Abstract
Peach latent mosaic viroid (PLMVd) is a chloroplast-replicating RNA that propagates in its natural host, peach (Prunus persica), as a complex mixture of variants, some of which are endowed with specific structural and pathogenic properties. This is the case of variant PC-C40, with an insertion of 12 to 13 nucleotides that folds into a hairpin capped by a U-rich loop, which is responsible for an albino-variegated phenotype known as peach calico (PC). We have applied a combination of ultrastructural, biochemical, and molecular approaches to dissect the pathogenic effects of PC-C40. Albino sectors of leaves infected with variant PC-C40 presented palisade cells that did not completely differentiate into a columnar layer and altered plastids with irregular shape and size and with rudimentary thylakoids, resembling proplastids. Furthermore, impaired processing and accumulation of plastid rRNAs and, consequently, of the plastid translation machinery was observed in the albino sectors of leaves infected with variant PC-C40 but not in the adjacent green areas or in leaves infected by mosaic-inducing or latent variants (including PC-C40Δ, in which the 12- to 13-nucleotide insertion was deleted). Protein gel blot and RT-PCR analyses showed that the altered plastids support the import of nucleus-encoded proteins, including a chloroplast RNA polymerase, the transcripts of which were detected. RNA gel blot and in situ hybridizations revealed that PLMVd replicates in the albino leaf sectors and that it can invade the shoot apical meristem and induce alterations in proplastids, bypassing the RNA surveillance system that restricts the entry of a nucleus-replicating viroid and most RNA viruses. Therefore, a non-protein-coding RNA with a specific structural motif can interfere with an early step of the chloroplast developmental program, leading ultimately to an albino-variegated phenotype resembling that of certain variegated mutants in which plastid rRNA maturation is also impaired. Our results highlight the potential of viroids for further dissection of RNA trafficking and pathogenesis in plants.
INTRODUCTION
With a genome composed of a small (246 to 401 nucleotides), single-stranded, circular RNA, viroids are a special class of non-protein-coding RNAs infecting plants, in which they frequently induce specific diseases (for reviews, see Diener, 2003; Tabler and Tsagris, 2004; Flores et al., 2005a; Ding and Itaya, 2007). Because viroids rely very much on their host machinery for replication and movement, they offer unique opportunities for studying RNA structure–function relationships in these two respects. Research on viroids has already contributed to unveiling major biological issues, including the discovery of hammerhead ribozymes (Hutchins et al., 1986; Prody et al., 1986) and the RNA-directed methylation of nuclear DNA (Wassenegger et al., 1994), and is currently helping to elucidate the mechanisms of RNA trafficking in plants (for review, see Ding et al., 2005). Viroids are grouped into two taxonomic families on the basis of their biological, structural, and biochemical properties (Flores et al., 2005b). Members of the family Pospiviroidae, type species Potato spindle tuber viroid (PSTVd) (Diener, 1972; Gross et al., 1978), adopt a rod-like or quasi-rod-like conformation with characteristic motifs, prominent among them a central conserved region, and replicate in the nucleus by an asymmetric rolling-circle mechanism (Branch et al., 1988; Harders et al., 1989; Bonfiglioli et al., 1996; Qi and Ding, 2003b). By contrast, members belonging to the family Avsunviroidae (Flores et al., 2000; Fadda et al., 2003), type species Avocado sunblotch viroid (ASBVd) (Symons, 1981; Hutchins et al., 1986), may adopt branched conformations, do not contain a central conserved region, and replicate in the chloroplast by a symmetric rolling-circle mechanism in which the oligomeric strands of both polarities self-cleave through hammerhead ribozymes (Bonfiglioli et al., 1994; Daròs et al., 1994; Lima et al., 1994; Bussière et al., 1999; Navarro et al., 1999).
