Abstract
Fatty acids are essential for membrane biosynthesis in all organisms and serve as signaling molecules in many animals. Here, we found that saturated very-long-chain fatty acids (VLCFAs; C20:0 to C30:0) exogenously applied in ovule culture medium significantly promoted cotton (Gossypium hirsutum) fiber cell elongation, whereas acetochlor (2-chloro-N-[ethoxymethyl]-N-[2-ethyl-6-methyl-phenyl]-acetamide; ACE), which inhibits VLCFA biosynthesis, abolished fiber growth. This inhibition was overcome by lignoceric acid (C24:0). Elongating fibers contained significantly higher amounts of VLCFAs than those of wild-type or fuzzless-lintless mutant ovules. Ethylene nullified inhibition by ACE, whereas C24:0 was inactive in the presence of the ethylene biosynthesis inhibitor (l-[2-aminoethoxyvinyl]-glycine), indicating that VLCFAs may act upstream of ethylene. C24:0 induced a rapid and significant increase in ACO (for 1-aminocyclopropane-1-carboxylic acid oxidase) transcript levels that resulted in substantial ethylene production. C24:0 also promoted Ser palmitoyltransferase expression at a later stage, resulting in increased sphingolipid biosynthesis. Application of C24:0 not only stimulated Arabidopsis thaliana root cell growth but also complemented the cut1 phenotype. Transgenic expression of Gh KCS13/CER6, encoding the cotton 3-ketoacyl-CoA synthase, in the cut1 background produced similar results. Promotion of Arabidopsis stem elongation was accompanied by increased ACO transcript levels. Thus, VLCFAs may be involved in maximizing the extensibility of cotton fibers and multiple Arabidopsis cell types, possibly by activating ethylene biosynthesis.
INTRODUCTION
Cotton (Gossypium hirsutum) fibers are single-celled trichomes derived from the ovule epidermis that serve as the mainstay of the modern textile industry. Their unicellular and linear structures make them an ideal model for studies of plant cell elongation and cell wall biosynthesis (Kim and Triplett, 2001). Through microarray transcriptome profiling of 11,692 cotton fiber UniESTs, we identified 778 cDNAs that were preferentially expressed during the fast fiber-elongating period; among them, 162 fiber-specific genes were mapped to 102 metabolic pathways, with ethylene biosynthesis being the most significantly upregulated pathway (Shi et al., 2006). In the biochemical pathway leading toward ethylene production, conversion of S-adenosylmethionine to 1-aminocyclopropane-1-carboxylic acid (ACC) catalyzed by ACC synthases (ACSs), and subsequent oxidation of ACC to form ethylene by ACC oxidases (ACOs), are the last two committed steps (Yang and Hoffman, 1984). Significantly higher transcript levels of ethylene signal transduction pathway genes are found in the long-fiber AA subgenome compared with the short-fiber DD subgenome or the nonfibered mutant n1n1 by a different microarray analysis (Yang et al., 2006). Recently, an ethylene-responsive cotton ascorbate peroxidase was reported to regulate fiber growth by modulating endogenous hydrogen peroxide homeostasis (Li et al., 2007).
Fatty acid biosynthesis and elongation is ranked as the second-most upregulated biochemical pathway during cotton fiber cell elongation, together with three other pathways (Shi et al., 2006). Interestingly, many cotton genes encoding nonspecific lipid transfer proteins and enzymes involved in various steps during fatty acid chain elongation are highly upregulated during early fiber development (Ji et al., 2003; Qin et al., 2005, 2007; Shi et al., 2006; Gou et al., 2007), indicating that the biosynthesis of very-long-chain fatty acids (VLCFAs) or their transport may be required for fiber cell elongation processes. A comprehensive lipid analysis has indicated that linolenic (18:3) and palmitic (16:0) acids are the most abundant fatty acids in developing cotton fibers (Wanjie et al., 2005). VLCFAs are generated in the endoplasmic reticulum by a four-step reaction cycle: condensation of malonyl-CoA with a long-chain acyl-CoA to yield 3-ketoacyl-CoA by 3-ketoacyl-CoA synthase (KCS); reduction of 3-ketoacyl-CoA to 3-hydroxyacyl-CoA by 3-ketoacyl-CoA reductase; dehydration of 3-hydroxyacyl-CoA to trans-2-enoyl-CoA by 3-hydroxyacyl-CoA dehydratase; and further reduction of trans-2-enoyl-CoA by trans-2-enoyl-CoA reductase (ECR) to form an elongated acyl-CoA (Fehling and Mukherjee, 1991). The condensing enzyme, KCS, may be regarded as the rate-limiting enzyme that also determines the substrate and tissue specificities of the reaction in higher plants (Lassner et al., 1996; Millar and Kunst, 1997).
In yeast, Elo1p is responsible for the elongation of C14 to C16, Elo2p is responsible for elongation up to C24, and Elo3p is essential for the conversion of C24 to C26 (Toke and Martin, 1996; Oh et al., 1997). Repeated rounds of two-carbon addition that elongated shorter fatty acids into VLCFAs of various chain lengths were recently realized in a cell-free system with purified membrane components, including Elop, two reductases, and one novel dehydratase from yeast (Denic and Weissman, 2007). Yeast mutants with both ELO2 and ELO3 deleted cannot produce fatty acids with chain lengths longer than C20, and deficiency in these genes is synthetically lethal (Oh et al., 1997). The viability of these cells is restored by heterologous expression of Arabidopsis thaliana FAE1-like genes that encode enzymes for the condensation reactions (Paul et al., 2006). In Arabidopsis, KCS1 and CER6 (CUT1) are responsible for the biosynthesis of VLCFAs with chain lengths longer than C22 (Millar et al., 1999; Todd et al., 1999; Fiebig et al., 2000).
VLCFAs are present predominantly as sphingolipids, essential components of plasma membrane microdomains (lipid rafts) that mediate membrane-localized signal transduction pathways in a wide range of eukaryotic organisms (Worrall et al., 2003; Futerman and Hannun, 2004). Hydroxylated C26 is a major VLCFA component of yeast sphingolipids (Oh et al., 1997), whereas hydroxylated C24 accumulates in Arabidopsis sphingolipids (Zheng et al., 2005). Downregulation of sphingolipid biosynthesis via the suppression of At LCB1, which encodes one subunit of Ser palmitoyltransferase, results in much reduced cell expansion with a marked reduction in final plant size (Zheng et al., 2005; Chen et al., 2006). Molecular studies of the Arabidopsis fiddlehead (fdh) mutant, which carries a disrupted putative KCS gene, indicate that functional FDH is required for trichome differentiation and also for the maintenance of leaf epidermis stability (Yephremov et al., 1999; Pruitt et al., 2000). Another putative condensing enzyme, HIC (for high carbon dioxide release), is involved in signal transduction leading to controlled stomatal patterning in response to elevated CO2 concentration (Gray et al., 2000).
Experimental data obtained in this work through systematic study of cotton ovules cultured in the presence or absence of inhibitors of either VLCFA or ethylene biosynthesis pathways, and functional characterization of four cotton KCS genes encoding a putative KCS as well as the Arabidopsis cut1 mutant, demonstrate that VLCFAs play an important role in cell elongation and expansion by activating ethylene biosynthesis and signaling.
