTS
In an effort to identify factors controlling directed cell movement of myxobacteria, fractions from wild-type M. xanthus DK1622 cells were applied to filter disks, placed on top of starving wild-type cells, and assessed for their affect on motility. One particular cellular fraction induced cells to coalesce into thick radial ridges that appeared to migrate toward the disk. The bioactive material was extracted with chloroform/methanol/water (9), and purified by silica gel G adsorption chromatography (10) and TLC (11). The compound was identified as PE, the predominant M. xanthus membrane phospholipid (12), based on its elution pattern during adsorption chromatography, its TLC Rf value of 0.6 and its reactivity with ninhydrin, a color indicator of free amino groups (data not shown).
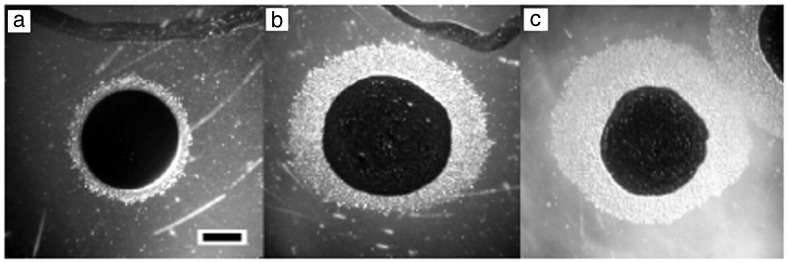
Gliding cells are motile only on surfaces, and it was necessary to construct a lipid gradient in agar that was suitably steep and stable to observe directed movement. A dansyl-labeled fluorescent PE derivative was applied in the center of an agar plate, the chloroform was evaporated, and the plate was incubated for 18 hr to permit PE diffusion. Agar slices were sectioned along a radial transect, and the fluorolipid was extracted with chloroform and quantified by using a luminescence spectrometer. A 10-fold drop in PE concentration was observed within the first 3 mm, and this was the area in which directed movement was examined (data not shown). When placed adjacent to a spot of evaporated chloroform alone, an M. xanthus colony expanded from its origin (demarked by India ink) evenly in all directions (Fig. 1a). However, cells within a gradient of PE purified from vegetative M. xanthus cells preferentially migrated up the gradient (Fig. 1b), and PE purified from developing M. xanthus induced a similar motility bias but at a 10-fold lower PE concentration (Fig. 1c). M. xanthus colonies did not respond to gradients of the fluorescent PE (data not shown).
Figure 1.
M. xanthus directed movement on PE concentration gradients. Test compounds originate at the top of each panel. Cell suspensions in 10% India ink (Higgins) were spotted about 3 mm from the test compounds. (a) Delivery solvent only. (b) One microgram of PE purified from vegetative M. xanthus cells. (c) PE (0.1 μg) purified from developing M. xanthus cells. (Bar: 1 mm.)
The asymmetric colonies could form one of two ways. The PE could cause the cells to travel faster up the gradient than down the gradient. In this case, the PE could facilitate more rapid gliding by altering the interaction of the cell with the agar surface (thigmotaxis) or acting as a transduced stimulus to accelerate the gliding motor (chemokinesis). In either case the velocity of the cells would be expected to accelerate on the PE gradient. Alternatively, cellular velocity could remain constant but the cells could prolong their movement along trajectories that move them up the gradient (chemotaxis). A good example is the chemotaxis of Escherichia coli, a process that is described by alternating behaviors known as runs and tumbles. During a run the bacterium swims in a line and punctuates that behavior with a period of erratic motion known as tumbling. After tumbling, the cell runs in a new, randomly chosen direction. Movement up a chemoattractant gradient occurs because cells that are oriented up the gradient run for a longer period of time than cells running down the gradient. Chemotaxis in E. coli is accomplished by a biased, random walk. This behavior is theoretically applicable to gliding bacteria. M. xanthus cells glide in the direction of the long cell axis and reverse their direction of movement every 6.8 min on average (13). By analogy with the enteric system, directed movement of M. xanthus could occur if cells suppressed direction reversals while moving up a gradient.