Viroid infection, which entails replication and movement, may interfere with the host regulation machinery and induce wide modifications in host gene expression (Itaya et al., 2002) that ultimately lead to the onset of macroscopic symptoms. Although downregulation of a specific tomato (Solanum lycopersicum) gene involved in cell expansion has been correlated with the stunting phenotype induced by severe PSTVd strains (Qi and Ding, 2003a), the primary molecular event has not been identified. Direct interaction of the mature viroid RNAs with a still unidentified host protein(s) and viroid-mediated silencing of a specific host gene(s) have been proposed as two possible primary events in viroid pathogenesis (for review, see Flores et al., 2005a). The first alternative is partially supported by the identification of pathogenic determinants modulating symptom expression in members of the families Pospiviroidae (Schnölzer et al., 1985; Visvader and Symons, 1986; Owens et al., 1996; Reanwarakorn and Semancik, 1998; Qi and Ding, 2003a) and Avsunviroidae. In the latter, these determinants have been mapped at a UUUC tetraloop in the in vivo branched conformation of Chrysanthemum chlorotic mottle viroid (Navarro and Flores, 1997; De la Peña et al., 1999; De la Peña and Flores, 2002) and in Peach latent mosaic viroid (PLMVd) (Hernández and Flores, 1992; Flores et al., 2006) at a 12- to 13-nucleotide insertion folding into a hairpin capped by a U-rich loop that is associated with an albino-variegated phenotype (peach calico [PC]) (Malfitano et al., 2003; Rodio et al., 2006). Pathogenic determinants could induce other portions of the molecule to rearrange, thereby creating new protein binding sites.
The second alternative (i.e., that RNA-mediated downregulation of specific host genes could be the primary pathogenic event) is based on the identification of viroid-derived small RNAs (vd-sRNAs) resembling the 21- to 24-nucleotide small interfering RNAs that accumulate in plants infected with members of both families (Itaya et al., 2001, 2007; Papaefthimiou et al., 2001; Martínez de Alba et al., 2002; Markarian et al., 2004; Martín et al., 2007). This hypothesis (Wang et al., 2004) links the pathogenic effects elicited by viroids and certain satellite RNAs with RNA-mediated transcriptional and/or posttranscriptional gene silencing (for reviews, see Baulcombe, 2004; Brodersen and Voinnet, 2006). In the proposed mechanism, the vd-sRNAs, generated by an enzyme of the RNase III class (Dicer; Dicer-like in plants) acting on the double-stranded RNA replicative intermediates or on the highly structured genomic RNA, direct host DNA methylation (Pelissier and Wassenegger, 2000) or mimic the effect of microRNAs (Papaefthimiou et al., 2001; Markarian et al., 2004; Wang et al., 2004), a special class of endogenous small regulatory RNAs that target host mRNAs for degradation or translation inhibition (for review, see Kidner and Martienssen, 2005). However, the targeted genomic DNA or mRNA remains to be identified and characterized (Flores et al., 2005a).
The movement and spread of viroids into host plants have been studied in members of the family Pospiviroidae (for review, see Ding et al., 2005). For PSTVd, interactions between viroid sequences or structural motifs and cellular factors have been involved in transport to the nucleus (Woo et al., 1999; Zhao et al., 2001), selective entry of the (+) RNA strand into the nucleolus (Qi and Ding, 2003b), cell-to-cell viroid movement via plasmodesmata (Ding et al., 1997), and long-distance viroid trafficking through the phloem (Palukaitis, 1987; Zhu et al., 2001, 2002; Zhong et al., 2007). Moreover, PSTVd does not invade the shoot apical meristem (SAM) of tomato and Nicotiana benthamiana (Zhu et al., 2001), thus suggesting that the surveillance system limiting the access of host and viral RNAs to the SAM (Foster et al., 2002) could also restrict viroid trafficking. Host proteins potentially involved in the RNA trafficking of representative members of family Pospiviroidae have also been identified (Gómez and Pallás, 2001, 2004; Owens et al., 2001; Martínez de Alba et al., 2003). By contrast, essentially no information is available in this respect for the family Avsunviroidae.