RESULTS
VLCFAs Promote Significant Cotton Fiber Cell Elongation
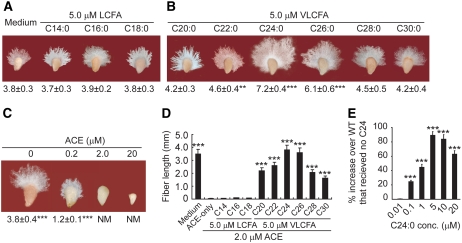
When saturated long-chain fatty acids (LCFAs; C14:0 to C18:0; 5 μM) were added to cultured wild-type cotton ovules, no growth promotion was observed over the 6-d experimental period, whereas saturated VLCFAs (C20:0 to C30:0; especially C24:0) at the same concentration promoted very significant levels of fiber cell elongation (Figures 1A and 1B). Addition of 2 μM chloroacetamide herbicide acetochlor (ACE), a structural analog of alachlor and an inhibitor of VLCFA biosynthesis (Götz and Böger, 2004; Trenkamp et al., 2004), to the culture medium completely blocked fiber elongation with minimal inhibition of ovule cell expansion (Figure 1C). Because 20 μM or higher concentrations of ACE inhibited ovule expansion severely (Figure 1C), 2 μM ACE was used for all subsequent ovule culture experiments, when applicable. The inhibition by ACE was overcome by 5 μM saturated C24:0 or C26:0 (Figure 1D). Other VLCFAs, including C20:0, C22:0, C28:0, and C30:0, only partially compensated for the inhibition by ACE (Figure 1D). LCFAs did not reverse the ACE-induced inhibition of fiber elongation (Figure 1D), suggesting that VLCFAs, particularly C24:0, play an important role during this process.
Figure 1.
VLCFA Modulates Fiber Cell Elongation.
(A) Exogenously applied LCFAs did not promote wild-type fiber cell elongation over the entire experimental period.
(B) Exogenously applied VLCFAs promoted significant fiber cell growth.
(C) ACE (2 μM) blocked cotton fiber cell elongation but not ovule cell expansion. NM, not measurable.
(D) VLCFAs (C20:0 to C30:0) reversed the inhibition of ACE on fiber growth.
(E) C24:0 promoted fiber growth at concentrations as low as 0.1 μM.
All ovules were collected at 1 DPA from wild-type plants and cultured for a total of 6 d with the addition of different chemicals as specified. All data reported here were obtained from three independent ovule culture experiments with 30 fibers measured for each treatment and are presented as means ± se. Significance values in (A), (B), (C), and (E) were obtained by comparison with the Medium group, and those in (D) were obtained by comparison with the ACE-only group. ACE-only, wild-type ovules cultured in the medium supplemented with 2 μM ACE; Medium, wild-type ovules cultured in the medium specified by Shi et al. (2006) containing methyl tert-butyl ether (MTBE; ∼0.8 μM). Statistical significance was determined using one-way analysis of variance software combined with Tukey's test. ** P < 0.01, *** P < 0.001.
Dose-response studies showed that concentrations as low as 0.1 μM C24:0 were able to stimulate fiber growth, with the best stimulation occurring at ∼5 μM (Figure 1E). Cotton ovules took up significant amounts of C24:0 or ACE at 3 h after the addition of these compounds into the culture medium (see Supplemental Figures 1 and 2 online). At the end of a 24-h culture period, almost twice as much C24:0 was recovered from ovules supplemented with 5 μM C24:0 compared with nontreated wild-type ovules, designated Medium-3 h and Medium-24 h (see Supplemental Figure 1 online). Application of C24:0 did not affect the rate of ACE uptake, as verified by gas chromatography–mass spectrometry (GC-MS) analyses (see Supplemental Figure 2 online). Ovule cells accumulated between 0.85 and 0.90 μM ACE at 3 h after 2 μM of this compound was added into the culture medium (see Supplemental Figure 2 online). These results indicated that the growth promotion or inhibition was a direct result of the chemicals supplemented in the culture.
Functional Characterization of Four Cotton KCS Genes
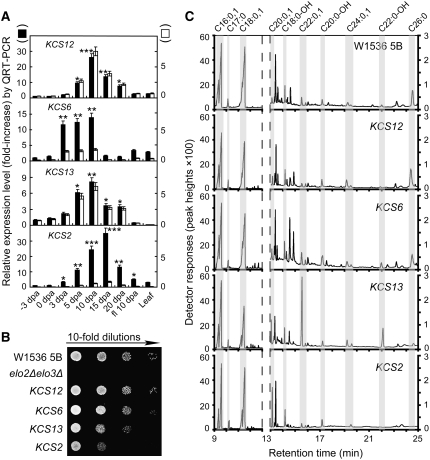
Quantitative real-time RT-PCR (QRT-PCR) analysis confirmed that four cotton KCS genes were highly upregulated during cotton fiber development (Figure 2A; see Supplemental Table 1 online). In general, 8- to 35-fold increases in transcript levels were recorded for different cotton KCS genes at the 10- or 15-d post anthesis (DPA) growth stage relative to that of 0 DPA ovules. Transcript levels for both KCS12 and KCS13/CER6 reached six to seven times the amount of mRNAs encoding the cotton Ubiquitin7 (UBQ7). Furthermore, compared with short-fiber germplasms, long-fiber cotton contained almost twice the amount of transcripts for all four KCSs at the fast-growing 10-DPA stage (see Supplemental Figure 3 online), indicating that cotton germplasms with longer final fiber lengths were potentially able to synthesize more VLCFAs. The functionality of these cotton genes was investigated by a genetic complementation assay in the yeast elo2Δ elo3Δ double-deletion mutant. Viable spores expressing active cotton KCSs were obtained by tetrad dissection, and their growth rates were examined on YPD plates. All four cotton KCS genes were able to restore the viability of the elo2Δ elo3Δ mutant, with KCS12 and KCS6 producing the wild-type growth rate (Figure 2B).
Figure 2.
All Four Fiber-Preferentially Expressed Cotton KCSs Encode Functional Enzymes That Complement the elo2Δ elo3Δ Yeast Mutant by Producing VLCFAs.
(A) QRT-PCR analyses of transcript levels of four KCSs at different growth stages (from −3 to 20 DPA) of fiber cells. PCR was performed in triplicate using independent RNA samples prepared from different ovule cultures. Closed bars with scales to their left side indicate fold increase relative to the −3-DPA value (arbitrarily set to 1) of the same KCS gene to show its fiber specificity. Open bars with scales to their right side indicate fold increase of each KCS relative to cotton UBQ7, a major housekeeping gene in the fiber transcriptome that maintains a relatively stable level throughout the growth period. See Figure 1 legend for statistical treatments. * P < 0.05.
(B) Genetic complementation of the elo2Δ elo3Δ double-deletion yeast mutant by different cotton KCSs.
(C) GC analyses of total fatty acids extracted from wild-type W1536 5B yeast cells or elo2Δ elo3Δ double-deletion mutants complemented by different cotton KCSs.
Note that different response scales are used to show peaks that appeared before 13 min (primarily LCFAs) and those that appeared after 13 min (VLCFAs) by GC. Similar results were obtained from three independent cell culture and fatty acid extractions for all cell lines, with quantitative analysis reported in Supplemental Table 2 online. The efficiency of the extraction process was monitored by external addition of fatty acid C17:0.
The elongase activities of these cotton enzymes were monitored by GC-MS using total fatty acids extracted from genetically complemented mutant yeast cells. Heterologous expression of cotton KCS12 and KCS6 resulted in VLCFA profiles almost identical to those of wild-type cells by producing significant amounts of C26:0 (Figure 2C; see Supplemental Table 2 online for quantitative analysis). Cells rescued by cotton KCS13/CER6 displayed a substrate preference for C20-CoA by accumulating high levels of saturated C22:0 and α-hydroxylated C22 fatty acids. Consistent with their poor growth phenotype, cells expressing cotton KCS2 contained large amounts of C20:0, with undetectable amounts of C26:0 and very low levels of C22:0 and C24:0 (Figures 2B and 2C), suggesting that C24:0 and C26:0 VLCFAs are required for normal-rate yeast cell growth. Several peaks with retention times around 13.5 and 14.5 to 15.0 min in Figure 2C could not be identified using current databases.