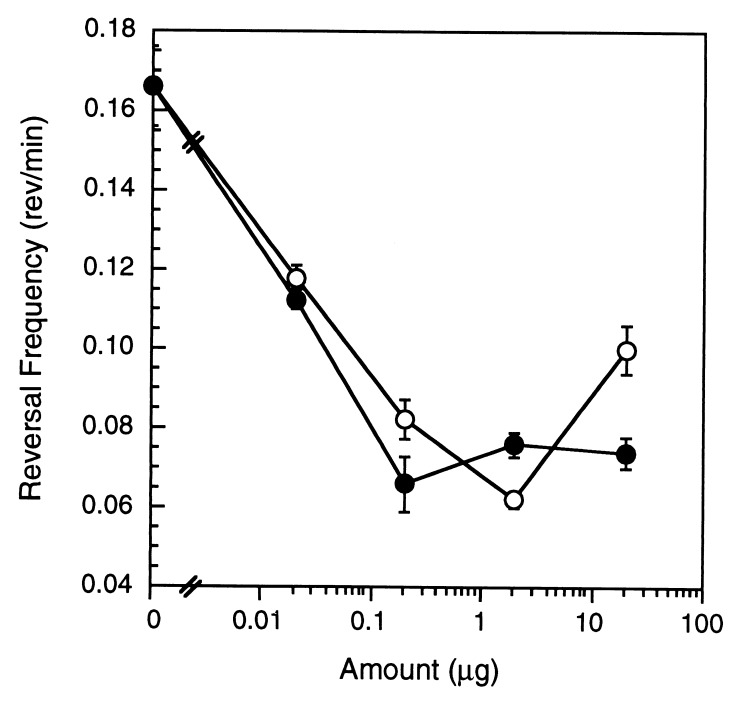
Stop-motion digital microscopy was used to measure the velocity of individual cells placed on an agar surface containing a uniform PE concentration. M. xanthus individuals traveled at a rate of 1 μm/min but moved 25% slower in the presence of uniform concentrations of M. xanthus PE equivalent to those encountered on the agar gradient. In marked contrast, the reversal frequency was suppressed with increasing amounts of M. xanthus vegetative and developmental PE (Fig. 2). These results are consistent with a chemotactic mechanism for directed movement.
Figure 2.
PE suppressed M. xanthus direction reversal frequency. M. xanthus reversal frequencies in the presence of uniform amounts of vegetative (○) and developmental (•) PE. Error bars are the SD of three replicates.
Classical chemotaxis of flagellated bacteria is defined by three additional characteristics: (i) the attractant is restricted to a narrow subset of molecular species (14), (ii) a signal transduction pathway leads from stimulus perception (15) to motor control (16, 17), and (iii) cells exhibit adaptation to the stimulus (18). We examined each of these criteria to determine whether M. xanthus cells display chemotaxis toward phospholipids.
Molecular Specificity.