To fill this gap and to further explore the molecular basis of viroid interference with host development, PLMVd seems particularly appropriate because variants with different pathogenicity, ranging from latency to PC induction, have been characterized (Hernández and Flores, 1992; Ambrós et al., 1998, 1999; Malfitano et al., 2003; Rodio et al., 2006). Moreover, the pathogenic determinant of PLMVd variants responsible for PC has been mapped (see above). Interestingly, this albino-variegated phenotype recalls that of several plant mutants (Chatterjee et al., 1996; Wang et al., 2000; Bellaoui et al., 2003; Bisanz et al., 2003; Bellaoui and Gruissem, 2004). Using the system formed by PLMVd and its natural host (peach [Prunus persica]) and taking a multidisciplinary approach, we now report that a key plant developmental process can be drastically influenced by a motif of the viroid genomic RNA and discuss a model for viroid pathogenesis based on the ability of certain non-protein-coding RNAs to alter the proplastid developmental program.
RESULTS
Characteristics of the PC Phenotype
Slash-inoculation of GF-305 peach seedlings with in vitro transcripts of certain PLMVd variants containing specific insertions in loop A causes PC. Variant PC-C40, characterized previously (Figure 1) (Malfitano et al., 2003), is particularly efficient in eliciting this stochastic symptom of albinism-variegation (Figure 2A) and has been used in this study. Like other PC-inducing variants assayed to date (Malfitano et al., 2003; Rodio et al., 2006), symptom expression was stimulated when, 6 weeks after inoculation, seedlings were chilled to induce dormancy prior to transferring back to the greenhouse to promote the emergence of new flushes. The albino-variegated phenotype was expressed on the entire growing stems and expanding leaves (Figure 2B) or was restricted to leaf sectors of varying area (Figure 2C). This phenotype is already visible on the small nonexpanded leaves of the new flushes emerging after the dormant period (Figure 2D) and is very different from the mosaic (Figure 2E) caused by some PLMVd variants that, like GDS6 (Ambrós et al., 1999), lack any insertion in loop A (see Supplemental Figure 1 online).
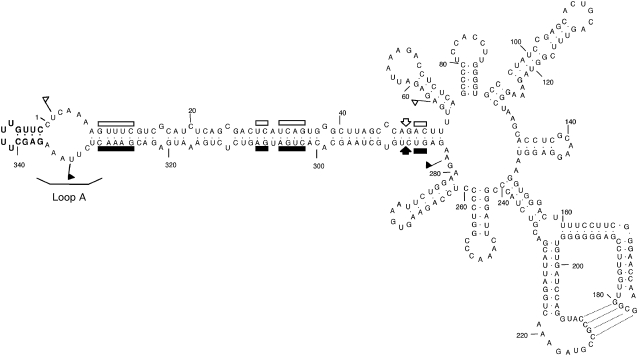
Figure 1.
The PLMVd Reference Variant Inducing the Albino-Variegated Phenotype Known as PC.
Primary and predicted secondary structures of the lowest free energy of PLMVd variant PC-C40 (Malfitano et al., 2003). The nucleotides of the characteristic insertion in loop A containing the PC determinant are highlighted in boldface. (+) and (−) self-cleaving domains are delimited by flags, residues conserved in most natural hammerhead structures are indicated by bars, and the self-cleavage sites are marked by arrows. Closed and open symbols refer to (+) and (−) polarities, respectively. Residues involved in a pseudoknot between positions 177 to 180 and 210 to 213, proposed on the basis of in vitro nuclease mapping and oligonucleotide binding shift assays (Bussière et al., 2000) and natural variability (Malfitano, 2000), are indicated by broken lines.
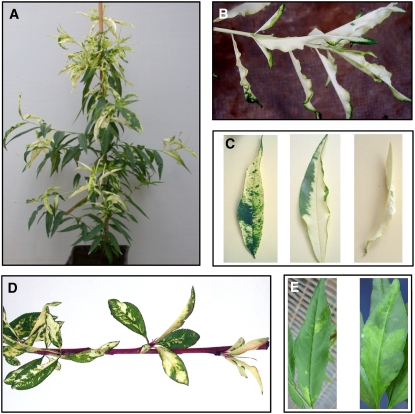
Figure 2.
PC and Other PLMVd-Induced Phenotypes in GF-305 Peach Seedlings.
