Abstract
A novel ribonucleoprotein complex enriched in nucleolar proteins was purified from yeast extracts and constituents were identified by mass spectrometry. When isolated from rapidly growing cells, the assembly contained ribonucleic acid (RNA) polymerase (pol) I, and some of its transcription factors like TATA-binding protein (TBP), Rrn3p, Rrn5p, Rrn7p, and Reb1p along with rRNA processing factors, like Nop1p, Cbf5p, Nhp2p, and Rrp5p. The small nucleolar RNAs (snoRNAs) U3, U14, and MRP were also found to be associated with the complex, which supports accurate transcription, termination, and pseudouridylation of rRNA. Formation of the complex did not depend on pol I, and the complex could efficiently recruit exogenous pol I into active ribosomal DNA (rDNA) transcription units. Visualization of the complex by electron microscopy and immunogold labeling revealed a characteristic cluster-forming network of nonuniform size containing nucleolar proteins like Nop1p and Fpr3p and attached pol I. Our results support the idea that a functional nucleolar subdomain formed independently of the state of rDNA transcription may serve as a scaffold for coordinated rRNA synthesis and processing.
Keywords: in vitro transcription, Saccharomyces cerevisiae, ribosome biogenesis, matrix assisted laser desorption ionization (MALDI) mass spectrometry, nucleolus
Introduction
The synthesis and processing of RNA require a complex network of protein–protein and protein–nucleic acids interactions. Immunofluoresence and EM observations suggest that all three eukaryotic nuclear RNA polymerases (pols) are involved in large transcription factories in distinct regions of the nucleus (Jackson et al. 1993; Wansink et al. 1993; Iborra et al. 1996; Pombo et al. 1999; Scheer and Hock 1999). The reactions leading to mature rRNA take place in the nucleolus, a specialized compartment of the nucleus. The nucleolus can be divided into three morphological subdomains: the fibrillar center, the dense fibrillar component, and the granular component (for reviews see Dechampesme et al. 1999; Leger-Silvestre et al. 1999; Scheer and Hock 1999). There is growing evidence that synthesis of ribosomal RNA by pol I occurs at the dense fibrillar component and fibrillar center interface. For instance, pol I activity has been localized near the surface of the fibrillar center (Weipoltshammer et al. 1996; Pombo et al. 1999). Furthermore, it was shown that nascent pre-rRNA transcripts are extended into the dense fibrillar component (Lazdins et al. 1997). Immunological studies, as well as pulse labeling experiments with bromo-uridine 5′-triphosphate short enough to ensure that most transcripts remain at their site of synthesis, also pointed towards the dense fibrillar compartment as the site of transcription (Hozak et al. 1994; Jackson and Cook 1995; Mosgoeller et al. 1998).
Interestingly, components involved in processing of rRNA, like the small nucleolar RNAs (snoRNAs) U14, U3, and MRP, or the protein Gar1p, were also shown to localize to the dense fibrillar component (Lazdins et al. 1997; Leger-Silvestre et al. 1997), indicating that processing activities are in close proximity to the transcription apparatus. Furthermore, the nucleolar protein fibrillarin colocalizes with pol I in mouse early embryos (Cuadros-Fernandez and Esponada 1996), but also with nascent rRNA transcripts (Garcia Blanco et al. 1995). Taken together, these results suggest a cooperative interface of rRNA synthesis and processing; however, the molecular basis of such an interaction has still to be elucidated.
Large pol I–containing protein machineries that combine several enzymatic entities have been reported in plants, mice, and frogs (Saez-Vasquez and Pikaard 1997; Seither et al. 1998; Albert et al. 1999). These putative pol I holoenzymes comprise all activities required for the initiation of transcription in vitro (Saez-Vasquez and Pikaard 1997; Seither et al. 1998). The holoenzyme isolated from Xenopus contained additional enzymatic features like histone acetyltransferase and protein kinase activities (Albert et al. 1999). However, all pol I–containing complexes described so far failed to show RNA processing activities.
A pol I–containing holoenzyme has yet to be reported in yeast. Initiation of transcription by pol I is dependent on the presence of the transcription initiation factors TATA-binding protein (TBP), core factor (CF), upstream activating factor (UAF), and Rrn3p, which have yet to be identified in a preassembled complex. The TATA-binding protein TBP (Cormack and Struhl 1992; Schultz et al. 1992), which could be isolated as part of a stable 240-kD protein complex (Milkereit et al. 1997), was shown to interact in vitro and in vivo both with components of the CF (Lin et al. 1996) and the UAF (Steffan et al. 1996, Steffan et al. 1998). CF is composed of three stably associated proteins encoded by the genes RRN6, RRN7, and RRN11 (Keys et al. 1994; Lalo et al. 1996; Lin et al. 1996), whereas UAF is a multiprotein complex consisting of at least five different proteins: Rrn5p, Rrn7p, Rrn10p (Keys et al. 1996), and the histones H3 and H4 (Keener et al. 1997). Binding of UAF to the promoter may be necessary to recruit CF and the initiation-competent form of pol I, which forms a stable complex with the transcription initiation factor Rrn3p (Yamamoto et al. 1996; Milkereit and Tschochner 1998), to the start site of rRNA synthesis. During in vitro transcription, Rrn3p was shown to be dissociated from pol I, resulting in inactivation of the initiation complex (Milkereit and Tschochner 1998).
Yeast pol I also requires other transcription factors for elongation and termination. These include Reb1p, which binds to the template and stops movement of pol I along the DNA in vitro (Lang et al. 1994), and a releasing activity that was necessary to release terminated transcripts from the template (Tschochner and Milkereit 1997).
Ribosomal RNA is processed by methylation, pseudouridylation, and cleavage to generate the mature 18S, 5.8S, and 25–28S rRNAs. Several proteins and many snoRNAs are required for these fundamental steps of rRNA maturation (for review see Venema and Tollervey 1995; Smith and Steitz 1997; Lafontaine and Tollervey 1998).
The snoRNAs can be divided into three classes (Balakin et al. 1996): box C+D, box H and ACA, and MRP snoRNAs. Box C+D snoRNAs are all associated with yeast fibrillarin Nop1p and two homologous proteins, Nop56p and Nop58p (Lafontaine and Tollervey 1998), and are either necessary to specify sites of ribose methylation or are involved in pre-RNA cleavage (for instance, U3). All box H and ACA snoRNAs interact with a protein complex that contains Gar1p, Nhp2p, Nop10p, and Cbf5p (Henras et al. 1998; Watkins et al. 1998), and are required for either sequence-specific pseudouridylation (Henras et al. 1998; Lafontaine et al. 1998; Watkins et al. 1998) or cleavage of immature rRNA (for instance, snR10, snR30) (Tollervey and Kiss 1997). The remaining snoRNA is the RNA component of the RNase holoenzyme MRP, which contains eight further protein subunits and is essential for pre-rRNA cleavage at one specific site (Tollervey and Kiss 1997; Chamberlain et al. 1998). Finally, other nucleolar proteins are involved in maturation of rRNA. These include Rrp5p, which is required for formation of both 18S and 5.8S rRNA (Venema and Tollervey 1996), and a multienzyme complex designated as exosome, which consists of at least five polypeptides (Rrp4p, Rrp41p, Rrp42p, Rrp43p, and Rrp44p) that are required for the 3′ processing of the 5.8S rRNA (Mitchell et al. 1997).
The goal of this study was to investigate how transcription and processing events are coupled. We have reported previously that all factors required for pol I–dependent transcription initiation copurify through several purification steps (Tschochner 1996; Milkereit et al. 1997). To test if synthesis and maturation of rRNA were linked, we attempted to isolate nucleolar subcomplexes active for transcription and processing of rRNA and to identify their constituent proteins. Mass spectrometry has recently become an efficient and highly sensitive technique for the identification of proteins in small amounts separated by SDS-PAGE (Shevchenko et al. 1996a). We have previously suggested the use of these powerful techniques for the characterization of mutiprotein complexes (Lamond and Mann 1997), and have already characterized a large number of nuclear and cytosolic complexes. We have now isolated a nucleolar RNP assembly that contains proteins required for transcription initiation, transcription termination, RNA processing, and ribosomal proteins as well as snoRNAs and mature rRNA, and is able to catalyze rRNA initiation, termination, and modification. This nucleolar subassembly could also be isolated from pol I–deficient cells and from cells inactive for transcription, suggesting that an organized structure independent of active rRNA synthesis provides a scaffold for the regular formation of the nucleolus.
Materials and Methods
Strains and Templates
Yeast wild-type strain BJ926, strain Gpy2 containing hemagglutinin (HA)-tagged A43 (generous gift of Drs. A. Setenac, C. Carles, M. Riva, and G. Peyroche, CEA/Saclay, Gif sur Yvette, France), strain YJV166 (generous gift of D. Tollervey, Institute of Cell and Molecular Biology [ICMB], Edinburgh, UK) (Venema and Tollervey 1996), which contained HA-tagged Rrp5p, and strain OG39-6d (Δ135; generous gift of Dr. O. Gadal, Biochemie-Zentrum Heidelberg, Heidelberg, Germany), which contained a disruption in the second largest pol I subunit A135, strain NOY797 and NOY844, which contained HA-tagged Rrn7p and Rrn5p, respectively (generous gift of Dr. M. Nomura, University of California Irvine, Irvine, CA) (Keener et al. 1997, Keener et al. 1998), were used for preparation of the extracts and subsequent fractionation. Plasmid pSES5 (Stewart and Roeder 1989; Tschochner 1996), which was linearized with EcoRV and circular pSIRT (Musters et al. 1989a), served as template for the initiation assays. The 3′ extended template pItailKS (Tschochner 1996) was used to generate transcripts with purified pol I. Antibodies directed against pol I subunits A190 and A43 were generous gifts of Drs. A. Sentenac, C. Carles, and M. Riva (CEA/Saclay), antibodies directed against Reb1p were kindly provided by Dr. W. Lang (Hutchinson Cancer Research Center, Seattle, WA), and antibodies generated against Fpr3p were kindly provided by Dr. J. Thorner (University of California Berkeley, Berkeley, CA).
In Vitro Transcription
Promoter-dependent and nonspecific transcription reactions were performed as described elsewhere (Milkereit et al. 1997).
Preparation of Whole Cell Extracts and Fractionation by Gel Filtration
Preparations of whole cell extracts (WCEs) on a small scale were performed as described (Grandi et al. 1993), with the exception that the lysis buffer contained 20% glycerol, 20 mM Hepes, pH 7.8, 150 mM potassium acetate, 50 mM NaCl, 1 mM MgCl2, 1 mM DTT, 1 mM PMSF, and 2 mM benzamidine, and four 20-min bead beatings were performed after zymolyase treatment. The resulting suspension was centrifuged at 100,000 g for 20 min in a table top centrifuge, and 0.15 ml of the supernatant (protein concentration 34 mg/ml) was loaded on a Superose-6 column (FPLC; Amersham Pharmacia Biotech). The column was processed in 10% glycerol, 20 mM Hepes, pH 7.8, 300 mM potassium acetate, 1 mM MgCl2, 1 mM DTT, 0.2 mM EDTA, 1 mM PMSF, and 2 mM benzamidine with a flow rate of 200 μl/min, and 0.6 ml fractions were collected. 25 μl of every second fraction was TCA-precipitated, separated by 10% SDS PAGE, and analyzed by Western blotting.