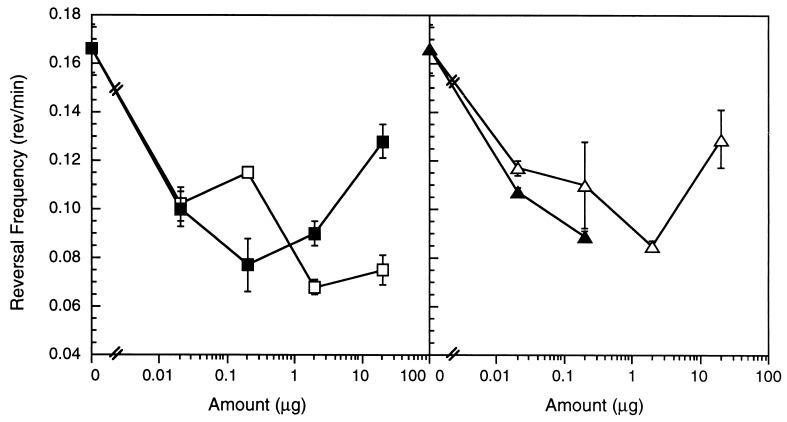
A number of synthetic PE species were assayed, including dilauroyl PE (di C12:0), dimyristoyl PE (di C14:0), dipalmitoyl PE (di C16:0), diheptadecanoyl PE (di C17:0), distearoyl PE (di C18:0), and dioleoyl PE (di C18:1 ω9c). Only dilauroyl PE and dioleoyl PE suppressed direction reversals (Fig. 3) and stimulated directed movement up a gradient (data not shown). Molecular specificity would be unlikely if directed movement was caused by a purely physical interaction between the lipid and the cell surface because PE species with similar amphipathicity should exhibit similar surfactant properties.
Figure 3.
Dilauroyl (diC12:0) and dioleoyl (diC18:1 ω9c) PE and their diacyl glycerol derivatives suppressed M. xanthus direction reversals. (Left) M. xanthus reversal frequencies in the presence of uniform concentrations of dilauroyl PE (□) and dilauroyl glycerol (■). (Right) M. xanthus reversal frequencies in the presence of uniform concentrations of dioleoyl PE (▵) and dioleoyl glycerol (▴). Error bars are the SD of three replicates.
To further evaluate molecular specificity, a variety of components of the biologically active PE species were tested, including dilauroyl glycerol, dioleoyl glycerol, lysolauroylphosphatidylethanolamine, lauric acid, phosphoglycerol, phosphoglycerolethanolamine, and phosphoethanolamine. Dilauroyl glycerol and dioleoyl glycerol were the only derivatives capable of suppressing M. xanthus reversal frequency (Fig. 3) and stimulating directed colony expansion (data not shown). Both diacyl glycerol species suppressed direction reversals to a similar extent as their parent compound although dioleoyl glycerol inhibited motility at concentrations greater than 0.2 μg (Fig. 3, Right). The observed molecular specificity for particular PE and diacyl glycerol species argues for a specific sensory system.
Sensory Transduction.
In enteric bacteria, a sensory transduction system couples the perception of the chemotactic stimulus to the rotation of the flagellar motor. A methyl-accepting chemoreceptor protein (MCP) binds the stimulus in the periplasm and transduces the information across the cytoplasmic membrane (15). The MCP-ligand complex inhibits autophosphorylation of a histidine kinase (CheA), a response regulator (CheY) is dephosphorylated (16), and the flagellar motor turns counterclockwise to produce a run (19). Methylation of the MCP allows adaptation to the stimulus (20) by increasing the rate of CheA phosphorylation 100-fold (21). M. xanthus contains sensory components similar to those found in the enteric bacteria. The M. xanthus FrzCD protein is an MCP homolog that curiously enough lacks the transmembrane and periplasmic segments used for chemoattractant binding (22). FrzCD does become methylated during M. xanthus development (23) and demethylated in the presence of chemorepellents such as isopropanol (24).