(A) PC symptoms elicited by PLMVd variant PC-C40 (Malfitano et al., 2003).
(B) Close view of an almost completely albino shoot from a PC-expressing plant inoculated with PLMVd variant PC-C40.
(C) Distinct leaf patterns of the albino-variegated phenotype incited by variant PC-C40.
(D) PC symptoms induced by variant PC-C40 in young expanding leaves of new flushes emerging after a dormant period.
(E) Typical leaf mosaic induced by PLMVd variant GDS6 (Ambrós et al., 1999).
Leaf and Cell Alterations Induced by Different PLMVd Variants
To gain a further insight into the processes leading to the typical PC phenotype, leaf tissues from GF-305 seedlings infected with variant PC-C40 were examined by light and electron microscopy and compared with tissues noninfected and infected with the natural latent variant PC-P1.159 (Rodio et al., 2006) and with the artificial latent variant PC-C40Δ, generated by site-directed deletion of the PC determinant from variant PC-C40 (Malfitano et al., 2003) (see Supplemental Figure 1 online). In some experiments, leaf tissues displaying the typical mosaic incited by variant GDS6 were also included as an additional control.
Light microscopy showed that, in contrast with noninfected leaves, which had typical epidermal, columnar palisade, and spongy mesophyll cell layers (Figure 3A), albino sectors from leaves at similar developmental stages infected with variant PC-C40 were thinner than normal because of the more compact structure and the limited expansion of palisade cells (Figure 3B). Green sections from leaves infected with variant PC-C40 or with the latent variants PC-P1.159 and PC-C40Δ, as well as with the mosaic-inducing variant GDS6, presented a normally differentiated palisade cell layer comparable to that observed in the noninfected control (Figure 3A).
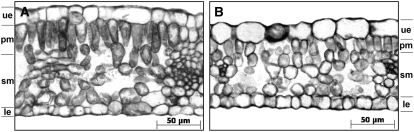
Figure 3.
Light Microscopy Images of Mature GF-305 Peach Leaves.
Transverse sections from a healthy leaf (A) and from the albino sector of a leaf infected by variant PC-C40 (B). In the latter, the palisade cells are not completely differentiated in a columnar layer (the height of which was approximately half that of green leaves from healthy and PLMVd-infected controls). ue, upper epidermis; pm, palisade mesophyll; sm, spongy mesophyll; le, lower epidermis.
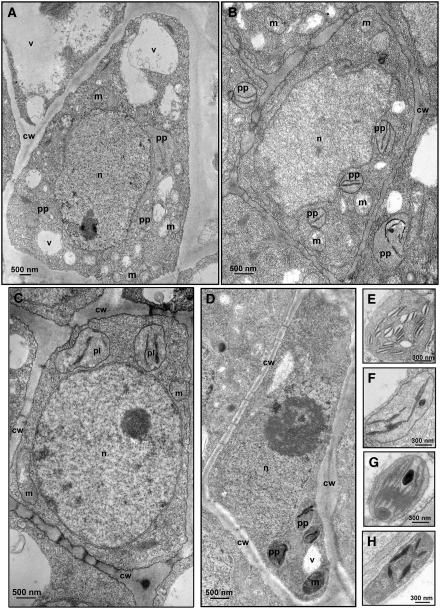
Transmission electron microscopy observations of mesophyll cells from the noninfected control showed mature chloroplasts with typical lamellar membrane organization. Grana and intergrana structures were clearly visible (Figures 4A and 4B) and remained still distinguishable, even if less well defined, in senescent cells (Figure 4C). Instead, albino sectors from leaves infected with variant PC-C40 showed plastids with irregular shape and size, bearing only rudimentary thylakoids dispersed in an electron-dense matrix (Figures 4D and 4E). These plastids resembled the proplastids of meristematic cells, although they were larger than typical proplastids and contained vesicles (Figure 4E) that sometimes enclosed electron-dense material of unknown nature (see Supplemental Figure 2 online). Structural alterations were less striking in green sectors from leaves infected with variant PC-C40 or from leaves of the same plants with no PC symptoms. In these tissues, the chloroplasts were apparently differentiated but thylakoids were not organized in grana and intergrana and showed wider interspaces (Figures 4F and 4G). Similar alterations were found in the chloroplasts of leaves infected by the latent variants PC-P1.159 and PC-C40Δ (see Supplemental Figure 3 online).