Preparation of the Nucleolar RNP Assembly
Fraction PA600 (protein concentration 2.5–5 mg/ml) was prepared as described previously (Tschochner 1996; Milkereit et al. 1997).
Gel Filtration of Fraction PA600 on Superose-6
50 μl of fraction PA600 was loaded on a Superose-6 column (SMART; Amersham Pharmacia Biotech) and processed in buffer BU300 (buffer BU supplemented with 300 mM potassium acetate) with a flow rate of 12.5 μl/min. Fractions of 0.05 ml were collected. 25 μl of each fraction was TCA-precipitated, separated by SDS-PAGE, and analyzed by Western blotting.
Purification of pol I
Homogenous pol I-A was a generous gift of C. Carles and colleagues (CEA/Saclay). pol I-p, a mixture of pol I–Rrn3p complex and of initiation inactive pol I, was purified as described previously (Milkereit et al. 1997). After purification on BioRex70, 50 μl of the pol I–containing fraction B2000 was loaded on a Superose-6 column (SMART; Amersham Pharmacia Biotech) and processed at a flow rate of 15 μl/min of buffer BU1500 (buffer BU containing 1,500 mM potassium acetate). Fractions of 50 μl were collected. The peak fraction of pol I (fraction 28), which contained both pol I–Rrn3p complex as well as monomeric pol I, was used for the assays as indicated.
Immunoprecipitation of HA-tagged pol I
4 ml of WCEs (12.5 mg/ml) derived from yeast strain Gpy2, which contained a HA-tagged (and His6-tagged) A43 subunit or control strains, was diluted sevenfold in buffer BU300, supplemented with 10 mg/ml milk powder, and immunoprecipitated with 0.025 ml anti-HA antibodies. Before the incubation with the yeast extracts, the antibodies were cross-linked to 0.05 ml protein A–Sepharose with 20 mM DMP (dimethyl-pimelidate) for 30 min at room temperature according to a protocol described elsewhere (Harlow and Lane 1988) and washed with 3× 0.5 ml PBS, 1× 0.5 ml 20 mM Hepes, pH 7.8, and 2× 0.5 ml BU300 with 10 mg/ml milk powder. After incubation with the extracts at 4°C for 2 h, the beads were washed with 2× 0.5 ml BU300 and 1× 0.5 ml 20 mM Hepes, pH 7.8, and resuspended in 0.05 ml SDS sample buffer. 20 mg of each WCE and 5 ml of the immunoprecipitation were analyzed by Western blotting. The identical conditions were chosen when immunoprecipitation was performed from nuclease-treated WCEs. Before immunoprecipitation, WCEs were incubated with 120 U of DNase I and 0.2 mg/ml RNase A for 30 min at 30°C. These conditions removed all of the RNA present in the WCE and 20 μg DNA of added salmon sperm DNA, as was judged by ethidium bromide–stained agarose gels (data not shown). (Note: endogenous DNA was not visible in WCEs). For the control experiments depicted in Fig. 1, we chose the pol I–deficient strain OG39-6d, which lacked both an epitope tag on one of its subunits, and furthermore, is the second largest pol I subunit. In control experiments performed with functional pol I, we had found that untagged pol I binds to the matrix as well, although at a lower degree. Although a significant difference was obtained in the amount of pol I–binding proteins recovered between the tagged versus the untagged pol I, the experiment as outlined in Fig. 1 makes this difference even more clear-cut.
Figure 1.
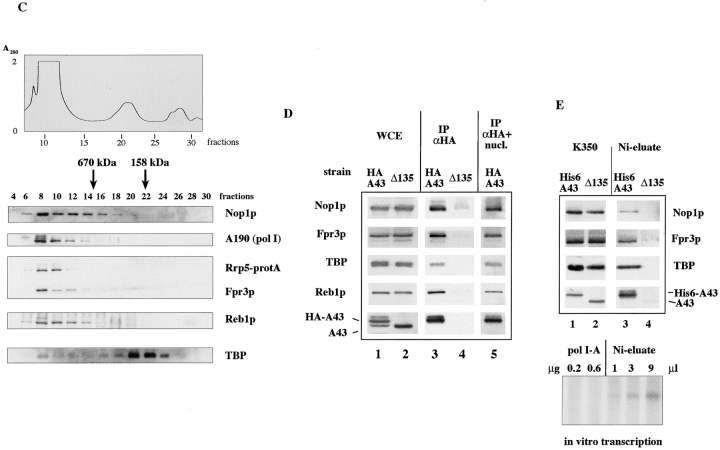
Copurification of nucleolar proteins. (A) Fractionation scheme of WCEs. WCE was prepared and fractionated as described (Tschochner 1996; Milkereit et al. 1997). The relative amounts of total protein in the single fractions are indicated in italics. Note that <1% of the proteins were found in fraction PA600. (B) Monitoring of different proteins through the established purification scheme. 20 μg of WCE, 8 μg of fractions K90, K350, and T0, and 4 μg of fraction PA600 were separated by 10% SDS-PAGE and analyzed by Western blotting with antibodies as indicated. (C) Gel filtration of WCEs in the presence of 300 mM acetate. 0.15 ml WCE (34 mg/ml) of yeast strain YJV166 (Venema and Tollervey 1996), which contained protein A–tagged Rrp5p, was generated in buffers with low ionic strength and fractionated on a Superose-6 column (FPLC) in the presence of 300 mM potassium acetate. 0.1 ml of each fraction was TCA precipitated and analyzed by Western blotting. Nop1p, yeast fibrillarin; A190, pol I subunit A190; Rrp5p-ProtA, protein A–tagged Rrp5p; Reb1p, pol I–specific transcription termination factor. (D) Immunoprecipitation of HA-tagged A43. Equal amounts of WCEs derived from strains Ypg2, which contained a HA- and His6-tagged A43 subunit, and OG39-6d (Δ135) in which A135 has been disrupted, were immunoprecipitated with antibodies directed against the HA tag. 20 μg of WCE (lanes 1 and 2) and 10% of the immunoprecipitations (lanes 3–5) were analyzed by Western blotting. Lane 5: same conditions as lane 3; however, treatment of the WCE with a mixture of RNase and DNase precluded the immunoprecipitation. (E) Affinity purification of His6-tagged A43. 8.5 mg of fraction K350 derived from yeast strain Gpy2 that contained a HA- and His6-tagged A43 subunit and the same amount of the Δ135 strain, which contained approximately the same amount of Nop1p, were nuclease-treated (see above), loaded on Ni-agarose (0.5 ml) in the presence of buffer A supplemented with 10 mM imidazole, washed with buffer BU300 including 10 mM imidazole, and eluted with buffer BU300 including 250 mM imidazole. 0.1% of the load and 2% of the elution step was analyzed by Western blotting (upper panel). Increasing volumes of the eluate derived from the His6-A43 strain were tested in transcription initiation (pol I-A, purified pol I which lacks initiation factors).
Mass Spectrometry and Protein Identification
0.25 ml of the pol I–containing fraction B2000 (4.5 mg/ml) was loaded on a Sephacryl S-300 column and processed in buffer BU600 at a flow rate at 0.4 ml/min. 0.75-ml fractions were collected and 0.4 ml of the fraction with the highest protein concentration, which also contained the highest amount of pol I, was separated by 10% SDS-PAGE and silver-stained. The single bands were excised, washed, in-gel reduced, S-alkylated, and subjected to tryptic digestion as described (Shevchenko et al. 1996a,Shevchenko et al. 1996b). The mixture of saturated alpha hydroxyccynnamic acid and nitrocellulose was used as a thin film matrix (Vorm et al. 1994). Peptide mass maps were recorded on a Bruker Reflex III Matrix Assisted Laser Desorption Ionization (MALDI) Time-of-Flight mass (Bruker-Daltonics). The list of peptide masses measured with accuracy, on average better than 50 ppm, was searched against a comprehensive sequence database NRDB without any restrictions on molecular weight or species of origin using Peptide Search software (Jensen et al. 1996).
Pseudouridylation
Analysis of pseudouridylation was performed as described previously (Grosjean et al. 1990; Szweykowska Kulinska et al. 1994; Simos et al. 1996). RNA was transcribed in vitro in the presence of [32P]GTP and [32P]UTP. Transcripts were separated on 6% polyacrylamide gels and the bands of the characteristic size were excised and eluted. Equal amounts of isolated radiolabeled transcripts were digested with RNaseT2 and further analyzed as described (Grosjean et al. 1990).
Northern Blot Analysis
0.5 ml of fraction K350, T0, and P600 was phenol/chloroform extracted, ethanol precipitated, and analyzed on an agarose gel to standardize their content of nucleic acids. Finally 0.25% of fraction P600 and 0.13% of fraction K350 and T0 were loaded on a 1.2% agarose/formaldehyde gel. The RNA from WCEs was obtained from 100-ml cultures that were grown to an OD600 of 0.7. 10-ml aliquots were centrifuged for 5 min at 3,000 rpm, then washed in 10 ml diethylpyrocarbonate-water and centrifuged again. The pellet was resuspended in 500 μl diethylpyrocarbonate-water and mixed with 200 μl baked glass beads, 200 μl sterile filtrated detergent mix (2% Triton X-100, 1% SDS, 100 mM NaCl, 10 mM Tris, pH 8, 1 mM EDTA), and 300 μl phenol/chloroform (1:1). The probe was vortexed nine times for 20 s and cooled on ice between the cycles. After centrifugation for 10 min at 4°C at 14,000 g, the supernatant was phenol/chloroform extracted. 2 μl of the hydrous phase was loaded on the gel. Further analysis of the RNA was according to a protocol described elsewhere (Sambrook et al. 1989). For quantification of the RNAs, 0.2% of each fraction was loaded and the gel was analyzed after Northern blotting with a PhosphorImager. The probes for U3, U14, and MRP snoRNAS were kindly provided by Dr. D. Tollervey (ICMB). They were the same as described in Beltrame and Tollervey 1992 and Morrissey and Tollervey 1997 (5′-CUAUAGAAAUGAUCCU-3′ for U3, 5′-TCACTCAGACATCCTAGG-3′ for U14, and 5′-AATAGAGGTACCAGGTCAAGAAGC-3′ for MRP, and 5′-ATGTCTGCAGTATGGTTTTAC-3′ for snR30).