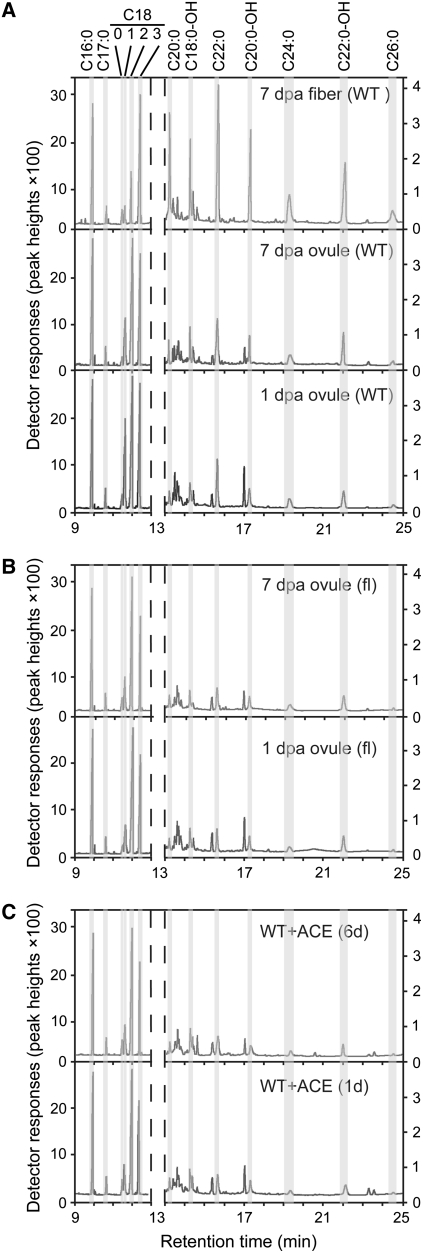
Elongating Fibers Contain High Levels of VLCFAs That Are Specifically Inhibited by Exogenous ACE
When total fatty acids were extracted from different cotton materials and analyzed by GC-MS, significantly higher levels of VLCFAs were observed only from 7-DPA wild-type cotton fibers (Figure 3A). Both wild-type ovules harvested at 1 or 7 DPA with fibers stripped off, and ovules of the fuzzless-lintless (fl) mutant harvested at the same growth stages, contained significantly lower levels of VLCFAs (from C20:0 to C26:0) compared with those of the fibers (Figures 3A and 3B). A close examination revealed that 7-DPA fiber cells contained approximately three to five times the amounts of all VLCFAs, with similar levels of C16:0 and C18:0. Fiber cells contained lower levels of C18:1 and C18:2, with significantly higher amounts of C18-OH and C18:3 compared with ovule samples (Figure 3; see Supplemental Table 3 online). This is consistent with the finding that developing fiber cells contained significantly higher levels of FAD3 transcripts encoding the desaturase responsible for the synthesis of linolenic acid (C18:3) from linoleic acid (C18:2) (see Supplemental Figure 4A online). C18:3 is one of the most abundant fatty acids in developing cotton fibers (Wanjie et al., 2005). Biosynthesis of VLCFAs (C20:0 to C26:0), but not that of LCFAs (C16:0 and C18:0), was inhibited when wild-type ovules were cultured in medium containing 2 μM ACE, as revealed by GC analysis (Figure 3C). These ovules produced fatty acid profiles almost identical to that of the fl mutant (Figures 3B and 3C).
Figure 3.
Fast-Elongating Cotton Fibers Contain Significantly Higher Levels of VLCFAs but Not LCFAs.
(A) GC analyses of LCFAs and VLCFAs from 7-DPA fiber cells (top panel), 7-DPA wild-type ovules with fibers stripped off (middle panel), and 1-DPA wild-type ovules (bottom panel).
(B) GC analysis of LCFAs and VLCFAs from 7-DPA (top panel) and 1-DPA (bottom panel) fl mutant ovules.
(C) GC analysis of LCFAs and VLCFAs from wild-type cotton ovules treated with 2 μM ACE for 6 d (top panel) or 1 d (bottom panel).
Similar results were obtained from three independent ovule cultures, with quantitative analysis reported in Supplemental Table 3 online. In general, ∼55% of the original C17:0 was recovered after the extraction processes in this and all other related experiments. See Figure 2 legend for different scaling of the chromatograms.
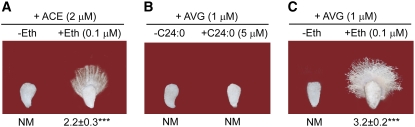
Ethylene Effectively Negates the Growth Inhibition of ACE
Because ethylene was reported to stimulate fiber cell elongation (Shi et al., 2006), we added 0.1 μM ethylene in wild-type ovule culture medium together with 2 μM ACE and measured fiber lengths after a 6-d culture. As shown in Figure 4A, ethylene effectively negated the inhibition by ACE. By contrast, the growth inhibition caused by l-[2-aminoethoxyvinyl]-glycine (AVG), the ethylene biosynthesis inhibitor, was only reverted by exogenous ethylene and not by C24:0 (Figures 4B and 4C), implying that ethylene may act downstream of VLCFA in regulating fiber cell elongation. Expression of the genes responsible for VLCFA as well as polyunsaturated C18 biosynthesis genes was not affected by 72 h of ethylene treatment (see Supplemental Figures 4B and 4C online). Except for C20:0-OH and C22:0-OH, ovules treated with 2 μM ACE for a 6-d period showed similar total fatty acid profiles by GC compared with ovules that were treated with ACE and ethylene simultaneously (see Supplemental Figure 5 online).
Figure 4.
Ethylene Reverses the Inhibitory Effect of ACE, Whereas C24:0 Is Inactive against Exogenously Applied AVG in Cotton Ovule Cultures.
(A) Exogenous ethylene reversed the ACE effect on fiber growth.
(B) C24:0 was inactive in the presence of the ethylene biosynthesis inhibitor AVG.
(C) Ethylene negated the inhibitory effect of AVG.
Wild-type ovules were harvested on the day of flowering and were cultured in the presence of various chemicals for a total of 6 d before being analyzed. See Figure 1 legend for statistical analysis.
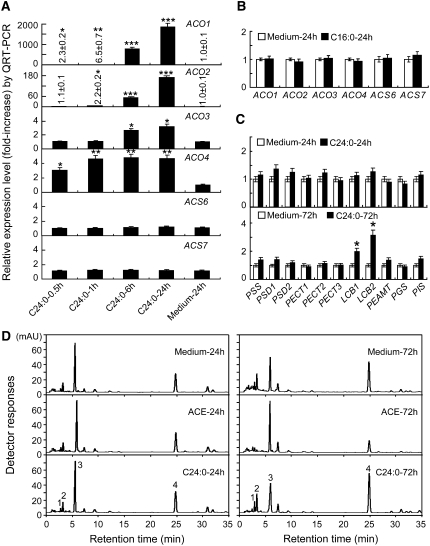
C24:0 Rapidly Promotes the Expression of Ethylene Biosynthesis Genes
QRT-PCR analysis indicated that the transcript levels of all four cotton ACOs currently identified increased significantly shortly after the application of 5 μM C24:0 (Figure 5A). Transcript levels of ACO1 and ACO4 increased significantly at 0.5 h after C24:0 was added to the culture. ACO2 expression was induced significantly in 1 h and ACO3 in 6 h (Figure 5A). Greater than 2000-fold ACO1 and 170-fold ACO2 mRNAs were recovered from ovules treated with C24:0 over a 24-h period. Almost no transcripts of these two genes were detected in ovules harvested at this stage if C24:0 was not included in the medium. Expression of two ACS genes was not induced by C24:0 (Figure 5A). Application of C16:0 in the culture for as long as 24 h did not induce the expression of cotton ACOs and ACSs (Figure 5B).
Figure 5.
Exogenous C24:0 Promotes the Accumulation of ACO Transcripts Rapidly While Inducing Sphingolipid Biosynthesis at a Later Stage.
(A) C24:0 promoted significant accumulation of ACO transcripts but not ACS transcripts.
(B) C16:0 did not affect transcript levels of ACSs and ACOs.
(C) C24:0 promoted the accumulation of transcripts encoding several sphingolipid biosynthesis genes (LCBs) when it was supplemented in the culture for >24 h.