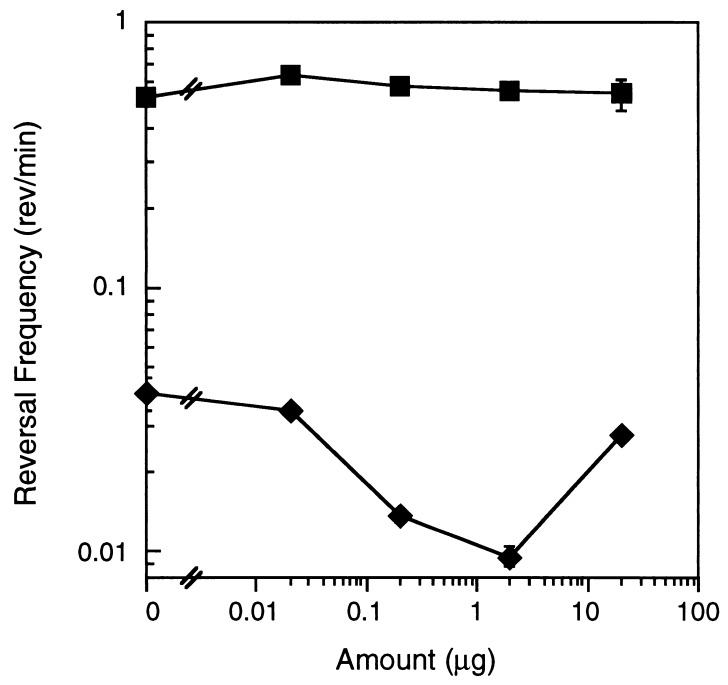
As an alternative to a receptor-mediated sensory transduction system, PE could intercalate into the M. xanthus membrane and interact directly with the gliding motor to produce less frequent reversals by a mechanism that does not involve chemoreception and sensory transduction. This type of perception pathway should bypass the Frz sensory system, which presumably feeds into the motor in a different manner. To explore this possibility, we used an M. xanthus mutant in the Frz sensory transduction pathway, which has a hyper-reversal phenotype. frzCD224 is truncated within the C-terminal MCP methylation domain. This mutant has a constitutive reversal period of 2.2 min, but unlike wild-type cells that can vary the interval between reversals widely, it exhibits a repetitive reversal cycle that results in little net movement of cells (13). If the PE motility effect is caused by a physical perturbation of the motor one might expect PE to bypass the Frz sensory transduction system and alter the typical mutant behavior. The frzCD224 reversal frequency was unaffected by the addition of dilauroyl PE (Fig. 4). This result suggests that the lipid does not bypass the signal transduction machinery and interact directly with the gliding motor.
Figure 4.
Two alleles of an M. xanthus MCP homolog responded differently to dilauroyl PE. Reversal frequencies of M. xanthus frzCD alleles, frzCD224 (■) and frzCD null mutant (⧫), in the presence of uniform amounts of dilauroyl PE. Error bars are the SD of three replicates.
To determine whether the Frz proteins form the sensory cascade through which PE is detected we examined the behavior of two frz mutants containing null mutations. The frzCD null allele contains an N-terminal insertion and has an exceedingly long reversal period of 20 min (0.05 rev/min reversal frequency). The other mutation is in frzE, which produces a bifunctional protein with homology to both CheA and CheY (25). Both frzCD and frzE mutants were defective in migration up gradients of dilauroyl PE but did respond to about 1/3 the extent of wild-type colonies (data not shown). These results provide further evidence that PE taxis is a transduced response.
Sensory Adaptation.