EM
Fractions PA600, PA600 Δ135, and B2000 were diluted 100, 100, and 40 times, respectively, in buffer A (20 mM Hepes, pH 7.8, 2 mM MgCl2, 90 mM KOAc, 20% glycerol) to form the clusters. To prevent cluster formation, buffer A had to be supplemented with KOAc to reach a final concentration >100 mM. 6-nm colloidal gold particles were coupled to antibodies as described by the manufacturer (Aurion). A slight excess of antibodies was allowed to interact with the gold particles to form a stable gold solution at high ionic strength. The solution was centrifuged three times at 45,000 g and the fluffy pellet was resuspended each time in buffer B (5 mM carbonate buffer, pH 9.7) to remove the excess of unbound antibodies. Finally, the solution was centrifuged under the same conditions but the pellet was resuspended in buffer B supplemented with 0.01% Tween 20.
The immunogold labelings were performed by mixing 1 μl of fraction B2000, 4 μl of buffer A, and 1 μl of the gold-coupled antibody solution diluted to reach a normalized density of gold particles. The incubations were left for 2 h at room temperature then diluted with 38 μl of buffer A. 5 μl of each solution was deposited on a glow-discharged carbon-coated grid, washed with buffer C (10 mM Tris, pH 7.5, 0.01% Tween 20), and negatively stained with a 2% uranyl acetate solution. Micrographs were recorded on Kodak SO163 films at a magnification of 45,000 with a Phillips CM120 transmission electron microscope operating at 100 kV.
Results
A Macromolecular Complex Containing pol I, pol I–dependent Transcription Factors, and Other Nucleolar Proteins
We have described previously the simple derivation of an efficient promoter-dependent pol I–specific in vitro transcription system starting from yeast WCEs (Tschochner 1996; Milkereit et al. 1997). All of the components that were required for in vitro initiation of transcription at the ribosomal gene promoter copurified in a single chromatographic fraction (fraction PA600; Fig. 1 A). The copurification of the entire array of pol I–dependent transcription factors through the chromatographic steps and through a precipitation step with low ionic strength suggested that all the factors were associated in a large complex. It seemed unlikely that all the necessary transcription factors (>24 different polypeptides) would copurify and precipitate under the same conditions by virtue of having the same reduced solubility in low ionic strength buffers. Therefore, we assumed that this precipitation step might be specific for factors involved in rRNA synthesis and for proteins possibly required for the biogenesis of ribosomes.
Indeed, when the distribution of the two exclusively nucleolar proteins, Nop1p and Fpr3p (Benton et al. 1994), was monitored through our established purification scheme (Fig. 1 B), we found that both proteins precipitated during dialysis and were enriched in the transcriptionally active fraction PA600. Although this fraction contained <1% of the total proteins (Fig. 1 A), Nop1p and Fpr3p were enriched >25 times during the purification procedure as judged by comparative Western blot analysis (data not shown). Significant amounts of pol I and of factors involved in pol I–dependent transcription such as the TBP, Rrn7p and Rrn5p (components of CF and UAF, respectively), or the transcription termination factor Reb1p were also precipitated within fraction PA600 (Fig. 1 B). On the other hand, preparations of pol I, TBP, and Reb1p that were further purified remained soluble in the same low ionic strength buffer (data not shown), suggesting that a physical interaction between pol I and the transcription factors with other precipitable components is required for coprecipitation. Interestingly, all nonnucleolar proteins analyzed so far, like the vacuolar carboxypeptidase y (data not shown) or nuclear proteins, neither copurified nor coprecipitated with the nucleolar factors (Fig. 1 B). For instance, no specific component of the pol II transcription machinery, such as pol II, transcription factor IIB, and transcription factor TFIIS, precipitated in low ionic strength buffers, which underlines the specificity of the precipitation for nucleolar factors (Fig. 1 B). Therefore, we favor the assumption that the coprecipitated nucleolar factors might represent a functional or structural subunit of the nucleolus, rather than a casual aggregation of proteins with similar properties.
To ensure that the whole array of nucleolar factors was not adventitiously associated as a result of nonspecific aggregation in low ionic strength buffers, we monitored the chromatographic behavior of the pol I transcription factors and nucleolar proteins in cell extracts. WCEs were generated under conditions very similar to the ionic strength of the living cell. Proteins of a 100,000-g supernatant of such yeast WCEs were fractionated by size on a Superose-6 column in the presence of 300 mM potassium acetate, and the migration of pol I and other nucleolar factors was monitored. Yeast fibrillarin Nop1p and pol I eluted with an apparent molecular mass of >10 MD from the column (Fig. 1 C). Many other nucleolar proteins, such as Fpr3p and Rrp5p (Venema and Tollervey 1996), or proteins that play a role in pol I–dependent transcription initiation or termination, coeluted with Nop1p (Fig. 1 C).
Physical interaction between pol I and other nucleolar proteins in WCEs was verified by coimmunoprecipitation assays. When whole yeast cell extracts from strains containing a HA-tagged A43 subunit were immunoprecipitated with antibodies directed against the HA epitope, nucleolar proteins like Nop1p and Fpr3p as well as the transcription factors TBP and Reb1p coprecipitated together with pol I (Fig. 1 D, lanes 3 and 5). No significant coprecipitation of the nucleolar factors was observed in control experiments with yeast strains lacking an epitope-tagged pol I subunit (Fig. 1 D, lane 4; see also Materials and Methods). To rule out that oligomeric nucleic acids might mediate the observed association between pol I and other nucleolar proteins, the immunoprecipitation experiment was repeated with WCEs from which nucleic acids had been removed by treatment with nucleases; the nucleolar proteins still coprecipitated with pol I (Fig. 1 D, lane 5).
Affinity chromatography of His6-tagged pol I on Ni-agarose was used to show that the coelution of pol I, initiation factors, and nucleolar factors (Fig. 1 E, upper panel) was sufficient to evoke an initiation active complex (Fig. 1 E, lower panel). In contrast, high amounts of highly purified pol I-A were not able to initiate transcription.
The coprecipitation and coelution of pol I and all tested nucleolar factors supported the idea that these factors interact in solution, and consequently, might be able to form a large macromolecular assembly.
Analysis of the Resolubilized Complex and Identification of Components by Mass Spectrometry
After precipitation with low ionic strength, the nucleolar proteins could be resolubilized in buffers with moderate salt concentrations (i.e., 600 mM potassium acetate). Gel filtration experiments confirmed that even under these elevated ionic strength conditions, a proportion of a large pol I–containing complex remains associated. To determine the size of the putative assembly, the resolubilized fraction PA600, which contained the putative nucleolar assembly, was clarified and applied to a Superose-6 column. As published previously, most of the pol I eluted from the column in fractions corresponding to a molecular mass between 600–1,500 kD (Milkereit et al. 1997). However, a significant proportion of pol I eluted approximately with the exclusion volume from the column (Fig. 2 A, fraction 18). A significant amount of the other nucleolar proteins tested, such as Nop1p and Fpr3p, as well as the different pol I–dependent transcription factors Rrn3p, TBP, and Reb1p, were also detected in the same fraction (Fig. 2 A). The observation that not all of pol I and the other nucleolar components comigrated is possibly due to a partial disruption of the macromolecular assembly. Perhaps the salt conditions necessary to solubilize the precipitated components additionally promote disintegration of the assembly into smaller subcomplexes, which can be separated on the sizing column.
Figure 2.
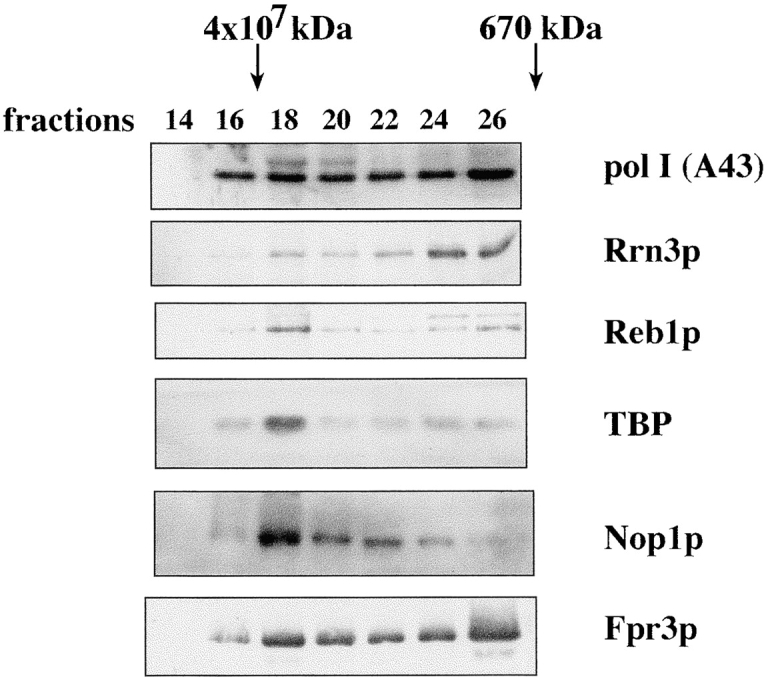
Analysis of fraction PA600 by gel filtration on Superose-6. Coprecipitated proteins were resolubilized in buffer BU600, which contained 600 mM potassium acetate diluted to an ionic strength of 300 mM potassium acetate, centrifuged, and 50 μl was applied to a Superose-6 column (SMART; Amersham Pharmacia Biotech) and processed with a flow rate of 12.5 μl/min in buffer BU300. Fractions of 50 μl were collected and 25 μl of each was analyzed by Western blotting.
The putative nucleolar assembly present in fraction PA600 could be divided into two fractions, designated B600 and B2000 (Fig. 1 A), by chromatography on a BioRex 70 column (Milkereit et al. 1997). When fraction B2000, which was significantly enriched in pol I, was dialyzed against low ionic strength buffers, pol I and the nucleolar proteins present in fraction B2000 were coprecipitated, suggesting that some of the components in this fraction are particularly involved in holding the multidinous factors together (see also Fig. 6 C).
Figure 6.
The nucleolar assembly isolated from pol I–deficient cells can recruit pol I to result in a transcriptionally active complex. (A) The nucleolar components copurify and coprecipitate when prepared from yeast strains not active in pol I–dependent transcription. (Upper panel) 8 μg of fraction PA600 derived from stationary cells (lane 3), from strain delta 135, which was disrupted in the A135 pol I subunit (strain OG39-6d) (lane 2), and from growing cells was analyzed by Western blotting with antibodies as indicated. (Lower panel) 1.25 μg of each PA600 fraction was analyzed in promoter-dependent transcription. (B) Purified pol I can be reassociated into a functionable assembly. Purified pol I-p (7 μl), which also contained pol I/Rrn3p complex but lacked TBP-cpl and fraction B600 (see Milkereit et al. 1997; Milkereit and Tschochner 1998) and thus was not able to initiate transcription on the rDNA promoter by itself, was combined with 12.5 μg (lanes 1–3) or without (lanes 4–6) fraction PA600 derived from strain Δ135 lacking A135 and adjusted to 1.5 M potassium acetate. The fractions were dialyzed against buffer BU100 containing 100 mM potassium acetate and centrifuged. Precipitated components were resolubilized in buffer BU600 containing 600 mM acetate and analyzed in promoter-dependent transcription. (C) In vitro assay to analyze the pol I–recruiting activity. Purified pol I coprecipitates with components present in fraction B2000 (Δ135) rather than with constituents in fraction B600 (Δ135). 0.5 μg pol I-A was combined with either 6 μg fraction PA600, 4.8 μg fraction B600, or fraction B2000, all derived from the pol I–deficient strain Δ135 in a total volume of 25 μl, adjusted to 1.5 M potassium acetate, dialyzed against buffer BU100, and centrifuged. Precipitates were resolubilized in 15 μl BU600 and analyzed in nonspecific RNA synthesis.