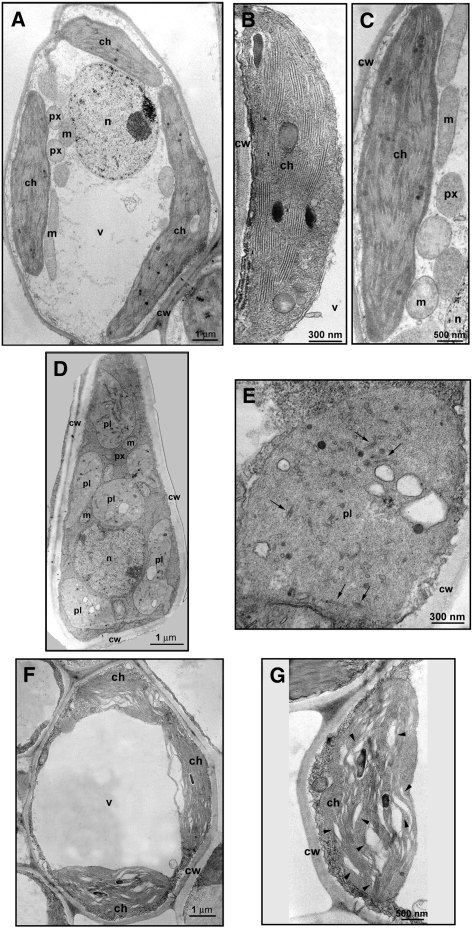
Figure 4.
Transmission Electron Microscopy Images of Cells and Plastids of Mature GF-305 Peach Leaves.
(A) to (C) Mesophyll cell (A) and mature (B) and senescent (C) chloroplasts of a healthy control.
(D) and (E) Mesophyll cell (D) and plastid resembling a proplastid and containing vesicles (E) observed in the albino sector of a leaf infected by variant PC-C40. Black arrows in (E) indicate rudimentary thylakoids.
(F) and (G) Mesophyll cell (F) and altered chloroplast (G) from a green sector of a leaf infected by variant PC-C40. Arrowheads in (G) point to enlarged thylakoid interspaces.
ch, chloroplast; cw, cell wall; m, mitochondrion; n, nucleus; pl, plastid; px, peroxisome; v, vacuole.
Light microscopy and transmission electron microscopy examinations were also extended to albino leaf sectors of a GF-305 seedling infected with a natural PLMVd population of PC-inducing variants not including the variant PC-C40 (Rodio et al., 2006). The same modifications of the palisade cell layer and plastids were observed, indicating that these ultrastructural and cellular alterations are closely associated with the PC phenotype irrespective of the PC-inducing variant.
PC Phenotype Is Associated with Impaired Plastid Transcription and Translation
Histological and cellular alterations similar to those found in the albino sectors of PC-affected leaves have been observed previously in certain albino-variegated mutants of several plant species in which chloroplast development is blocked at a very early stage. In some of these mutants (Barkan, 1993; Bellaoui et al., 2003; Bisanz et al., 2003; Bellaoui and Gruissem, 2004; Bollenbach et al., 2005), impairment of plastid translation resulting from altered processing of plastid rRNA is regarded as the major biochemical defect. Maturation of plastid rRNAs in flowering plants depends on the ordered processing of the primary transcript from a single operon that also encodes three tRNAs. The initial excision of the tRNAs and additional cleavages generate the mature 16S and 5S rRNAs and a 23S–4.5S intermediate (p23S-4.5S). The subsequent processing of this intermediate leads to the 23S and 4.5S rRNAs, which require further nucleolytic activities to produce their mature 5′ and 3′ termini (Bollenbach et al., 2005).