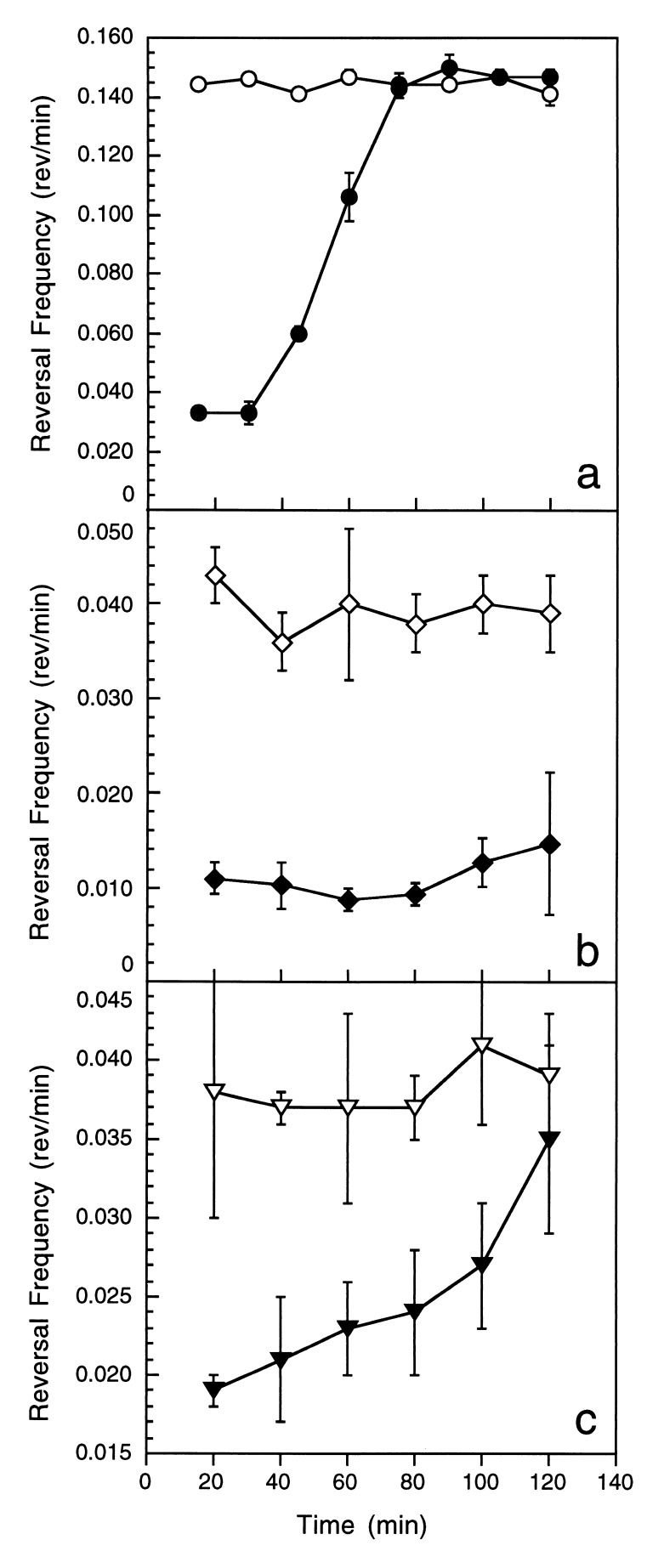
When exposed to a uniform concentration of a chemoattractant, enteric bacteria temporarily decrease their frequency of directional change but will return to their prestimulus behavior upon prolonged exposure (18). This phenomenon of sensory adaptation is essential for assessing changes in the stimulus concentration. To determine whether M. xanthus adapts to PE, cells on an agar surface were incubated with a uniform concentration of dilauroyl PE for an extended period of time. Cells suppressed reversals during the first 15 min of exposure and subsequently adapted to the prestimulus reversal frequency (6.8-min reversal period) in about an hour (Fig. 5a). Enteric bacteria respond to the attractant within 1 sec and adapt during the next 3 sec, though recovery time for a saturating stimulus can be much longer (18). Given the fact that the M. xanthus reversal period is 6.8 min on average, an adaptation period of 1 hr for a saturating stimulus is consistent with the scale differences relative to the E. coli system. A critical feature of the enteric system is that adaptation occurs within the time frame during which rotational brownian motion carries a cell more than 90 degrees off track. Although gliding cells are not subject to Brownian effects, they are influenced by the contours over which they glide and thus may bend from a linear trajectory in response to surface irregularities. We examined the tracks of M. xanthus cells, stimulated by 2 μg of dilauroyl PE, to determine whether 90-degree bends occur within the time frame of adaptation. Time was measured from a reversal until the path of a cell curved 90 degrees from its original orientation. On average, bends occurred 22 min ± 10 after a reversal event, a value well within the time period of adaptation to a saturating stimulus. These results may indicate fundamental differences in adaptation between enteric bacteria and myxobacteria.