To identify more proteins of the putative nucleolar assembly, polypeptides in fraction B2000 were then analyzed by mass spectrometry. After a further purification step by gel filtration on Sephacryl S-300 (Milkereit et al. 1997), the fraction with the highest amount of pol I was separated by SDS-PAGE and silver-stained (Fig. 3 A). The prominent bands were screened by peptide mass fingerprinting, since the yeast genome had been sequenced at the time of these experiments. MALDI mass spectrometry was performed on a small part of the supernatant of in-gel digested proteins (see Fig. 3 B). The resulting peptide mass lists were searched in a comprehensive database and the results were evaluated as described (Jensen et al. 1996; Shevchenko et al. 1996a). Table lists the identification of known yeast proteins from the gel in Fig. 3 A. Interestingly, all proteins identified so far turned out to be localized in the nucleolus. Among the identified proteins were the pol I subunits in the range between 14–190 kD, factors that have been shown to be involved in the processing of rRNA like Rrp5p (Venema and Tollervey 1996), Cbf5p (Lafontaine et al. 1998), and Nhp2p (Henras et al. 1998), and factors that very likely play a role in ribosomal biogenesis and/or processing of rRNA, like Fpr3p and Fpr4p (Benton et al. 1994; Dolinski et al. 1997). Two gene products of unknown function were also identified (designated as open reading frames), which also were localized to the nucleolus (Milkereit, P., A. Podtelejnikov, M. Mann, and H. Tschochner, unpublished data). Interestingly, some ribosomal proteins (RPS10B, RPS7A, YL8b) were also found in this fraction.
Figure 3.
Identification of proteins of the assembly by mass spectrometry. (A) pol I present in fraction PA600 was further purified on BioRex70 and separated on a Sephacryl S-300 column in the presence of 300 mM potassium acetate as described previously (Milkereit et al. 1997). The peak fraction in terms of protein content, which also contained most of pol I (data not shown), was separated by SDS-PAGE. Single bands of silver-stained gels were excised, in-gel digested with trypsin, and analyzed by MALDI mass spectrometry followed by database searching with the resulting peptide masses. Due to high mass accuracy of MALDI peptide maps, proteins were identified unambiguously. (B) MALDI mass spectrum of peptides recovered after tryptic in-gel digestion of single protein band. A search with detected masses in nonredundant protein database showed that 18 peaks (P) matched to calculated tryptic peptide masses of yeast FK506-binding nuclear protein (Fpr3) within an accuracy better than 50 ppm and covered 33% of the sequence. The peaks marked by M correspond to tryptic autolysis products.
Table 1.
Results of Protein Identification by High Mass Accuracy MALDI Peptide Mapping
| Protein | Mol wt | No. of peptidesmatched | Sequencecoverage |
|---|---|---|---|
| kd | % | ||
| Rrp5p | 194 | 61 | 42.6 |
| A190 | 188 | 38 | 33.5 |
| ORF | 117 | 17 | 19 |
| A135 | 137 | 15 | 15 |
| ORF | 82 | 32 | 44 |
| Cbf5 | 55.2 | 10 | 25 |
| Fpr4 | 42.9 | 12 | 37 |
| Fpr3 | 46.5 | 18 | 33 |
| A49 | 42.7 | 21 | 63 |
| A43 | 36.3 | 17 | 47 |
| A40 | 38 | 20 | 61 |
| A34.5 | 34.2 | 7 | 23 |
| A23 | 18 | 6 | 44 |
| Rps10b | 28.7 | 13 | 49 |
| Rps7a | 29.3 | 10 | 32 |
| A27 | 25.2 | 10 | 51 |
| YL8 | 27.7 | 12 | 51 |
| Nhp2 | 18.9 | 7 | 42 |
| A14/14.5 | 17.9 | 6 | 44 |
Functional Properties of the Isolated Nucleolar Assembly
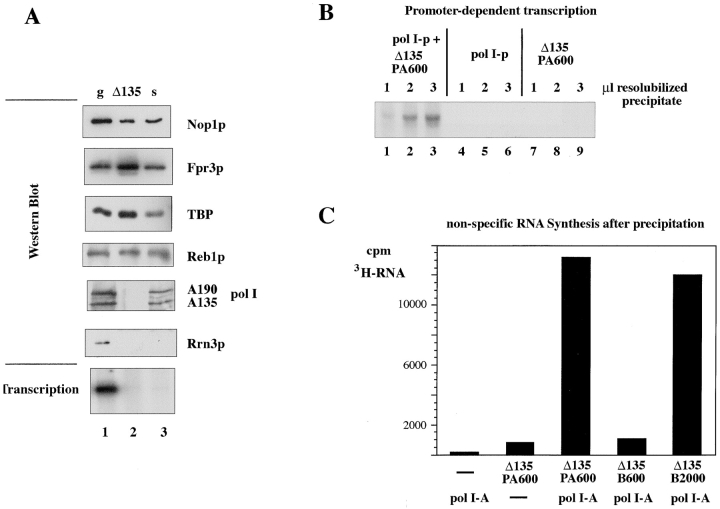
Functional analysis of fraction PA600, which contains the putative nucleolar assembly, supported the proposal that we have precipitated a cooperative macromolecular complex of the nucleolus. When the precipitated proteins were redissolved in buffers containing 600 mM potassium acetate, they demonstrated efficient activity in accurate inititation of transcription (Fig. 4 A, lane 3) (Milkereit et al. 1997). In contrast, neither the supernatant T0, with the soluble remaining pol I (Fig. 4 A, lane 2), nor purified pol I (designated pol I-p; Fig. 4 A, lane 4), was able to initiate transcription at the ribosomal gene promoter, indicating that other factors are required in order for pol I to function in transcription initiation, and that all these necessary factors were gathered in the precipitate (fraction PA600).
Figure 4.
In vitro activities of the isolated nucleolar assembly. (A) Fraction PA600 is capable of initiating and terminating rDNA promoter-dependent transcription. 16 μg of fraction T0 and 1.25 μg of fraction PA600 that contained both approximately equal amounts of pol I and 0.8 μg of purified pol I (pol I-A) were used in promoter-dependent transcription assays. In reactions depicted in lanes 1–3, 100 ng of plasmid pSES5, which contained the rDNA promoter but did not contain the termination site, served as a template. The plasmid was linearized with EcORV before adding to the assays analyzed in lanes 2 and 3. Each 100 ng of uncut plasmid pSIRT (Musters et al., 1989b), which contained both the rDNA promoter and the termination site, was used in reactions analyzed in lanes 4–6. (B) Fraction PA600 is capable of pseudouridylating RNA. 38 U of T7-pol in the absence (panel a) or presence (panel b) of 0.3 μg of the tRNA-specific pseudouridylation factor Pus1p, 5 μg of purified pol I (pol I-p) (panel c), and 12.5 μg of fraction PA600 (panel d) were used to synthesize RNA. DNA coding for intron containing Ile-tRNA (Szweykowska Kulinska et al. 1994; Simos et al. 1996) (panels a and b), template pItailKS (Tschochner and Milkereit 1997) containing an extended 3′ end, at which purified pol I-p can initiate transcription without promoter, and plasmid pSIRT (panel d) served as templates, respectively. Transcription was performed in the presence of [32P]UTP and [32P]GTP. Labeled transcripts were separated on a 7% polyacrylamide/7 M urea gel and transcripts corresponding to the expected size were excised from the gel and digested with RNase T2. The resulting nucleotides were analyzed by two-dimensional thin-layer chromatography as described (Grosjean et al. 1990).
Since the transcription termination factor Reb1p also coprecipitated with pol I, the initiation factors, and the other nucleolar proteins (Fig. 1 B), we proceeded to assay the putative protein assembly in fraction PA600 for a coordinated transcription termination activity. Indeed, fraction PA600 was also capable of terminating a correctly initiated transcript. In vitro transcription of a circular plasmid containing the pol I promoter and the native termination site resulted in a transcript of the expected size (Fig. 4 A, lane 6), whereas no defined transcripts could be detected when transcription was performed with circular plasmids that lacked the termination sequence (Fig. 4 A, lane 1). This result provides further evidence that the isolated nucleolar protein assembly contains different distinct functional properties, such as transcription initiation and termination, and that these functions might be linked in a cooperative fashion.
Cbf5p and Nhp2p, two other proteins detected in the isolated nucleolar assembly, were recently reported to be involved in pseudouridylation of rRNA (Henras et al. 1998; Lafontaine et al. 1998). These findings encouraged us to investigate whether the isolated nucleolar complex(es) can also modify their correctly synthesized and terminated RNA in vitro. Transcripts were synthesized by the putative nucleolar assembly using plasmid pSIRT as a template, which contained both the pol I promoter and a stretch of 700 bp from the 25S ribosomal DNA (rDNA). Accurately initiated and terminated transcripts were gel purified and their nucleotide composition was analyzed after RNase T2 digestion by two-dimensional thin-layer chromatography. As depicted in Fig. 4 B, panel d, the nucleolar assembly was capable of modifying uridines into pseudouridines, whereas purified pol I incorporated exclusively uridines into transcripts that had been nonspecifically initiated on a single-stranded DNA template (Fig. 4 B, panel c). tRNA synthesized in vitro by T7 pol in the absence (Fig. 4 B, panel a) or presence (Fig. 4 B, panel b) of the tRNA-specific pseudouridine synthase Pus1p, served as a control to identify pseudouridines within the nucleotides on the chromatograms.
Methylation is another common modification of mature rRNA. Fraction PA600, which contained the putative nucleolar assembly, was also able to methylate RNA since transcription in the presence of 14C-labeled S-adenosyl methionine showed the incorporation of 14CH3 in synthesized transcripts (Tschochner, H., unpublished observation).
Taken together, these results suggest that assembly of the different factors within the isolated nucleolar assembly might result in a cooperative enzymatic network that guides both synthesis and maturation of rRNA.