RNA samples were prepared from three independent ovule cultures in the presence or absence of 5.0 μM C24:0 or C16:0 for the indicated times. QRT-PCR experiments were performed using gene-specific primers, as reported in Supplemental Table 1 online. All 11 lipid biosynthesis genes available in our cotton transcriptome were included in this analysis. Medium-24 h, wild-type ovules cultured in the original medium with MTBE for 24 h.
(D) Liquid chromatography–MS analysis of o-phthaldialdehyde derivatives of cotton LCBs. Sphingolipids were extracted from 1-DPA wild-type ovules cultured in the medium (Medium-24 h), in the presence of 2 μM ACE (ACE-24 h), or in the presence of 5 μM C24:0 (C24:0-24 h) for 24 h (left panel) or 72 h (right panel). LCBs were released by hydrolysis from sphingolipids and converted to their o-phthaldialdehyde derivatives. Peak 1, t18:1(8Z)-glucose; peak 2, t18:1(8E)-glucose; peak 3, t18:1(8E); peak 4, 1,4-anhydro-t18:1(8E).
C24:0 Induces Sphingolipid Biosynthesis at a Later Stage
Among all 11 membrane lipid biosynthesis genes available in our cotton transcriptome, two LCB genes were activated at 72 h, but not at 24 h, after C24:0 was applied (Figure 5C). Analysis of LCB (sphingolipid long-chain base) classes indicated that there were no visible changes in the long-chain bases extracted from cotton ovules treated with C24:0 or ACE for 24 h (Figure 5D, left panel; see Supplemental Figures 6A and 6B online for structures of major LCB classes found in cotton fiber cells and for plant sphingolipid classifications, respectively). However, three of four major LCB peaks—t18:1(8Z)-glucose (peak 1), t18:1(8E)-glucose (peak 2), and 1,4-anhydro-t18:1(8E) (peak 4)—had substantially increased concentrations in samples treated with C24:0 for 72 h compared with the nontreated wild-type ovules for the same period (Figure 5D, right panel; see Supplemental Figure 6C online). In ovules treated with ACE for 72 h, the amount of t18:1(8E) (peak 3) increased, while those of t18:1(8Z)-glucose, t18:1(8E)-glucose, and 1,4-anhydro-t18:1(8E) (peaks 1, 2, and 4) decreased (Figure 5D, right panel). Our data seem to indicate that a sustained supply of sphingolipids derived from t18:1(8Z)-glucose, t18:1(8E)-glucose, and 1,4-anhydro-t18:1(8E) bases may also be required for fiber growth.
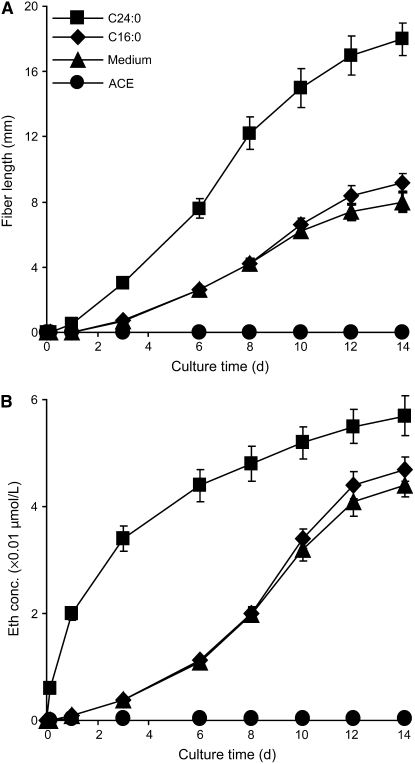
C24:0 Promotes and ACE Inhibits Ethylene Production in Ovule Culture
After 2 weeks of culture, wild-type ovules produced 7.4- ± 0.48-mm-long fibers in the untreated medium group, whereas the addition of C24:0 more than doubled the fiber length (18.7 ± 1.1 mm) (Figure 6A). A significant amount of ethylene was released only 3 h after the addition of C24:0 to the culture, with peak values recorded in ∼3 to 6 d (Figure 6B). C16:0 did not promote either significant fiber elongation or an increase in ethylene production over the untreated medium group for at least 8 to 10 d (Figure 6). ACE completely inhibited fiber growth as well as ethylene production (Figure 6). The correlation coefficient between ethylene production and fiber elongation was 0.992 in the wild-type medium control and 0.860 in C24:0-treated ovules.
Figure 6.
The Rate of Fiber Elongation Correlates with the Speed of Ethylene Production in Cultured Wild-Type Ovules after Various Treatments.
(A) Fiber growth curve obtained from cultured wild-type ovules after various treatments.
(B) Ethylene production from cultured wild-type ovules after various treatments.
Wild-type ovules were collected at 1 DPA and cultured for different periods as indicated. Data reported are means ± se of three independent experiments. A very small amount of ethylene always present in fl ovule culture that did not increase throughout the entire culture period was considered injury-related ethylene and was subtracted from all data reported here. For cultures of >6 d, we reinforced the culture with 50% of the initial amounts of C24:0, C16:0, or ACE on day 7.
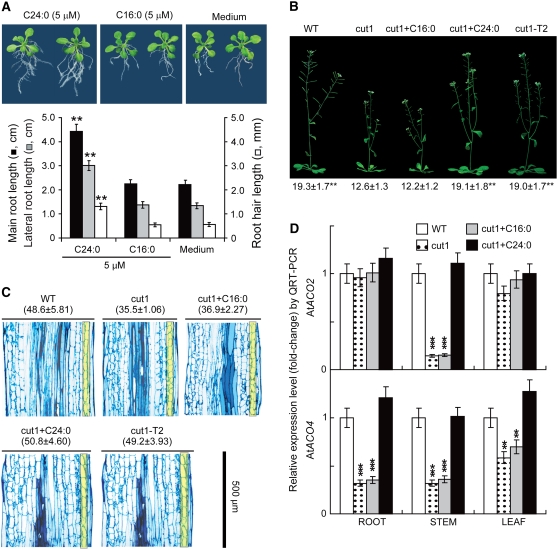
C24:0 Promotes Substantial Elongation of Multiple Arabidopsis Cell Types and Rescues the Dwarf Phenotype of the Arabidopsis cut1 Mutant
When 14-d-old Arabidopsis seedlings grown on solid half-strength Murashige and Skoog medium with or without supplementation of 5 μM C24:0 or C16:0 were examined, significantly longer root systems, including main roots, lateral roots, and root hairs, were observed from plants treated with C24 but not from C16-treated plants (Figure 7A). The aboveground parts of plants (hypocotyls, petioles, and leaves) treated with C24:0 were not as elongated (Figure 7A). Wild-type architecture (leaf size and mature stem length) was observed in the Arabidopsis cut1 mutant when C24:0 was applied exogenously through the root system or when Gh KCS13/CER6, a homolog of Arabidopsis CER6 (CUT1), was transformed into the cut1 mutant background (Figure 7B). C24:0 application or heterologous expression of Gh KCS13/CER6 resulted in 25 to 30% elongation of stem cells (Figure 7C) over those of the nontreated cut1 mutant. C24:0 treatment induced a significant increase in At ACO2 (At1g63280) transcript levels in cut1 stems (Figure 7D, top panel) and in At ACO4 (At1g05010) transcript levels in cut1 root, stem, and leaf cells (Figure 7D, bottom panel). Although a total of six ACOs were found in the Arabidopsis genome, QRT-PCR analysis showed that only At1g63280 and At1g05010 were expressed in vegetative tissue. Our data indicate that the regulatory linkage between VLCFA and ethylene biosynthesis may be a common mechanism in higher plants.
Figure 7.
VLCFA Results in Significant Extension of Several Arabidopsis Cell Types.
(A) Exogenous C24:0 promoted significant Arabidopsis root cell elongation. Top panel, photographs of 14-d-old seedlings grown in medium supplemented with C24:0 or C16:0. Medium, plants received MTBE with no C24:0 or C16:0 added. Bottom panel, statistics obtained from 30 individual seedlings representing three independent experiments. See Figure 1 legend for statistical procedures and annotations. Significance values were obtained by comparison with the Medium group.