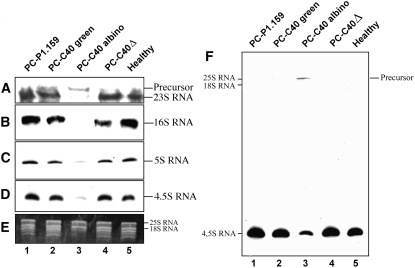
To explore whether alterations in this complex biochemical pathway could also be involved in eliciting the PC phenotype, RNA gel blot hybridizations with cDNA probes specific for peach plastid rRNAs were performed (Figures 5A to 5D). Mature plastid 23S, 16S, 5S, and 4.5S rRNAs generated very weak signals in RNA preparations from the albino leaf sectors (Figures 5A to 5D, lane 3), although the signals were clearly visible when the membranes were overexposed. By contrast, the accumulation of these rRNAs in green areas of the same symptomatic leaves (Figures 5A to 5D, lane 2) was similar to that found in leaves from plants infected with the latent variants PC-P1.159 and PC-C40Δ and in leaves from noninfected plants (Figures 5A to 5D, lanes 1, 4, and 5). Interestingly, in RNA preparations from the albino leaf sectors, the probe specific for the 23S rRNA revealed a larger RNA that was absent in the other RNA preparations (Figure 5A, compare lane 3 with the other lanes). This larger RNA most likely corresponds to the p23S-4.5S intermediate because it was also detected with the probe specific for the 4.5S rRNA (Figure 5F). These results strongly support the notion that, besides accumulation, processing of plastid rRNAs is altered in the albino leaf sectors infected by PC-C40 and undermine the possibility of changes in plastid stability impeding the extraction of intact rRNA from such plastids. Equal loading of RNA preparations was assessed by ethidium bromide staining of the larger bands corresponding to the nucleus-encoded 25S and 18S rRNAs (Figure 5E). The smaller bands were absent in the ethidium bromide profile of the RNA preparation from the albino sectors, consistent with the low accumulation of plastid rRNAs observed by RNA gel blot hybridization (Figure 5E, lane 3).
Figure 5.
Accumulation Levels of Plastid rRNAs in GF-305 Peach Leaves.
(A) to (D) RNA gel blot analyses of RNA preparations hybridized with cDNA probes specific for 23S, 16S, 5S, and 4.5S rRNAs after electrophoresis on denaturing agarose (1.2%) gels for detecting 23S and 16S rRNAs ([A] and [B]) or on denaturing polyacrylamide (5%) gels for detecting 5S and 4.5S rRNAs ([C] and [D]). Only the autoradiogram sections corresponding to each of the four rRNAs, and to a larger precursor observed exclusively in lane 3, are presented.
(E) Ethidium bromide pattern of the upper section of the polyacrylamide gel showing equal intensity of the larger bands generated by the nucleus-encoded 25S and 18S rRNAs. The absence of the smaller bands in the RNA preparation from the albino leaf sector is consistent with the low accumulation of plastid rRNAs detected by RNA gel blot hybridization.
(F) Overexposure of the membrane corresponding to (D) showing the presence of a larger RNA that most likely corresponds to the p23S-4.5S intermediate.