The Isolated Nucleolar Complex Contains snoRNAs
RNA coprecipitated with the nucleolar proteins within the macromolecular assembly. Since several snoRNA-associated proteins that are involved in rRNA processing, like Nop1p, Rrp5p, Nhp2p, and Cbf5p, could be detected in the isolated nucleolar complex, we wondered whether the isolated RNA contained snoRNAs. Indeed, Northern blot analysis revealed the presence of all the tested snoRNAs in the precipitated nucleolar assembly. Among the snoRNAs analyzed were MRP, the Box C and D snoRNAs U3 and U14 (Fig. 5), and the box H and ACA snoRNA snR30 (data not shown). Furthermore, substantial amounts of mature 18S rRNA (Fig. 5) were also detected. In contrast, RNAs synthesized by pols II and III, like the mRNAs for actin and the translation factor translation elongation factor eEF1 alpha-A chain, and the tRNAs for glutamic acid and leucine, were absent in the isolated nucleolar complex (Fig. 5). The amount of precipitated snoRNAs varied between 8% for MRP and 33% for U14, which corresponded to a 4- and 14-fold enrichment, respectively, if the quantities of snoRNAs were related to the amount of proteins present in the corresponding fractions. In contrast, no tRNAs were specifically enriched; <2% of them were precipitated. Taken together, these results provide additional evidence that the purification procedure described herein is highly specific for components organized in the nucleolus, and suggest that RNAs involved in the biogenesis of ribosomes are also present in the isolated nucleolar RNP assembly.
Figure 5.
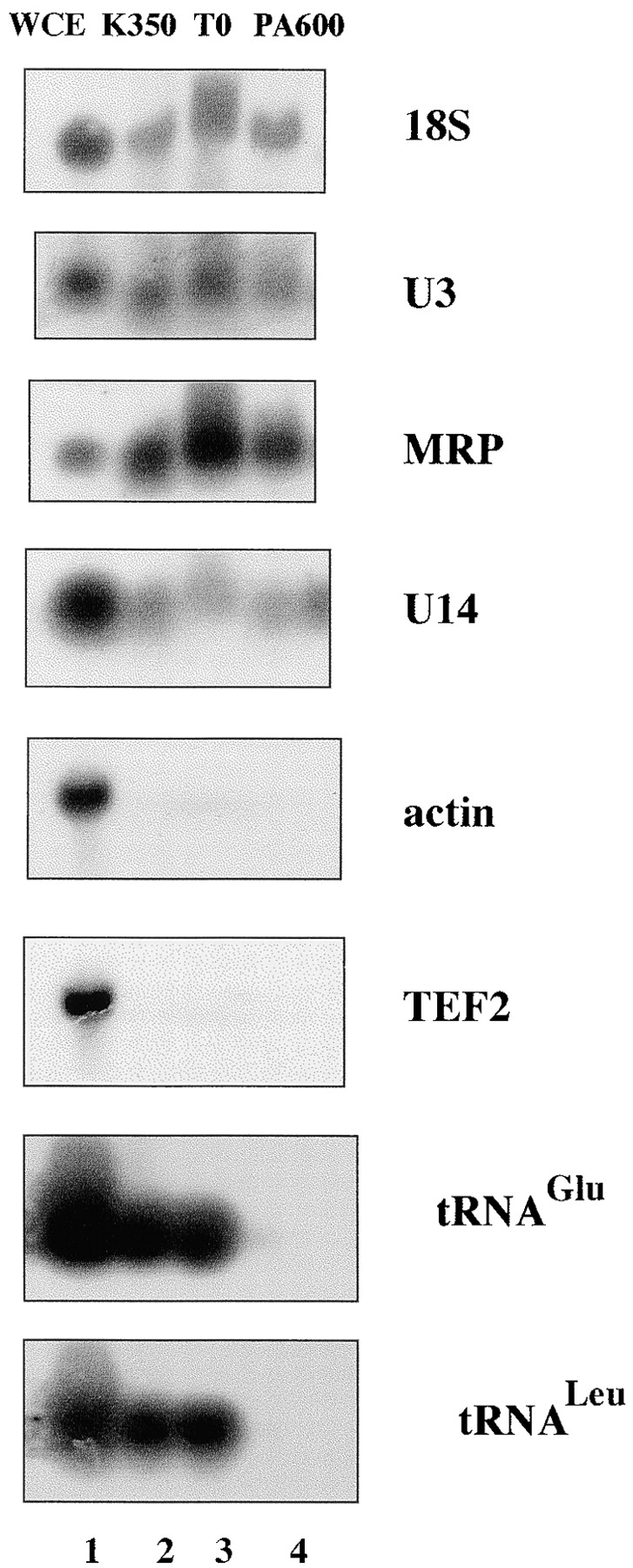
The isolated complex contains nucleolar RNA. WCEs and fractions generated according to the established purification scheme were prepared and analyzed by Northern blotting with specific oligonucleotides as described in Materials and Methods.
Pol I and Ongoing RNA Synthesis Is Not Essential for the Nucleolar Assembly; However, pol I Can Be Recruited to the Complex
The various structural components and functional properties of this nucleolar RNP assembly suggested the existence of an active multienzyme complex, in which synthesis and processing of rRNA might take place in a cooperative fashion. To investigate whether the isolated assembly was solely held together by a cooperative transcription-processing event, the structural integrity of the assembly was investigated in conditions in which pol I–dependent transcription has stopped. Thus, cell extracts were prepared from stationary cells in which rRNA synthesis was known to be completely downregulated (Clarke et al. 1996; Milkereit and Tschochner 1998), and from the yeast strain OG39-6d (Δ135), which is lacking the second largest pol I subunit, and thus, is deficient in pol I–dependent transcription. In this strain, the essential rRNAs were synthesized by pol II transcribing one rDNA unit under the control of a pol II–dependent promoter (Nogi et al. 1991). The same purification procedure used to isolate the nucleolar RNP complex from wild-type nonstationary cells was applied to both cell extracts (stationary wild-type cells and Δ135 strain). Similar amounts of proteins and RNA coprecipitated in each of the three preparations to result in fraction PA600g, PA600s, and Δ135, respectively (data not shown). Western blot analysis confirmed that both pol I–specific transcription factors, such as TBP or Rrn10p (a component of UAF) and termination factor Reb1p, and other nucleolar factors, like Nop1p and Fpr3p, were still associated together. In contrast to wild-type cells, pol I–dependent rRNA synthesis was completely abolished in the complex(es) isolated from stationary cells and the mutant strain (Fig. 6 A). In the mutant strain, neither the two largest pol I subunits nor the pol I–associated transcription factor Rrn3p could be detected in the nucleolar complex (Fig. 6 A, lane 2). Depletion of the pol I subunit A135 resulted both in a complete disruption of pol I and in the dissociation of most of the pol I subunits from the nucleolar complex, although other transcription initiation factors still assembled within the nucleolar substructure. It is important to note that fraction PA600 Δ135 was neither enriched in pol II nor did it display pol II–dependent transcription activity (data not shown). This result indicates that although the rRNA was synthesized by pol II, in this mutant strain, the rDNA transcribing pol II machinery was not integrated into the nucleolar assembly, and therefore, underlines the specificity of our purification procedure.
In stationary cells, all tested transcription and nucleolar factors, with the exception of Rrn3p, which was demonstrated to be essential for pol I to initiate transcription (Yamamoto et al. 1996), were present in the nucleolar assembly (Fig. 6 A, lane 3). Lack of pol I–associated Rrn3p was shown to be responsible for the loss of initiation activity, and therefore probably resulted in the loss of rRNA synthesis in stationary cells (Milkereit and Tschochner 1998). Again, although transcriptionally inactive, it was possible to purify the nucleolar assembly, providing further evidence that a cooperative function between rDNA transcription and subsequent rRNA processing steps is not required to keep the complex(es) assembled.
We further investigated whether the pol I–deficient nucleolar assembly was still able to recruit purified pol I and to restore the transcriptional activity. Highly purified pol I preparations like pol I-A and pol I-p, which contained both pol I–Rrn3p complex and free pol I, did not precipitate when dialyzed against buffers with low ionic strength (see Fig. 6 C). In contrast, coprecipitation of pol I was observed when dialysis was performed in the presence of the nucleolar complex(es) that had been derived from the mutant Δ135 strain, which completely lacked pol I (Fig. 6B and Fig. C). After resolubilization of the coprecipitated pol I and the nucleolar assembly, >85% of the applied nonspecific pol I activity in RNA synthesis was found in the pellet (Fig. 6 C). Even more striking, after coprecipitation, the pol I–Rrn3p complex regained its ability to cooperate with the remaining transcription initiation factors to restore an active initiation complex (Fig. 6 B, lanes 1–3), whereas in control reactions, neither precipitated mutant PA600 (Fig. 6 B, lanes 7–9) nor purified pol I (Fig. 6 B, lanes 4–6) showed any transcriptional activity. The purified pol I–Rrn3p complex used for this experiment lacked both the 240-kD TBP-containing complex and the necessary activity in fraction B600, and was therefore incapable of initiating transcription by itself (data not shown; see also Milkereit et al. 1997). To recover a functional transcription complex, all remaining factors required for in vitro transcription initiation, with the exception of pol I–Rrn3p, had to be present in the isolated nucleolar assembly derived from the mutant strain.
This result indicates that the isolated nucleolar substructure provides a scaffold for the assembly of a variety of nucleolar factors involved in rRNA synthesis and processing, independent of whether pol I–dependent RNA synthesis is turned on or off.
Based on this finding, we developed an in vitro assay to test various fractions for their ability to coprecipitate pol I. Further fractionation of PA600 Δ135, which contained the putative nucleolar assembly on BioRex70, revealed that the pol I–recruiting activity could be separated from the majority of proteins. Fraction B2000 Δ135 of the BioRex column, which contained ∼18% of the applied proteins, was able to coprecipitate pol I, whereas the same amount (or a threefold excess [data not shown]) of fraction B600 (Δ135) could not significantly affect the solubility of pol I in low salt buffers (Fig. 6 C). We take this as evidence for a feasible enrichment of a structural module that is capable of interacting with pol I and might serve as the structural backbone to associate with other nucleolar proteins, and thus, might be involved in building up the entire isolated nucleolar subcomplex.
Electron Microscopic Analysis of the Nucleolar Assembly
The biochemical evidence of large pol I–containing complexes present in fractions PA600 and B2000 prompted us to investigate the assemblies by EM. When the wild-type PA600 fraction was prepared in the presence of a buffer containing >600 mM KOAc, a homogeneous distribution of particles was observed (data not shown). The dilution of this fraction in the same buffer containing 90 mM KOAc resulted in the formation of loose supramolecular structures 0.1–0.3 μm in size (Fig. 7A and Fig. B). These assemblies, hereafter identified as clusters, consisted of a barely visible protein network associated with a large number of globular particles 10–15 nm in size located within or at the periphery of these structures.
Figure 7.