(B) Wild-type lengths of stems were produced in Arabidopsis cut1 mutant when C24:0 was applied exogenously through the root system or when Gh KCS13/CER6 was transformed in the mutant background. WT, wild-type Landsberg erecta ecotype plants; cut1+C16:0, cut1 plants that received 50 mL of 5 μM C16:0 supplementation daily from the day of bolting for each 8-inch pot; cut1+C24:0, cut1 plants that received the same amount of 5 μM C24:0; cut1-T2, homozygous transgenic cut1 plants that expressed the Gh KCS13/CER6 gene. The same amount of water containing MTBE was applied to the medium. Photographs were taken of 40-d-old soil-grown plants. Average stem lengths (centimeters) measured from 30 individual plants from three independent experiments are reported beneath representative plants.
(C) Comparisons of longitudinal cell lengths from mature stem sections of wild-type, cut1, cut1+C16, cut1+C24, and cut1-T2 plants. Longitudinal sections were prepared from Arabidopsis stems, and average cell length (micrometers, means ± se; n = 5), reported at the top of each section in parentheses, was calculated by dividing the section length (500 μm) by the number of cells in the cortex layer (shown in yellow). The width of each section roughly reflects the actual thickness of the stem.
(D) C24:0 induced a specific and significant increase in expression of At ACO genes in cut1 root, stem, and leaf cells. The cut1 mutant plants were watered with 50 mL of 5 μM C24:0 or C16:0 for 3 d starting from the day of bolting and were harvested separately as roots, stems, and leaves for RNA extraction. QRT-PCR analyses were performed in triplicate using RNA samples prepared from independent plant materials and gene-specific primers, as listed in Supplemental Table 1 online. Wild-type plants and T2 transgenic plants received the same amount of MTBE (∼0.8 μM). We followed www.ncbi.nlm.nih.gov for nomenclature for At ACOs.
DISCUSSION
VLCFA or Its Immediate Derivatives May Serve as Signal Molecules to Activate Ethylene Biosynthesis
Through in vitro application and systematic analysis, we found that saturated VLCFAs, particularly C24:0, promoted significant cotton fiber cell elongation (Figure 1). Furthermore, application of this amphipathic compound to the root systems of cut1 Arabidopsis plants resulted in wild-type stem length and leaf size (Figure 7), suggesting that signal(s) derived from VLCFA may be perceived far from the site of application. In agreement with this possibility, a sharp and significant increase in ACO expression, with resultant ethylene release, was recorded in cotton ovules cultured in the presence of C24:0 for <1 h (Figures 5A and 6), suggesting that VLCFAs may control the rate of cell extension by regulating ethylene biosynthesis. As indicated earlier (Li et al., 2007), the hydrogen peroxide signaling pathway may act, at least in part, downstream of ethylene to induce cell expansion. The VLCFA effect was not attributed directly to its serving as a precursor to membrane lipids, because no obvious change in sphingolipid composition was recorded at 24 h after C24:0 or ACE was included in ovule culture systems (Figure 5D).
Ethylene plays a positive role in cotton fiber cell elongation (Shi et al., 2006) and in Arabidopsis root growth and root hair development (Stepanova et al., 2005). Similar conclusions can be inferred from studies of the irregularly positioned long-root-hair Arabidopsis nrp1-1 nrp2-1 (histone chaperones) double mutants, because 3 of the 20 double mutants specifically upregulated genes encoding putative ethylene-response element binding proteins (EREBPs) (Zhu et al., 2006). Elevated transcript levels of several AP2/EREBP transcription factors, including WXP1, SHN, and WIN1, in transgenic Arabidopsis and alfalfa (Medicago sativa) plants increased leaf wax content (Aharoni et al., 2004; Broun et al., 2004), indicating that wax production, which is downsteam of VLCFA biosynthesis, may be regulated by the ethylene signaling pathway as well. Consistent with the suggestion that VLCFA acts upstream of ethylene, overexpression of WIN1 in transgenic Arabidopsis plants did not affect KCS13/CER6 (CUT1) expression (Broun et al., 2004). Recently, ethylene was found to modulate stem cell division in the quiescent center of Arabidopsis root tips (Ortega-Martínez et al., 2007).
VLCFAs were involved in signal transduction pathways in various cellular processes. For example, long-chain acyl-CoA and unsaturated VLCFA act as ligands for nuclear transcription factors that modulate target gene expression (Hertz et al., 1998; de Urquiza et al., 2000). In both human and Chinese hamster cells, VLCFAs serve as extracellular signals that activate specific G-protein complexes (Briscoe et al., 2003; Itoh et al., 2003). Elevated C18:1 levels, generated in a ssi2 act1 double mutant that blocked its metabolism (Kachroo et al., 2003), were required for information flow between salicylic acid– and jasmonic acid–dependent defense pathways in Arabidopsis chloroplasts. Plastidial fatty acid levels regulate resistance gene–dependent defense signaling (Chandra-Shekara et al., 2007). Overproduction of the Arabidopsis fatty acid amide hydrolase, which hydrolyzes N-acylethanolamines to form free fatty acids and ethanolamine, leads to significantly enlarged cell size and enhanced seedling growth (Wang et al., 2006).
ACOs May Constitute a Rate-Limiting Step in Ethylene Biosynthesis
The availability of ACC synthesized from S-adenosylmethionine by ACS is generally considered to be the rate-limiting step in the ethylene biosynthesis pathway (Yang and Hoffman, 1984). Our data, however, suggest that ethylene biosynthesis might be regulated at the level of ACO activity, as the transcripts of all four ACOs analyzed in this work accumulated rapidly after C24:0 treatment (Figure 5A), which resulted in robust ethylene production (Figure 6B); by contrast, ACS transcript levels were not affected (Figure 5A). Previously, three of the ACO genes were found to be specifically upregulated during the fast fiber-elongation stage (Shi et al., 2006). ACO activity was regarded as a limiting factor for ethylene biosynthesis in Rumex palustris under hypoxic conditions (Vriezen et al., 1999). Also, ACO activity, but not ACC availability, was found to be important for ethylene production during gravity-induced stem growth in hybrid aspen (Populus spp) trees (Andersson-Gunneras et al., 2003).
Biosynthesis of C20:0 to C26:0 VLCFAs in Cotton May Be Performed by Different KCSs
Heterologous expression of different cotton KCSs in the yeast elo2Δ elo3Δ double-deletion mutant resulted in significantly different fatty acid profiles. Wild-type levels of C26 were observed from mutant cells expressing KCS12 and KCS6, whereas large amounts of C22 and C20 were recovered from cells carrying Gh KCS13/CER6 or Gh KCS2, respectively (Figure 2C), indicating that the cotton enzymes have discrete substrate specificities. Gh KCS12 and Gh KCS6 are similar to yeast Elo2p and Elo3p, which are involved in the synthesis of VLCFAs from C20 to C26 (Oh et al., 1997). Gh KCS2 is homologous with a plant FAE-type KCS reported to use C18 as the substrate responsible for the production of mainly C20 and C20:1 (Millar and Kunst, 1997; Paul et al., 2006). Gh KCS13/CER6 shares 85 and 55% amino acid sequence identity with At CER6 (At CUT1) and Tropaeolum majus NasFAE, respectively. The latter enzyme was found to produce C22 from C20 (Mietkiewska et al., 2004), similar to the finding in this work. The enzyme activities of At CER6 (At CUT1) were never biochemically characterized. However, in CUT1-suppressed plants, the C24 wax components accumulated substantially, indicating that CUT1 was required for the elongation of C24 VLCFAs (Millar et al., 1999). Detailed analysis is required to solve this obvious discrepancy among enzymes of different plants.