Figure 5.
Wild-type and frz M. xanthus adapted differently to PE stimulation. Wild-type (a), frzCD null mutant (b), and frzE mutant (c) M. xanthus cells were incubated for 2 hr in the presence (closed symbols) and absence (open symbols) of 2 μg PE C12. Reversal frequencies were determined at 20-min intervals. Error bars are the SD of three replicates.
Mutants containing the null frzCD and frzE alleles responded to PE gradients, though much more weakly than wild-type cells. To determine what role FrzCD and FrzE play in response to PE, the excitation and adaptation response of the mutants was examined. Although wild-type cells in the presence of dilauroyl PE suppressed reversal frequency 2- to 4-fold (Figs. 3 and 5A), the frzCD null mutant suppressed reversal frequency about 4-fold (Fig. 4). However, the mutant exhibited no adaptation over 120 min of observation (Fig. 5b). The frzE mutant suppressed reversals 2-fold when initially exposed to dilauroyl PE and demonstrated an intermediate level of adaptation between wild-type and frzCD cells (Fig. 5c). Based on these results, the observed defect in directed movement up a PE gradient appears to be more a consequence of a defect in adaptation than excitation.
It could be argued that the observed adaptive response was caused by metabolism of the lipid rather than sensory adaptation because of the unusually long time required to restore the original reversal frequency. Although lipid metabolism has not been examined with M. xanthus, Myxococcus virescens can oxidize lipids (26). However, the Frz mutants present a compelling argument that the observed response is caused by adaptation rather than lipid consumption. One would not expect the frzCD and frzE mutants to differ in their pattern of lipid oxidation from an otherwise isogenic wild-type strain. These results argue that adaptation to PE is a transduced response and that the Frz system plays a role in adaptation.
DISCUSSION
We provide evidence that certain PE molecules are chemoattractants for M. xanthus by comparing the directed movement system with specific features of the enteric chemotaxis system. This comparison is complicated by the fact that the two types of bacteria translocate by completely different mechanisms. Enteric bacteria swim in liquid by using force generated from rotating flagella, while myxobacteria glide on solid surfaces by a mechanism that is as yet unknown. Nevertheless, three fundamental similarities point toward unifying principles. First, M. xanthus biased movement was correlated with a suppression of directional reversals to achieve longer runs when exposed to PE. Second, M. xanthus cells responded to PE molecules with particular fatty acids. Of the synthetic PE species we examined, only dilauroyl PE and dioleoyl PE, and their diacyl glycerol derivatives, induced a motility response. Third, M. xanthus exhibited adaptation to the attractant. These same three elements, suppression of directional change in the presence of an attractant, molecular specificity of the attractant, and sensory adaptation form the basis of enteric chemoattraction.
There are two puzzling differences between the E. coli response to attractants and the M. xanthus response, one in the length of time required for adaptation and the other dealing with the structure of the sensory system. When E. coli cells are exposed to a uniform, high concentration of an attractant, cells respond to the attractant within a second and adapt several seconds later, longer if the receptors become saturated (18). Ligand binding increases MCP methylation levels (27) and results in the restoration of the tumble state (20). M. xanthus requires over an hour to fully adapt. Although this lengthy period of adaptation may be justified in relation to the low frequency of cellular reversals and the slow rate of M. xanthus motility, the proposed mechanism of enteric adaptation seems too rapid to account for the M. xanthus response. In E. coli, four MCPs feed into a single common transduction system that controls both excitation and adaptation (15, 28, 29, 30). Because frzCD and frzE null mutants suppressed reversals in response to PE, the frz system is not essential for excitation and a second as yet unidentified transduction system is implicated in M. xanthus. As further evidence for a second series of Che homologs, mutations in the M. xanthus FrzG methyl esterase failed to eliminate FrzCD demethylation and implicated an undiscovered redundant homolog (7). Our data suggest the presence of more than one complete signal transduction pathway. Because frz mutants did not adapt to PE, M. xanthus sensory adaptation may be a process of competition between two pathways, one dedicated to PE excitation and the frz system dedicated to sensory adaptation. The interaction of multiple transduction systems may help slow sensory adaptation to levels consistent with the slow motility rate of M. xanthus.