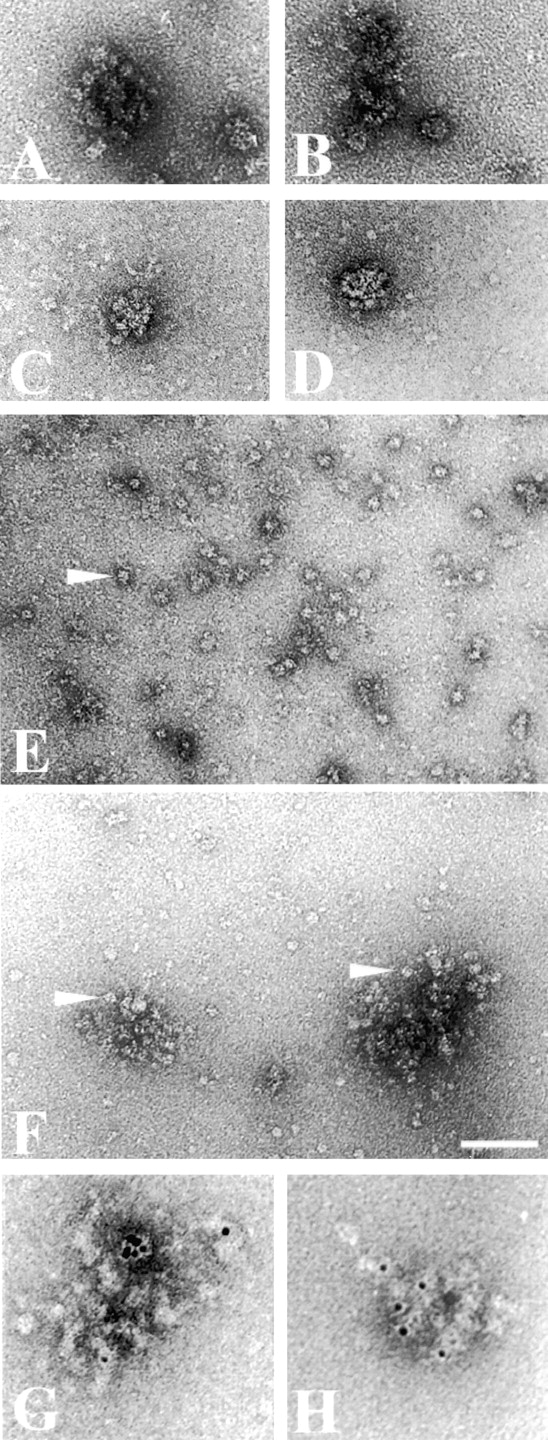
Electron microscopic analysis of the nucleolar assembly. (A and B) Clusters observed in fraction PA600 diluted 100 times in buffer B (see Material and Methods). (C and D) Clusters observed in fraction PA600 Δ135 diluted in the same conditions as above. (E) Field of fraction B2000 diluted in high ionic strength buffer. Pol I molecules are homogeneously dispersed on the carbon film (see also Schultz et al. 1990; Milkereit et al. 1997). The arrowhead points towards a pol I particle. (F) Field of fraction B2000 diluted in low–ionic strength buffer. The low ionic strength induces the formation of the clusters. The arrowheads point towards particles of the size of pol I. (G) Cluster labeled with the immunogold probe directed against Fpr3p. (H) Cluster labeled with the immunogold probe directed against Nop1p. Bar: 100 nm (A–F); and 50 nm (G and H).
The fraction PA600 Δ135, which contained the presumably nucleolar assembly from the pol I–disrupted strain, also formed supramolecular assemblies in low ionic strength buffers. These structures were, however, different from those formed from the wild-type strain, since the size of the particles was significantly smaller (≈7.5 nm) and since the particles seemed to be more closely packed (Fig. 7C and Fig. D). These data suggest that an underlying complex can form in the absence of pol I, and that pol I can be recruited to this complex by specific protein–protein interactions according to our biochemical data.
Due to the lower background of possible contaminations and the less complex protein composition, we decided to further investigate the pol I–containing assemblies in fraction B2000. This fraction has the same coprecipitation properties as the wild-type fraction PA600, is capable of recruiting pol I (see above), and similar clusters were observed upon lowering the ionic strength. In the presence of >600 mM KOAc, a homogeneous distribution of particles was observed. Digital analysis of EM images of these particles identified these structures as pol I molecules similar in size and shape to the previously analyzed highly purified pol I complexes (Schultz et al. 1990).
When the ionic strength of the buffer was lowered to 90 mM KOAc, the distribution of the pol I molecules was drastically altered and the vast majority of the molecules was recruited into the clusters. A preliminary image analysis of the particles associated with the clusters showed the characteristic size and shape of pol I molecules. Altogether, these observations indicate that the pol I molecules are assembled into large supramolecular assemblies.
To demonstrate that the nucleolar proteins Nop1p and Fpr3p are also included in the clusters, we analyzed the large pol I–containing complexes by immunogold labeling. Gold-coupled antibodies directed against the nucleolar proteins Nop1p and Fpr3p were incubated with fraction B2000 before cluster formation. The amount of labeled structures was determined for each specific probe and compared with a negative control formed by a gold-coupled nonspecific antibody. The density of the probes was carefully normalized by direct counting of gold particles adsorbed on a carbon film in order to have the same amount of gold particles in the incubation solution. In these conditions, the nonspecific antibody labeled ∼11% of the clusters, whereas the probes directed against the nucleolar proteins Fpr3p and Nop1p labeled 62 and 57% of the clusters, respectively (Fig. 7G and Fig. H). This significant difference was obtained in duplicate and indicated that the nucleolar proteins Fpr3p and Nop1p are part of the large assemblies.
These EM observations clearly show that a large macromolecular assembly, containing Fpr3p and Nop1p, is able to recruit several pol I molecules into a modular structure. The number of polymerases included in these structures depends on the size of the assemblies, precluding the possibility of a stoichiometric complex. These pol I–containing structures differ from protein aggregates by the fact that their thickness is globally monomolecular, and that in some cases, an underlying protein network can be distinguished between the pol I molecules and by the sequential assembly of the structure.
Discussion
From our results, we suggest the existence of a nucleolar RNP scaffold capable of forming an interface for the association of pol I and components involved in rDNA transcription and rRNA processing in vitro. Furthermore, this assembly is independent of rRNA synthesis. The assembly of these nucleolar factors when derived from growing cells results both in active in vitro transcription initiation and termination complexes, and in the ability to modify rRNA.
Specific Purification of the RNP Complex
The simple derivation and striking enrichment of one or several highly organized nucleolar multienzyme complexes, which comprise a whole variety of in vitro activities, underlines the specificity of the purification procedure, although the nature of the coprecipitation of the nucleolar factors remained obscure. Precipitation was not due to aggregation of chromatin in low salt buffers, a well-known practical problem during purification of nuclear scaffolds and matrices under physiological conditions (for review see Jackson and Cook 1995). Removal of aggregated chromatin and DNA from our extracts was accomplished by high-speed centrifugation and chromatography on diethylaminoethyl Sepharose (data not shown) before coprecipitation of the nucleolar factors occurred. Two further lines of evidence suggest that DNA is not involved in the precipitation process: (a) no ethidium bromide stained DNA could be detected in our nucleolar preparation; and (b) precipitation was also observed in the presence of 50–200 μg/ml ethidium bromide (data not shown), which has been reported to inhibit DNA–protein interactions (Lai and Herr 1992).
Nucleolar structures seem to be very sensitive to changes in the surrounding salt conditions. For example, Zatsepina et al. 1997 have reported that the nucleolus of cultured cells unraveled upon hypotonic shock and could be reformed when the cells were returned to isotonic medium. Changing the appropriate salt conditions might also cause the aggregation of a nucleolar multiprotein complex (fraction PA600), which is normally soluble under physiological conditions. A similar purification procedure using coprecipitation of proteins in buffers with low ionic strength to yield active rDNA transcribing fractions has been described independently (Riggs et al. 1995).
Components of the Nucleolar Assembly
Treatment of the complex with ethidium bromide, RNase, or DNase did not dissociate the assembled components, although incubation with RNase resulted in a significant loss of RNA (data not shown). Neither the genetic disruption of an essential pol I subunit (Fig. 6) nor incubation of the soluble complex with an excess of antibodies directed against Nop1p or against the pol I core enzyme was able to affect the formation of the nucleolar complex (data not shown). On the other hand, further fractionation of fraction PA600 derived from the pol I–lacking mutant on BioRex70 (Fig. 6 C) or on MonoS (data not shown) revealed fractions that preferentially coprecipitated soluble, purified pol I. We take these findings as evidence that neither RNA, DNA, nor pol I are required to keep the nucleolar substructure assembled. The proteinaceous factors holding the substructure together remain to be determined.
The identification of a variety of other proteins, in addition to pol I, in the isolated proteinaceous fraction confirms the existence of a macromolecular assembly of nucleolar components. All of the proteins identified so far could be localized to the nucleolus. Furthermore, some of the determined proteins, like Nop1p, Rrp5p, Nhp2p, and Cbf5p, have already been described to be components of higher organized complexes involved in rRNA processing (see Introduction) or in the assembly of mature ribosomes. For example, Rrp5p was shown to be required for formation of both 18S and 5.8S rRNA and the downregulation of yeast immunophilin Fpr3p, which is exclusively distributed to the nucleolus (Benton et al. 1994) and leads to an accumulation of 35S precursor rRNA, indicating its involvement in rRNA processing (Benton, B.M., and J. Thorner, personal communications).
In contrast to the well-characterized pol II holoenzyme, the components identified in the described macromolecular network do not form a stoichiometric complex. It seems more likely that a variable amount of different functional and structural subcomplexes can be gathered in this subnucleolar structure. For instance, several populations of pol I (initiation-competent as well as elongation-active pol I, pol I monomers, and dimers) could be identified in the assembly (Milkereit et al. 1997; Milkereit and Tschochner 1998). An uneven composition of the assembly was also confirmed by our electron microscopic analysis. For instance, nucleolar assemblies of increasing sizes were observed by EM after separation of the assemblies on velocity gradients (data not shown).
The nonstoichiometric distribution of the components as well as the structural data of the isolated complex based on EM favors the idea that proper nucleolar function might depend on the coalescence of supramolecular assemblies through structural interactions.
Interaction of rDNA-transcription and rRNA-processing Components
Our results provide the first biochemical evidence for a direct linkage between transcription and processing machineries and support previous suggestions (Brand et al. 1979; Veinot Drebot et al. 1988; Allmang and Tollervey 1998) and morphological observations describing a possible connection of both processes. For instance, the terminal balls, characteristic of eukaryotic rRNA transcription units in chromatin spreads, which are associated with the 5′ end of the growing rRNA chain, have been suggested to be rRNA processing complexes. Evidence supporting this idea comes from studies in which microinjected rDNA lacking the rRNA processing signal in the 5′ external spacer was transcribed without forming these 5′-terminal structures (Mougey et al. 1993).
Another important observation suggesting that processing might occur as the rRNA is synthesized was reported recently. Garcia Blanco et al. 1995 observed that when nucleoli were spread out into spherical bodies and visualized by fluorescent microscopy, the sites of rRNA synthesis and the location of pol I could be determined within these bodies. Each spherical body was tightly associated with a region containing fibrillarin, which can be taken as a marker of rRNA processing (Garcia Blanco et al. 1995), suggesting the interaction of rRNA synthesis and processing domains. It is conceivable that the superassembly of many of these domains composes the structural unit of the nucleolus.