In summary, we find that the biosynthesis of VLCFAs is specifically upregulated during early cotton fiber development stages. In both cotton and Arabidopsis plants, exogenously applied C24:0 stimulates significant elongation growth. VLCFAs may exert their growth-promoting effect through enhanced ethylene biosynthesis and signaling. The scenario outlined here suggests that the current paradigm for the mechanism of cell elongation should be reassessed.
METHODS
Plant Materials
Upland cotton (Gossypium hirsutum cv Xuzhou 142) and a fl mutant, originally discovered in a cotton field in China (Zhang and Pan, 1992), were grown in a soil mixture in a fully automated greenhouse as reported previously (Ji et al., 2003). Ovules were excised from bolls on cotton plants at the day of flower opening (defined as 1 DPA) and were used directly for in vitro cultures (Shi et al., 2006).
Fiber cells from three field-grown cotton germplasm lines were used for RNA extraction and for quantitative determination of Gh KCS transcript levels in relation to final fiber lengths: Arcot-Y1 (long-fiber), final fiber length of 31.8 ± 1.4 mm; Arcot-Y2 (intermediate-fiber), 27.19 ± 1.2 mm; and Nan-dan-ba-di-da-hua (short-fiber), 21.01 ± 1.1 mm.
For the root cell elongation assay, Arabidopsis thaliana seeds (Columbia ecotype) were surface-sterilized by 0.1% HgCl2 before being sown in 90-mm plastic Petri dishes containing solid half-strength Murashige and Skoog medium supplemented with 5 μM C24 or C16. Seedlings were grown in fully automated Conviron growth chambers with a 16/8-h light/dark cycle at 23/21°C in 70% humidity (Gong et al., 2004). Fourteen-day-old seedlings were used for photographs and also for measurement with a dissecting microscope (Leica MZ APO). Homozygous cut1 seeds (SALK CS6242; Landsberg erecta) were obtained from the SALK mutant collections (http://www.Arabidopsis.org/abrc), which originated from L. Kunst (Millar et al., 1999), and were used for stem length analysis. The full-length coding region of Gh KCS13/CER6 (DQ122189) was cloned in pCAMBIA1305 under the control of a 1.5-kb At CUT1 promoter (the 1.2-kb sequence reported by Kunst et al. [2000] plus a 300-bp upstream region) and transformed into the cut1 genetic background. Homozygous (T2) Arabidopsis plants (cut1-T2) were used for stem length and QRT-PCR analyses.
In Vitro Ovule Culture and Kinetic Studies of Fiber Growth after Different Treatments
Cotton ovules were collected immediately after flower opening. They were sterilized and cultured in medium containing different amounts of ACE (2-chloro-N-[ethoxymethyl]-N-[2-ethyl-6-methyl-phenyl]-acetamide; 770 g/L [Monsanto]) and various concentrations of saturated LCFAs, VLCFAs, ethylene, and AVG (l-[2-aminoethoxyvinyl]-glycine, hydrochloride; >95.0%) at 30°C in darkness as described (Beasley and Ting, 1974; Shi et al., 2006). Gaseous ethylene was added in 2-liter glass containers to specified concentrations, disregarding the volume of the 50-mL culture flasks sitting inside. The large container was sealed gas-tight, while the culture flasks were left open for gas exchange in this experiment. Cerotic acid (C26:0) was purchased from Dr. Ehrenstorfer GmbH, and all other fatty acids were purchased from Sigma-Aldrich. Fatty acids were dissolved in MTBE (>99.0%) to a stock concentration of 5 mM. ACE was dissolved in absolute ethanol and AVG was dissolved in water to 10 mM before being added to culture medium to specified concentrations when the medium cooled to 40 to 50°C. The lengths of fibers were measured with a dissecting microscope (Leica MZ APO).
QRT-PCR
Cotton ovules harvested at specified times were first frozen in liquid nitrogen before being ground to fine powder with a mortar and pestle using a modified hot borate method (Ji et al., 2003). Total RNA was extracted from wild-type or fl mutant cotton ovules after various treatments, and cDNA was reverse-transcribed from 5 μg of total RNA as reported (Shi et al., 2006). Primers for QRT-PCR analysis are listed in Supplemental Table 1 online. Cotton UBQ7 (accession number AY189972) and At UBQ5 were used as internal controls in respective PCR experiments.
Functional Complementation of the Yeast elo2Δ elo3Δ Double-Deletion Mutant by Cotton KCSs
Saccharomyces cerevisiae diploid strain W1536 ELO3/elo3Δ, ELO2/elo2Δ (Mat a/α; ade2Δ/ade2Δ, ade3Δ/ade3Δ, leu2-3/leu2-112, his3-11/his3-11, trp1-1/trp1-1, ura3-1/ura3-1, ELO3/elo3Δ∷knaMX4, elo2Δ∷knaMX4/ELO2), produced in our laboratory as reported (Qin et al., 2007), was transformed with individual Gh KCS cDNAs ligated into a TRP1-marked pYADE4 under the control of the ADH promoter. The transformants were selected on Sc-Trp plates and sporulated on solid medium as described (Kastaniotis et al., 2004). Ascospores were digested with zymolase (Seikagaku) and dissected using a Singer MSM manual dissection microscope (Singer Instrument). Separated spores were grown on YPD (1% yeast extract, 2% peptone, and 2% d-glucose) plates for 5 d. Wild-type and mutant spores containing pYADE4Gh KCSs were selected by replica-plating on YPD-G418 plates (YPD supplemented with 300 μg geneticin/mL) and FAA plates (synthetic complete medium containing 2% [w/v] d-glucose and 0.05% [w/v] 5′-fluoroanthranilic acid). The genotype of the strain carrying knaMX4 cassettes was confirmed by PCR analysis. Cells were plated by 10-fold dilutions and were grown on medium for 2 d at 30°C before photographs were taken.
Fatty Acid Extractions
Total fatty acids (LCFAs and VLCFAs) were extracted from 1 g fresh weight of W1536 5B wild-type yeast or elo2Δ elo3Δ double-deletion mutant yeast complemented by individual cotton KCSs as described (Paul et al., 2006) or from 1 g fresh weight of cotton ovules (harvested at 1 or 7 DPA) and 7-DPA fibers. For C24:0 uptake studies, total fatty acids were extracted from 1-DPA wild-type ovules cultured in medium with no C24:0 supplementation for 3 h (Medium-3 h) or 24 h (Medium-24 h) or with 5 μM C24:0 added for the same period (C24:0-3 h or C24:0-24 h). All cotton samples were ground to fine powder with a mortar and pestle after removing the surface waxes by immersion in chloroform:methanol (2:1, v/v) for 1 min (Millar et al., 1999). Fatty acids were extracted by ethanol:water:diethyl ether:pyridine:ammonium hydroxide (7.0 n) (15:15:5:1:0.018, v/v) three times (Markham et al., 2006), dried under nitrogen gas, and derivatized by heating to 85°C in methanolic HCl (3 n) for 5 h. After cooling to room temperature, fatty acid methyl esters were extracted three times with hexane and then concentrated to 0.5 mL. We injected 1 μL into a GC apparatus equipped with a DB-225 MS column (J&W). C17 fatty acid (heptadecanoic acid; Sigma-Aldrich), which does not exist in plants, was added before extraction to monitor sample recovery and also for quantitative purposes.
GC-MS Analysis of ACE Uptake by Cotton Ovules
Wild-type cotton ovules (1 DPA) were cultured for 3 h in the absence of both ACE and C24:0 (Medium-3 h), in medium supplemented with 2 μM ACE (ACE-3 h), or in medium supplemented with 2 μM ACE plus 5 μM C24:0 (C24:0+ACE-3 h). Homogenized cotton ovules (1 g) from various treatments were extracted in acetone (>99.5%) three times. The extracts were concentrated to 0.5 mL, and 1 μL was injected into the GC apparatus equipped with an HP-5 column (J&W). The peak corresponding to the ACE standard (1 μL of 2 μM commercial ACE injected directly into the GC apparatus; designated ACE2-μM in the figures) by retention time was verified with an HP 5973 mass selective detector (Agilent).