This work was initiated by the discovery that PE extracts of vegetative and developing cells stimulated directed movement. This observation indicates that the M. xanthus cell membrane contains one or more discrete PE species that serve as chemoattractants. Fatty acid profiles of vegetative and developmental PE suggest that the major chemotactic PE molecule(s) produced by M. xanthus remain to be discovered. Laurate was absent from both vegetative and developmental PE samples (<0.5%). Oleate accounted for 8.64% of the fatty acids in the vegetative PE and, based on the dose-response curve with dioleoyl PE, can account for only a portion of the chemotactic activity in the vegetative extract. However, oleate decreased to 1.54% in the more active developmental PE sample, suggesting the presence of an additional developmental PE chemoattractant. Most of the M. xanthus PE species are not commercially available, and the chemical structures of some fatty acid constituents remain unknown. Together these problems make it more difficult to determine the chemical nature of the active species. Because of differences in fatty acid chain length and saturation between dilauroyl and dioleoyl PE and because species with intermediate structures were ineffective, more than one biologically active species may exist in M. xanthus. E. coli uses two separate chemoreceptors to distinguish the amino acids serine and aspartate (31, 32). Separate receptors for dilauroyl and dioleoyl PE might explain the inefficacy of intermediate structures in M. xanthus.
We report on a phospholipid serving as a prokaryotic chemoattractant. Chemotaxis toward 1,2-diacylglycerol (33), lysophosphatidylcholine (34), and lysophosphatidic acid (35), has been previously reported in eukaryotic human leukocytes, human monocytes, and Dictyostelium discoideum amoebae, respectively. Intriguingly, each eukaryotic cell is a bacterial predator, like M. xanthus, and denotes a possible relationship between predation, surface translocation, and chemotaxis toward slowly diffusing surfactants. Furthermore, in each eukaryotic system a phospholipase-cleaved phospholipid derivative is responsible for directing movement. We have demonstrated that dilauroyl and dioleoyl glycerol can suppress M. xanthus direction reversals, which may implicate a similar involvement of phospholipase processing in M. xanthus signal transduction.
Acknowledgments
We are grateful to David Harrell for his excellent technical assistance and Dr. Harry Dailey for the use of his luminescence spectrometer. This work was supported by National Science Foundation Grants MCB9601077 and BIR9413235.
ABBREVIATIONS
- PE
phosphatidylethanolamine
- MCP
methylated chemotaxis protein
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. Burchard R. Annu Rev Microbiol. 1981;35:497–529. doi: 10.1146/annurev.mi.35.100181.002433. [DOI] [PubMed] [Google Scholar]
- 2.Shimkets L J. Microbiol Rev. 1990;54:473–501. doi: 10.1128/mr.54.4.473-501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagen D C, Bretscher A P, Kaiser D. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 4.Downard J, Ramaswamy S V, Kil K. J Bacteriol. 1993;175:7762–7770. doi: 10.1128/jb.175.24.7762-7770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lev M. Nature (London) 1954;173:501. [Google Scholar]
- 6.Dworkin M, Eide D. J Bacteriol. 1983;154:437–442. doi: 10.1128/jb.154.1.437-442.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi W, Kohler T, Zusman D R. Mol Microbiol. 1993;9:601–611. doi: 10.1111/j.1365-2958.1993.tb01720.x. [DOI] [PubMed] [Google Scholar]