Finally, a genetic link between a member of the transcription apparatus and one of the processing machinery has also been reported. Overexpression of transcription initiation factor Rrn3p was shown to suppress a mutation in CBF5, a gene required for pseudouridylation of rRNA (Cadwell et al. 1997).
Formation of the Nucleolus
Despite the increasing evidence describing the presence of functional domains in the nucleolus, relatively little is known about the forces that keep these supramolecular assemblies together. It is possible that the RNP assembly described herein represents a structural and operative subunit of the nucleolus. Many of these subunits might join through functional or structural interactions, such as active rRNA synthesis, attachment to the nuclear scaffold (Jackson and Cook 1995), or assembly on a rDNA gene. This model could explain the following different morphological observations, which at first glance, seem to be contradictory.
On the one hand, it has been suggested that nucleolar formation is driven by the procedure of building a ribosome, which implies that a coherent network of the particular synthesis, assembly, and processing reactions finally result in the appearance of a whole organelle. A regularly shaped nucleolus seems to be the consequence of active pol I–dependent gene transcription. For instance, inhibition of pol I–dependent transcription by actinomycin D blocked the formation of the nucleolus (Jordan et al. 1996). Furthermore, in pol I–deficient yeast strains in which rRNA was synthesized by pol II (Nogi et al. 1991), the crescent-shaped nucleolus is apparently disrupted into several granules (Oakes et al. 1993). A fragmented nucleolus was also observed when the rRNA genes were reduced to one single copy, suggesting that either the resulting low efficiency of rRNA synthesis or the organization of the rDNA strongly influences the formation of the nucleolus (Oakes et al. 1998).
In contrast, formation of a regular nucleolar structure was observed in pol I–inactive cells during early Xenopus development, which indicated that the organization of a defined nucleolar structure can be independent of the transcription process, but dependent on the presence of unprocessed rRNA (Verheggen et al. 1998).
It is conceivable that different functional domains of the nucleolus are arranged in a supramolecular assembly that provides the structural module of the nucleolus no matter whether transcription is off or on. For instance, pol I could be immobilized in the nucleolus through interaction with the constitutive proteins of the nucleolus. Such an organization might result in the appearance of a regularly shaped nucleolus and suggest that the transcription apparatus might pull through the DNA template rather than moving along the rDNA. Congruent with this idea is the observation that the pol I–dependent transcription machinery is associated to the nuclear scaffold independent of the presence of rDNA (Jackson and Cook 1995; Hozak 1996; Weipoltshammer et al. 1996).
Consistent with the idea of a superior organization made of single functional and structural subunits is the observation that inhibition of pol I–dependent rRNA synthesis by treatment with actinomycin D causes a redistribution of rDNA genes into clusters at the periphery of the regular nucleolus, whereas the major components of the rRNA transcription machinery including pol I and TBP still colocalize within these transcriptional inactive clusters (Jordan et al. 1996). Analogously, in yeast cells lacking pol I, the nucleolus is disrupted into granules, termed mini nucleolar bodies, which retain the ability to process pre-rRNA and assemble ribosomes (Oakes et al. 1993, Oakes et al. 1998). These mini nucleolar bodies are reminiscent of the prenucleolar bodies that occur at late telophase in cycling cells, and which are transcriptionally inactive and move to sites of active transcription after mitosis (Bell et al. 1992; Jimenez Garcia et al. 1994). Prenucleolar bodies are devoid of rDNA but contain nucleolar components, such as fibrillarin, nucleolin, and snoRNAs, and are prerequisites for forming a proper nucleolus (Jimenez Garcia et al. 1994). Remarkably, the isolated RNP complex described herein has biochemical and functional properties similar to these prenucleolar bodies. Furthermore, a reversible formation of prenucleolar-like bodies from intact nucleoli within the cell could be observed when cells were exposed to low ionic strength buffers (Zatsepina et al. 1997). The conditions used were similar to those that precipitate the RNP complex in our analysis, indicating that a conformational change in some nucleolar component(s) due to low salt conditions might affect the macromolecular network both of the nucleolus and of the RNP assembly described herein.
Our findings describe the isolation of a multifunctional nucleolar network, the identification of its protein components by modern mass spectrometric methods, and provides a concept of how the nucleolus might be structurally organized. The observations support the existence of distinct structural and functional modules capable of assembling and forming the nucleolus. This structural organization of the single modules might be necessary for the different multienzyme complexes to work synergistically to produce mature rRNA. Whether or not these structures exactly predominate in a living cell is still open to debate. However, the structural evidence for an assembly of the components that parallels acquisition of a complete rRNA synthesis activity, including initiation and termination of transcription as well as modification of rRNA, makes a strong case as to the functionality of this assembly.
Acknowledgments
We thank Drs. A. Sentenac, C. Carles, M. Riva, G. Peyroche, and J. Thorner for helpful discussions, and Dr. J. Thorner for his permission to mention data prior to publication. The excellent technical assistance of E. Draken and I. Eckstein is gratefully acknowledged. We are very grateful to Dr. I. Haas, Dr. J. Ortiz, Dr. Aled Edwards, Dr. Mike Sayre, and K. Fisher for helpful discussions and critical reading.
This work was supported by funds of the Deutsche Forschungsgemeinschaft, by a Human Frontier Science Program grant, the Institute National de la Santé et de la Recherche Médicale, the Centre National pour la Recherche Scientifique, the Hôpital Universitaire de Strasbourg (HUS), the Association pour la Recherche sur le Cancer, and the Danish National Science Foundation.
Footnotes
Abbreviations used in this paper: CF, core factor; HA, hemagglutinin; MALDI, matrix assisted laser desorption ionization; pol, RNA polymerase; rDNA, ribosomal DNA; snoRNA, small nucleolar RNA; TBP, TATA-binding protein; UAF, upstream activating factor; WCE, whole cell extract.
References
- Albert A.C., Denton M., Kermekchiev M., Pikaard C.S. Histone acetyltransferase and protein kinase activities copurify with a putative RNA polymerase I holoenzyme self-sufficient for promoter-dependent transcription. Mol. Cell. Biol. 1999;19:796–806. doi: 10.1128/mcb.19.1.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Tollervey D. The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J. Mol. Biol. 1998;278:67–78. doi: 10.1006/jmbi.1998.1693. [DOI] [PubMed] [Google Scholar]
- Balakin A.G., Smith L., Fournier M.J. The RNA world of the nucleolustwo major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- Bell P., Dabauvalle M.C., Scheer U. In vitro assembly of prenucleolar bodies in Xenopus egg extract. J. Cell Biol. 1992;118:1297–1304. doi: 10.1083/jcb.118.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M., Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO (Eur. Mol. Biol. Organ.) J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton B.M., Zang J.H., Thorner J. A novel FK506- and rapamycin-binding protein (FPR3 gene product) in the yeast Saccharomyces cerevisiae is a proline rotamase localized to the nucleolus. J. Cell Biol. 1994;127:623–639. doi: 10.1083/jcb.127.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand R.C., Klootwijk J., Sibum C.P., Planta R.J. Pseudouridylation of yeast ribosomal precursor RNA. Nucleic Acids Res. 1979;7:121–134. doi: 10.1093/nar/7.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell C., Yoon H.J., Zebarjadian Y., Carbon J. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol. Cell. Biol. 1997;17:6175–6183. doi: 10.1128/mcb.17.10.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain J.R., Lee Y., Lane W.S., Engelke D.R. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke E.M., Peterson C.L., Brainard A.V., Riggs D.L. Regulation of the RNA polymerase I and III transcription systems in response to growth conditions. J. Biol. Chem. 1996;271:22189–22195. doi: 10.1074/jbc.271.36.22189. [DOI] [PubMed] [Google Scholar]
- Cormack B.P., Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- Cuadros-Fernandez J.M., Esponada P. Immunocytochemical localization of the nucleolar protein fibrillarin and RNA polymerase I during mouse early embryogenesis. Zygote. 1996;4:49–58. doi: 10.1017/s0967199400002884. [DOI] [PubMed] [Google Scholar]
- Dechampesme A.-M., Koroleva O., Leger-Silvestre I., Gas N., Camier S. Assembly of 5S ribosomal RNA is required at a specific step of the pre-rRNA processing pathway. J. Cell Biol. 1999;145:1369–1380. doi: 10.1083/jcb.145.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinski K., Muir S., Cardenas M., Heitmann J. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. USA. 1997;94:13093–13098. doi: 10.1073/pnas.94.24.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Blanco M.A., Miller D.D., Sheetz M.P. Nuclear spreadsI. Visualization of bipartite ribosomal RNA domains. J. Cell Biol. 1995;128:15–27. doi: 10.1083/jcb.128.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Doye V., Hurt E.C. Purification of NSP1 reveals complex formation with ‘GLFG’ nucleoporins and a novel nuclear pore protein NIC96. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Droogmans L., Giege R., Uhlenbeck O.C. Guanosine modifications in runoff transcripts of synthetic transfer RNA-Phe genes microinjected into Xenopus oocytes. Biochim. Biophys. Acta. 1990;1050:267–273. doi: 10.1016/0167-4781(90)90179-6. [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane D. AntibodiesA Laboratory Manual 1988. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: pp. 1–726 [Google Scholar]
- Henras A., Henry Y., Bousquet Antonelli C., Noaillac Depeyre J., Gelugne J.P., Caizergues Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozak P. The nucleoskeleton and attached activities. Exp. Cell Res. 1996;229:267–271. doi: 10.1006/excr.1996.0370. [DOI] [PubMed] [Google Scholar]
- Hozak P., Cook P.R., Schofer C., Mosgoller W., Wachtler F. Site of transcription of ribosomal RNA and intranucleolar structure in HeLa cells. J. Cell Sci. 1994;107:639–648. doi: 10.1242/jcs.107.2.639. [DOI] [PubMed] [Google Scholar]
- Iborra F.J., Pombo A., Jackson D.A., Cook P.R. Active RNA polymerases are localized within discrete transcription “factories' in human nuclei. J. Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Jackson D.A., Cook P.R. The structural basis of nuclear function. Int. Rev. Cytol. 1995;162A:125–149. doi: 10.1016/s0074-7696(08)61230-9. [DOI] [PubMed] [Google Scholar]