GC-MS Analysis of Fatty Acid Composition
Fatty acid methyl esters prepared as described above were concentrated to 0.5 mL. Samples of 1 μL were analyzed by GC using an Agilent 6890N series GC system with a DB-225MS column (J&W) that operated with hydrogen carrier gas and a splitless inlet (injection temperature, 280°C), according to the manufacturer's instructions. All fatty acids and hydroxylated fatty acids were identified by the Agilent 6890N GC system coupled to an HP 5973 mass selective detector using both the National Institute of Standards and Technology and Wiley databases.
Lipid Extraction and Liquid Chromatography–Coupled MS Analysis of LCBs
Sphingolipids were extracted from 3 g of cotton ovules treated with 5 μM C24:0 or 2 μM ACE or from control ovules that received no treatments using ethanol:water:diethylether:pyridine:4.2 n ammonium hydroxide (15:15:5:1:0.018, v/v). We followed Markham et al. (2006) for the hydrolysis and liberation of LCBs from sphingolipids. LCBs were converted to o-phthaldialdehyde derivatives as reported (Merrill et al., 2000) and were dissolved in 0.5 mL of methanol. Samples of 10 μL were separated on a 4.6- × 150-mm XBD-C18 column and identified by Agilent 6410 triple quadrupole liquid chromatography–MS. No internal standard was added to monitor sample recovery in these experiments.
Ethylene Measurements
A gas chromatograph (GC-14C; Shimadzu) equipped with a flame-ionization detector with a 30-m HP-PLOT column (J&W) was used to measure the amounts of ethylene produced after various treatments, as reported (Shi et al., 2006). Approximately 20 freshly collected 1-DPA wild-type ovules were cultured in 20 mL of liquid medium in 50-mL glass flasks in darkness for various amounts of time until day 14 at 30°C. Air samples (100 μL) from each flask were removed and injected into the column for ethylene measurements.
Preparation of Arabidopsis Stem Sections and Histological Analysis
For analysis of stem cell length, the part of fully elongated stems immediately below the first pair of cauline leaves was harvested from wild-type Arabidopsis plants and the cut1 mutant that received C24:0, C16:0, or no fatty acid supplementation. Longitudinal sections were prepared from five individual Arabidopsis stems for each treatment, and they were fixed overnight in 2% paraformaldehyde and 2.5% glutaraldehyde in PBS, pH 7.2, at 4°C. Specimens were dehydrated in an ethanol series (30, 50, 70, 80, 90, 95, and 100%) and embedded in Spurr's resin according to the manufacturer's instructions. The tissue was sectioned at 4-μm thickness on a Leica RM 2265 microtome. Sections were observed under bright-field optics using a Leica DMR microscope after staining with 0.05% toluidine blue.
Statistical Analysis
Whenever applicable, data were evaluated by one-way analysis of variance software combined with Tukey's test to obtain statistical significance values.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU001741 (Gh KCS2), EF688567 (Gh KCS6), AJ608934 (Gh KCS12), DQ122189 (Gh KCS13/CER6), EU001742 (Gh ECR1), EU001743 (Gh ECR2), EF688568 (Gh PSS), EF688573 (Gh PSD1), EF688574 (Gh PSD2), EF688570 (Gh PECT1), EF688571 (Gh PECT2), EF688572 (Gh PECT3), EF688569 (Gh PEAMT), EF690291 (Gh PGS), EF690290 (Gh PIS), EF690288 (Gh LCB1), and EF690289 (Gh LCB2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Cotton Ovules Took Up C24:0 from the Culture Medium.
Supplemental Figure 2. ACE Was Actively Transported into Cotton Ovules in 3 h after It Was Added to the Medium, and C24:0 Treatment Did Not Affect the Rate of ACE Taken Up.
Supplemental Figure 3. Abundance of Cotton KCS Transcripts in Long-, Intermediate-, and Short-Fiber Cotton Germplasms.
Supplemental Figure 4. QRT-PCR Quantification of Cotton Genes Encoding Enzymes Involved in C18:3 and VLCFA Biosynthesis.
Supplemental Figure 5. Ovules Treated with Ethylene and ACE Simultaneously for 6 d Contained Similar Amounts of VLCFAs Compared with Those That Received ACE Only.
Supplemental Figure 6. Schematic Drawing and Quantification of Major LCBs from Cotton Fiber Cells.
Supplemental Table 1. Primers Used for QRT-PCR Analyses.
Supplemental Table 2. Quantitative Analysis of Fatty Acid Composition from Wild-Type Yeast and elo2Δ elo3Δ Double-Deletion Mutant Cells Genetically Complemented with Different Cotton KCSs, as Reported in Figure 2.
Supplemental Table 3. Quantitative Analysis of Fatty Acid Composition from Wild-Type Cotton Fiber Cells, Wild-Type or fl Mutant Ovule Cells, and ACE-Treated Wild-Type Cotton Ovules, as Reported in Figure 3.
Supplementary Material
Acknowledgments
This work was supported by grants from the China National Basic Research Program (Grant 2004CB117302), the National Natural Science Foundation of China (Grant 30470171), the Sigrid Jusélius Foundation of Finland, and the Academy of Finland. We thank Xiong-Ming Du of the Cotton Institute, Chinese Academy of Agricultural Sciences, for the cotton germplasms and Wolf-Hubert Kunau of Ruhr University for the pYADE4 vector.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yu-Xian Zhu (zhuyx@water.pku.edu.cn).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., van Arkel, G., and Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16 2463–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson-Gunneras, S., Hellgren, J.M., Björklund, S., Regan, S., Moritz, T., and Sundberg, B. (2003). Asymmetric expression of a poplar ACC oxidase controls ethylene production during gravitational induction of tension wood. Plant J. 34 339–349. [DOI] [PubMed] [Google Scholar]
- Beasley, C.A., and Ting, I.P. (1974). The effects of plant growth substances on in vitro fiber development from unfertilized cotton ovules. Am. J. Bot. 61 188–194. [Google Scholar]
- Briscoe, C.P., et al. (2003). The orphan G protein-coupled receptor GPR40 is activated by medium and long chain fatty acids. J. Biol. Chem. 278 11303–11311. [DOI] [PubMed] [Google Scholar]
- Broun, P., Poindexter, P., Osborne, E., Jiang, C.-Z., and Riechmann, J.L. (2004). WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 4706–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra-Shekara, A.C., Venugopal, S.C., Barman, S.R., and Kachroo, P. (2007). Plastidial fatty acid levels regulate resistance gene-dependent defense signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104 7277–7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., Han, G., Dietrich, C.R., Dunn, T.M., and Cahoon, E.B. (2006). The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell 18 3576–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic, V., and Weissman, J.S. (2007). A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 130 663–677. [DOI] [PubMed] [Google Scholar]
- de Urquiza, A.M., Liu, S., Sjöberg, M., Zetterström, R.H., Griffiths, W., Sjövall, J., and Perlmann, T. (2000). Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science 290 2140–2144. [DOI] [PubMed] [Google Scholar]
- Fehling, E., and Mukherjee, K.D. (1991). Acyl-CoA elongase from a higher plant (Lunaria annua): Metabolic intermediates of very-long-chain acyl-CoA products and substrate specificity. Biochim. Biophys. Acta 1082 239–246. [DOI] [PubMed] [Google Scholar]
- Fiebig, A., Mayfield, J.A., Miley, N.L., Chau, S., Fischer, R.L., and Preuss, D. (2000). Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12 2001–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futerman, A.H., and Hannun, Y.A. (2004). The complex life of simple sphingolipids. EMBO Rep. 5 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, W., et al. (2004). Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 135 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz, T., and Böger, P. (2004). The very-long-chain fatty acid synthase is inhibited by chloroacetamides. Z. Naturforsch. [C] 59 549–553. [DOI] [PubMed] [Google Scholar]