- 8.Tieman S, Koch A, White D. J Bacteriol. 1996;178:3480–3485. doi: 10.1128/jb.178.12.3480-3485.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bligh E G, Dyer W J. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton J G, Comai K. Lipids. 1988;23:1146–1149. doi: 10.1007/BF02535281. [DOI] [PubMed] [Google Scholar]
- 11.Yamanaka S, Fudo R, Kawaguchi A, Komogata K. J Gen Appl Microbiol. 1988;34:57–66. [Google Scholar]
- 12.Orndorff P E, Dworkin M. J Bacteriol. 1980;141:914–927. doi: 10.1128/jb.141.2.914-927.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blackhart B, Zusman D. Proc Natl Acad Sci USA. 1985;82:8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mesibov R, Adler J. J Bacteriol. 1972;112:315–326. doi: 10.1128/jb.112.1.315-326.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedblom M L, Adler J. J Bacteriol. 1980;144:1048–1060. doi: 10.1128/jb.144.3.1048-1060.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borkovich K A, Simon M I. Cell. 1990;63:1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- 17.Welch M, Oosawa K, Aizawa S, Eisenbach M. Proc Natl Acad Sci USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg H C. Cold Spring Harbor Symp Quant Biol. 1988;53:1–9. doi: 10.1101/sqb.1988.053.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Larsen S H, Reader R W, Kort E N, Lao W, Adler J. Nature (London) 1974;249:74–77. doi: 10.1038/249074a0. [DOI] [PubMed] [Google Scholar]
- 20.Goy M F, Springer M S, Adler J. Cell. 1978;15:1231–1240. doi: 10.1016/0092-8674(78)90049-1. [DOI] [PubMed] [Google Scholar]
- 21.Borkovich K A, Alex L A, Simon M I. Proc Natl Acad Sci USA. 1992;89:6756–6760. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBride M, Weinberg R, Zusman D R. Proc Natl Acad Sci USA. 1989;86:424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBride M J, Zusman D R. J Bacteriol. 1993;175:4936–4940. doi: 10.1128/jb.175.15.4936-4940.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBride M J, Kohler T, Zusman D R. J Bacteriol. 1992;174:4246–4257. doi: 10.1128/jb.174.13.4246-4257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCleary W R, Zusman D R. Proc Natl Acad Sci USA. 1990;87:5898–5902. doi: 10.1073/pnas.87.15.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loebeck M E, Klein H P. J Gen Microbiol. 1956;14:281–289. doi: 10.1099/00221287-14-2-281. [DOI] [PubMed] [Google Scholar]
- 27.Kort E N, Goy M F, Larsen S H, Adler J. Proc Natl Acad Sci USA. 1975;72:3939–3943. doi: 10.1073/pnas.72.10.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke S, Koshland D E., Jr J Biol Chem. 1979;254:9695–9702. [PubMed] [Google Scholar]
- 29.Kondoh H, Ball C B, Adler J. Proc Natl Acad Sci USA. 1979;76:260–264. doi: 10.1073/pnas.76.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manson M D, Blank V, Brade G, Higgins C F. Nature (London) 1986;321:253–256. doi: 10.1038/321253a0. [DOI] [PubMed] [Google Scholar]
- 31.Adler J. Science. 1969;166:1588–1597. doi: 10.1126/science.166.3913.1588. [DOI] [PubMed] [Google Scholar]
- 32.Springer M S, Goy M F, Adler J. Proc Natl Acad Sci USA. 1977;74:3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright T M, Hoffman R D, Nishijima J, Jakoi L, Snyderman R, Shin H S. Proc Natl Acad Sci USA. 1988;85:1869–1973. doi: 10.1073/pnas.85.6.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinn T, Parthasarathy S, Steinberg D. Proc Natl Acad Sci USA. 1988;85:2805–2809. doi: 10.1073/pnas.85.8.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jalink K, Moolenaar W H, Van Duijn B. Proc Natl Acad Sci USA. 1993;90:1857–1861. doi: 10.1073/pnas.90.5.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]