- Jackson D.A., Hassan A.B., Errington R.J., Cook P.R. Visualization of focal sites of transcription within human nuclei. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen O.N., Podtelejnikov A., Mann M. Delayed extraction improves specificity in database searches by matrix-assisted laser desorption/ionization peptide maps. Rapid Commun. Mass Spectrom. 1996;10:1371–1378. doi: 10.1002/(SICI)1097-0231(199608)10:11<1371::AID-RCM682>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Jimenez Garcia L.F., Segura Valdez M.L., Ochs R.L., Rothblum L.I., Hannan R., Spector D.L. NucleologenesisU3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol. Biol. Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P., Mannervik M., Tora L., Carmo-Fonseca M. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J. Cell Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J., Dodd J.A., Lalo D., Nomura M. Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc. Natl. Acad. Sci. USA. 1997;94:13458–13462. doi: 10.1073/pnas.94.25.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J., Josaitis C., Dodd J., Nomura M. Reconstitution of yeast RNA polymerase I transcription in vitro from purified factors. J. Biol. Chem. 1998;273:33795–33802. doi: 10.1074/jbc.273.50.33795. [DOI] [PubMed] [Google Scholar]
- Keys D.A., Vu L., Steffan J.S., Dodd J.A., Yamamoto R.T., Nogi Y., Nomura M. RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae . Genes Dev. 1994;8:2349–2362. doi: 10.1101/gad.8.19.2349. [DOI] [PubMed] [Google Scholar]
- Keys D.A., Lee B.S., Dodd J.A., Nguyen T.T., Vu L., Fantino E., Burson L.M., Nogi Y., Nomura M. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 1996;10:887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- Lafontaine D., Tollervey D. Birth of the snoRNPsthe evolution of the modification-guide snoRNAs. TIBS (Trends Biochem. Sci.). 1998;23:383–388. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- Lafontaine D.L.J., Bousquet-Antonelli C., Henry Y., Caizergues-Ferrer M., Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J.-S., Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalo D., Steffan J.S., Dodd J.A., Nomura M. RRN11 encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. J. Biol. Chem. 1996;271:21062–21067. doi: 10.1074/jbc.271.35.21062. [DOI] [PubMed] [Google Scholar]
- Lamond A., Mann M. Cell biology and the genome projects - a concerted strategy for characterizing multiprotein complexes by using mass spectrometry. Trends Cell Biol. 1997;7:139–142. doi: 10.1016/S0962-8924(97)01031-3. [DOI] [PubMed] [Google Scholar]
- Lang W.H., Morrow B.E., Ju Q., Warner J.R., Reeder R.H. A model for transcription termination by RNA polymerase I. Cell. 1994;79:527–534. doi: 10.1016/0092-8674(94)90261-5. [DOI] [PubMed] [Google Scholar]
- Lazdins I.B., Delannoy M., Sollner-Webb B. Analysis of nucleolar transcription and processing domains and pre-rRNA movements by in situ hybridization. Chromosoma. 1997;105:481–495. doi: 10.1007/BF02510485. [DOI] [PubMed] [Google Scholar]
- Leger-Silvestre I., Gulli M.P., Noaillac-Depeyre J., Faubladier M., Sicard H., Caizergues-Ferrer M., Gas N. Ultrastructural changes in the Schizosaccharomyces pombe nucleolus following the disruption of the gar2+ gene, which encodes a nucleolar protein structurally related to nucleolin. Chromosoma. 1997;105:542–552. doi: 10.1007/BF02510491. [DOI] [PubMed] [Google Scholar]
- Leger-Silvestre I., Trumtel S., Noaillac-Depeyre J., Gas N. Functional compartmentalization of the nucleus in the budding yeast Saccharomyces cerevisiae . Chromsoma. 1999;108:103–113. doi: 10.1007/s004120050357. [DOI] [PubMed] [Google Scholar]
- Lin C.W., Moorefield B., Payne J., Aprikian P., Mitomo K., Reeder R.H. A novel 66-kilodalton protein complexes with Rrn6, Rrn7, and TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae . Mol. Cell. Biol. 1996;16:6436–6443. doi: 10.1128/mcb.16.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P., Tschochner H. A specialized form of RNA polymerase I, essential for initiation and growth dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:3692–3703. doi: 10.1093/emboj/17.13.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P., Schultz P., Tschochner H. Resolution of RNA polymerase I into dimers and monomers and their function in transcription. Biol. Chem. 1997;378:1433–1443. doi: 10.1515/bchm.1997.378.12.1433. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. The exosomea conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Morrissey J.P., Tollervey D. U14 small nucleolar RNA makes multiple contacts with the pre-ribosomal RNA. Chromosoma. 1997;105:515–522. doi: 10.1007/BF02510488. [DOI] [PubMed] [Google Scholar]
- Mosgoeller W., Schofer C., Wesierska Gadek J., Steiner M., Muller M., Wachtler F. Ribosomal gene transcription is organized in foci within nucleolar components. Histochem. Cell Biol. 1998;109:111–118. doi: 10.1007/s004180050208. [DOI] [PubMed] [Google Scholar]
- Mougey E.B., O'Reilly M., Osheim Y., Miller O.L., Jr., Beyer A., Sollner Webb B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993;7:1609–1619. doi: 10.1101/gad.7.8.1609. [DOI] [PubMed] [Google Scholar]
- Musters W., Knol J., Maas P., Dekker A.F., van Heerikhuizen H., Planta R.J. Linker scanning of the yeast RNA polymerase I promoter Nucleic Acids Res. 17 1989. 9661 9678a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Yano R., Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl. Acad. Sci. USA. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M., Nogi Y., Clark M.W., Nomura M. Structural alterations of the nucleolus in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Mol. Cell. Biol. 1993;13:2441–2455. doi: 10.1128/mcb.13.4.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M., Aris J.P., Brockenbrough J.S., Wai H., Vu L., Nomura M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae . J. Cell Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A., Jackson D.A., Hollinshead M., Wang Z., Roeder R.G., Cook P.R. Regional specialization in human nucleivisualization of discrete sites of transcription by RNA polymerase III. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs D.L., Peterson C.L., Wickham J.Q., Miller L.M., Clarke E.M., Crowell J.A., Sergere J.C. Characterization of the components of reconstituted Saccharomyces cerevisiae RNA polymerase I transcription complexes. J. Biol. Chem. 1995;270:6205–6210. doi: 10.1074/jbc.270.11.6205. [DOI] [PubMed] [Google Scholar]
- Saez-Vasquez J., Pikaard C.S. Extensive purification of a putative RNA polymerase I holoenzyme from plants that accurately initiates rRNA gene transcription in vitro. Proc. Natl. Acad. Sci. USA. 1997;94:11869–11874. doi: 10.1073/pnas.94.22.11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular CloningA Laboratory Manual 1989. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 86. pp. 1–18 [Google Scholar]
- Scheer U., Hock R. Structure and function of the nucleolus. Curr. Opin. Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- Schultz M.C., Reeder R.H., Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell. 1992;69:697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- Schultz P., Celia H., Riva M., Darst S.A., Colin P., Kornberg R.D., Sentenac A., Oudet P. Structural study of the yeast RNA polymerase A. Electron microscopy of lipid-bound molecules and two-dimensional crystals. J. Mol. Biol. 1990;216:353–362. doi: 10.1016/S0022-2836(05)80326-2. [DOI] [PubMed] [Google Scholar]
- Seither P., Iben S., Grummt I. Mammalian RNA polymerase I exists as a holoenzyme with associated basal transcription factors. J. Mol. Biol. 1998;275:43–53. doi: 10.1006/jmbi.1997.1434. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Jensen O.N., Podtelejnikov A.V., Sagliocco F., Wilm M., Vorm O., Mortensen P., Shevchenko A., Boucherie H., Mann M. Linking genome and proteome by mass spectrometrylarge-scale identification of yeast proteins from two dimensional gels Proc. Natl. Acad. Sci. USA. 93 1996. 14440 14445a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels Anal. Chem. 68 1996. 850 858b [DOI] [PubMed] [Google Scholar]
- Simos G., Tekotte H., Grosjean H., Segref A., Sharma K., Tollervey D., Hurt E. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- Smith C.M., Steitz J.A. Sno storm in the nucleolusnew roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Steffan J.S., Keys D.A., Dodd J.A., Nomura M. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiaeTBP is required for upstream activation factor dependent recruitment of core factor. Genes Dev. 1996;10:2551–2563. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- Steffan J.S., Keys D.A., Vu L., Nomura M. Interaction of TATA-binding protein with upstream activation factor is required for activated transcription of ribosomal DNA by RNA polymerase I in Saccharomyces cerevisiae in vivo. Mol. Cell. Biol. 1998;18:3752–3761. doi: 10.1128/mcb.18.7.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S.E., Roeder G.S. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae . Mol. Cell. Biol. 1989;9:3464–3472. doi: 10.1128/mcb.9.8.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szweykowska Kulinska Z., Senger B., Keith G., Fasiolo F., Grosjean H. Intron-dependent formation of pseudouridines in the anticodon of Saccharomyces cerevisiae minor tRNA(Ile) EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:4636–4644. doi: 10.1002/j.1460-2075.1994.tb06786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Kiss T. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- Tschochner H. A novel RNA polymerase I-dependent RNase activity that shortens nascent transcripts from the 3′ end. Proc. Natl. Acad. Sci. USA. 1996;93:12914–12919. doi: 10.1073/pnas.93.23.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner H., Milkereit P. RNA polymerase I from S. cerevisiae depends on an additional factor to release terminated transcripts from the template. FEBS Lett. 1997;410:461–466. doi: 10.1016/s0014-5793(97)00636-4. [DOI] [PubMed] [Google Scholar]
- Veinot Drebot L.M., Singer R.A., Johnston G.C. Rapid initial cleavage of nascent pre-rRNA transcripts in yeast. J. Mol. Biol. 1988;199:107–113. doi: 10.1016/0022-2836(88)90382-8. [DOI] [PubMed] [Google Scholar]
- Venema J., Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae . Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- Venema J., Tollervey D. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:5701–5714. [PMC free article] [PubMed] [Google Scholar]
- Verheggen C., Le Panse S., Almouzni G., Hernandez Verdun D. Presence of pre-rRNAs before activation of polymerase I transcription in the building process of nucleoli during early development of Xenopus laevis . J. Cell Biol. 1998;142:1167–1180. doi: 10.1083/jcb.142.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorm O., Roepstorff P., Mann M. Improved resolution and very high sensitivity in MALDI TOF of matrix surfaces made by fast evaporation. Anal. Chem. 1994;66:3281–3287. [Google Scholar]
- Wansink D.G., Schul W., van der Kraan I., van Steensel B., van Driel R., de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J. Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N.J., Gottschalk A., Neubauer G., Kastner B., Fabrizio P., Mann M., Luhrmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weipoltshammer K., Schofer C., Wachtler F., Hozak P. The transcription unit of ribosomal genes is attached to the nuclear skeleton. Exp. Cell Res. 1996;227:374–379. doi: 10.1006/excr.1996.0287. [DOI] [PubMed] [Google Scholar]
- Yamamoto R.T., Nogi Y., Dodd J.A., Nomura M. RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:3964–3973. [PMC free article] [PubMed] [Google Scholar]
- Zatsepina O.V., Dudnic O.A., Todorov I.T., Thiry M., Spring H., Trendelenburg M.F. Experimental induction of prenucleolar bodies (PNBs) in interphase cellsinterphase PNBs show similar characteristics as those typically observed at telophase of mitosis in untreated cells. Chromosoma. 1997;105:418–430. doi: 10.1007/BF02510478. [DOI] [PubMed] [Google Scholar]