- Gou, J.Y., Wang, L.J., Chen, S.P., Hu, W.L., and Chen, X.Y. (2007). Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis. Cell Res. 17 422–434. [DOI] [PubMed] [Google Scholar]
- Gray, J.E., Holroyd, G.H., van der Lee, F.M., Bahrami, A.R., Sijmons, P.C., Woodward, F.I., Schuch, W., and Hetherington, A.M. (2000). The HIC signaling pathway links CO2 perception to stomatal development. Nature 408 713–716. [DOI] [PubMed] [Google Scholar]
- Hertz, R., Magenheim, J., Berman, I., and Bar-Tana, J. (1998). Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4α. Nature 392 512–516. [DOI] [PubMed] [Google Scholar]
- Itoh, Y., et al. (2003). Free fatty acids regulate insulin secretion from pancreatic β cells through GPR40. Nature 422 173–176. [DOI] [PubMed] [Google Scholar]
- Ji, S.J., Lu, Y.C., Feng, J.X., Wei, G., Li, J., Shi, Y.H., Fu, Q., Liu, D., Luo, J.C., and Zhu, Y.X. (2003). Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Res. 31 2534–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, A., Lapchyk, L., Fukushige, H., Hildebrand, D., Klessig, D., and Kachroo, P. (2003). Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell 15 2952–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastaniotis, A.J., Autio, K.J., Sormunen, R.T., and Hiltunen, J.K. (2004). Htd2p/Yhr067p is a yeast 3-hydroxyacyl-ACP dehydratase essential for mitochondrial function and morphology. Mol. Microbiol. 53 1407–1421. [DOI] [PubMed] [Google Scholar]
- Kim, H.J., and Triplett, B.A. (2001). Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 127 1361–1366. [PMC free article] [PubMed] [Google Scholar]
- Kunst, L., Clemens, S., and Hooker, T. (2000). Expression of the wax-specific condensing enzyme CUT1 in Arabidopsis. Biochem. Soc. Trans. 28 651–654. [PubMed] [Google Scholar]
- Lassner, M.W., Lardizabal, K., and Metz, J.G. (1996). A jojoba β-ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant Cell 8 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.B., Qin, Y.M., Pang, Y., Song, W.Q., Mei, W.Q., and Zhu, Y.X. (2007). A cotton ascorbate peroxidase is involved in hydrogen peroxide homeostasis during fibre cell development. New Phytol. 175 462–471. [DOI] [PubMed] [Google Scholar]
- Markham, J.E., Li, J., Cahoon, E.B., and Jaworski, J.G. (2006). Separation and identification of major plant sphingolipid classes from leaves. J. Biol. Chem. 281 22684–22694. [DOI] [PubMed] [Google Scholar]
- Merrill, A.H., Caligan, T.B., Wang, E., Peters, K., and Ou, J. (2000). Analysis of sphingoid bases and sphingoid base 1-phosphates by high-performance liquid chromatography. Methods Enzymol. 312 3–9. [DOI] [PubMed] [Google Scholar]
- Mietkiewska, E., Giblin, E.M., Wang, S., Barton, D.L., Dirpaul, J., Brost, J.M., Katavic, V., and Taylor, D.C. (2004). Seed-specific heterologous expression of a Nasturtium FAE gene in Arabidopsis results in a dramatic increase in the proportion of erucic acid. Plant Physiol. 136 2665–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.A., Clemens, S., Zachgo, S., Giblin, E.M., Taylor, D.C., and Kunst, L. (1999). CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11 825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.A., and Kunst, L. (1997). Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 12 121–131. [DOI] [PubMed] [Google Scholar]
- Oh, C.S., Toke, D.A., Mandala, S., and Martin, C.E. (1997). ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272 17376–17384. [DOI] [PubMed] [Google Scholar]
- Ortega-Martínez, O., Pernas, M., Carol, R.J., and Dolan, L. (2007). Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317 507–510. [DOI] [PubMed] [Google Scholar]
- Paul, S., Gable, K., Beaudoin, F., Cahoon, E., Jaworski, J., Napier, J.A., and Dunn, T.M. (2006). Members of the Arabidopsis FAE1-like 3-ketoacyl-CoA synthase gene family substitute for the Elop proteins of Saccharomyces cerevisiae. J. Biol. Chem. 281 9018–9029. [DOI] [PubMed] [Google Scholar]
- Pruitt, R.E., Vielle-Calzada, J.P., Ploense, S.E., Grossniklaus, U., and Lolle, S.J. (2000). FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc. Natl. Acad. Sci. USA 97 1311–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, Y.M., Pujol, F.M., Hu, C.Y., Feng, J.X., Kastaniotis, A.J., Hiltunen, J.K., and Zhu, Y.X. (2007). Genetic and biochemical studies in yeast reveal that the cotton fibre-specific GhCER6 gene functions in fatty acid elongation. J. Exp. Bot. 58 473–481. [DOI] [PubMed] [Google Scholar]
- Qin, Y.M., Pujol, F.M.A., Shi, Y.H., Feng, J.X., Liu, Y.M., Kastaniotis, A.J., Hiltunen, J.K., and Zhu, Y.X. (2005). Cloning and functional characterization of two cDNAs encoding NADPH-dependent 3-ketoacyl-CoA reductase from developing cotton fibers. Cell Res. 15 465–473. [DOI] [PubMed] [Google Scholar]
- Shi, Y.H., Zhu, S.W., Mao, X.Z., Feng, J.X., Qin, Y.M., Zhang, L., Cheng, J., Wei, L.P., Wang, Z.Y., and Zhu, Y.X. (2006). Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova, A.N., Hoyt, J.M., Hamilton, A.A., and Alonso, J.M. (2005). A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, J., Post-Beittenmiller, D., and Jaworski, J.G. (1999). KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 17 119–130. [DOI] [PubMed] [Google Scholar]
- Toke, D.A., and Martin, C.E. (1996). Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J. Biol. Chem. 271 18413–18422. [DOI] [PubMed] [Google Scholar]
- Trenkamp, S., Martin, W., and Tietjen, K. (2004). Specific and differential inhibition of very-long-chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proc. Natl. Acad. Sci. USA 101 11903–11908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriezen, W.H., Hulzink, R., Mariani, C., and Voesenek, L.A.C.J. (1999). 1-Aminocyclopropane-1-carboxylate oxidase activity limits ethylene biosynthesis in Rumex palustris during submergence. Plant Physiol. 121 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.S., Shrestha, R., Kilaru, A., Wiant, W., Venables, B.J., Chapman, K.D., and Blancaflor, E.B. (2006). Manipulation of Arabidopsis fatty acid amide hydrolase expression modifies plant growth and sensitivity to N-acylethanolamines. Proc. Natl. Acad. Sci. USA 103 12197–12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanjie, S.W., Welti, R., Moreau, R.A., and Chapman, K.D. (2005). Identification and quantification of glycerolipids in cotton fibers: Reconciliation with metabolic pathway predictions from DNA databases. Lipids 40 773–785. [DOI] [PubMed] [Google Scholar]
- Worrall, D., Ng, C.K.Y., and Hetherington, A.M. (2003). Sphingolipids, new players in plant signaling. Trends Plant Sci. 8 317–320. [DOI] [PubMed] [Google Scholar]
- Yang, S.F., and Hoffman, N.E. (1984). Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 35 155–189. [Google Scholar]
- Yang, S.S., Cheung, F., Lee, J.J., Ha, M., Wei, N.E., Sze, S.H., Stelly, D.M., Thaxton, P., Triplett, B., Town, C.D., and Chen, Z.J. (2006). Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J. 47 761–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov, A., Wisman, E., Huijser, P., Huijser, C., Wellesen, K., and Saedler, H. (1999). Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11 2187–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T., and Pan, J. (1992). Genetic analysis of a fuzzless-lintless mutant in Gossypium hirsutum L. Jiangsu J. Agric. Sci. 7 13–16. [Google Scholar]
- Zheng, H., Rowland, O., and Kunst, L. (2005). Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17 1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Dong, A., Meyer, D., Pichon, O., Renou, J.-P., Cao, K., and Shen, W.-H. (2006). Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell 18 2879–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.