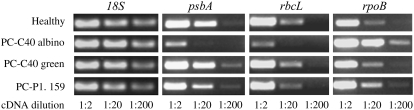
Similar to plant mutants in which plastid rRNA processing is also impaired, resulting in low levels of translation-competent plastid ribosomes and, consequently, of plastid-encoded proteins (Barkan, 1993; Bellaoui et al., 2003; Bisanz et al., 2003; Bollenbach et al., 2005), impairment of the plastid translation machinery was also expected in the albino sectors of PC-affected leaves. This would lead to limited accumulation and reduced activity of the eubacterium-like plastid-encoded polymerase (PEP), which cannot be synthesized in plastids lacking functional ribosomes (Hess et al., 1993; Zubko and Day, 2002). Consistent with this view, RT-PCR estimates using serial cDNA dilutions showed that the steady state levels of the PEP-dependent transcripts of the photosystem II D1 protein (encoded by the psbA gene) and the ribulose-1,5-bisphosphote carboxylase/oxygenase large subunit (encoded by the rbcL gene), which are two typical plastid-encoded proteins, were substantially lower in the albino sectors than in the green controls (the adjacent leaf sectors without PC symptoms or leaves infected by the latent variant PC-P1.159 or not infected). By contrast, the steady state levels of the nucleus-encoded 18S rRNA were similar in all samples (Figure 6). Furthermore, a typical transcript of the nucleus-encoded plastid polymerase (NEP), that of the plastid rpoB subunit of PEP, accumulated at higher levels in the albino tissues than in the green controls (Figure 6), supporting the notion that NEP is imported and active in the altered plastids. The overaccumulation in the albino tissues of the rpoB transcript, and most likely of other NEP-dependent transcripts, is not surprising because similar observations have been made in albino mutants with ribosome-deficient plastids or in which the plastid translation machinery is impaired as a consequence of reducing or abolishing the PEP activity (Hess et al., 1993; Hajdukiewicz et al., 1997; Krause et al., 2000; Zubko and Day, 2002; Legen et al., 2002; Nagashima et al., 2004). These data, together with the low accumulation of the plastid rRNAs shown before, strongly support the view that PEP is dysfunctional in the albino tissues.
Figure 6.
Transcript Accumulation in GF-305 Peach Leaves.
RT-PCR estimation of the nucleus-encoded 18S rRNA, the transcripts from the plastid-encoded psbA and rbcL genes, and the transcript from the nucleus-encoded rpoB gene. The cDNAs, generated by random-primed reverse transcription of equalized RNA preparations, were serially diluted (1:2, 1:20, and 1:200) before amplification with gene-specific primer pairs. The amplified products were analyzed by agarose gel electrophoresis and ethidium bromide staining. Only gel sections containing the amplified cDNAs are presented. Results are representative of three replicates conducted on two biologically independent samples.
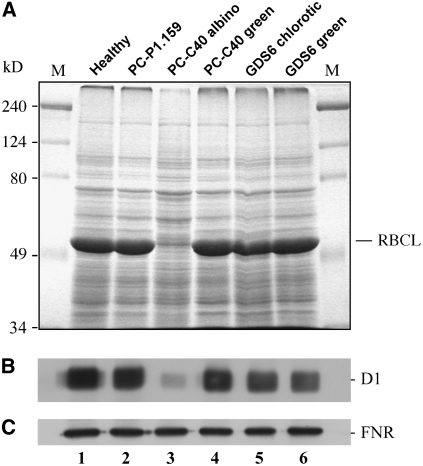
As anticipated, protein examination by SDS-PAGE and protein gel blot analysis showed that the two plastid-encoded proteins RBCL and D1 were almost undetectable in protein preparations from the albino sectors of PC-affected leaves (Figures 7A and 7B, lane 3). By contrast, but in agreement with the levels of their respective transcripts and the mature plastid rRNAs, the expression of psbA and rbcL genes in the green sectors of the same leaves was similar to that observed in leaves from noninfected plants (Figures 7A and 7B, compare lanes 4 and 1) and leaves infected by the latent variant PC-P1.159 (Figures 7A and 7B, lane 2). Furthermore, no effect on the accumulation of D1 and RBCL proteins with respect to the noninoculated control was detected in the chlorotic and nonchlorotic areas of leaves infected by the mosaic-inducing variant GDS6 (Figure 7, lanes 5 and 6), indicating that the mechanism underlying this symptom is most likely different from that involved in the PC phenotype. Equal loading of protein preparations was confirmed by the similar pattern and band intensity (except the very abundant RBCL subunit in green tissues that was almost undetectable in the albino leaf sectors) in gels stained with Coomassie blue (Figure 7A) or analyzed by protein gel blot for detecting the ferredoxin-NAD(P)+ oxidoreductase (FNR), a nucleus-encoded plastid protein that accumulates at similar levels in all of the samples (Figure 7C). These findings also support the view that protein stability is not compromised by a general degradation in the altered plastids and that their protein import machinery is functional because no extra band corresponding to a possible FNR precursor, bearing an N-terminal plastid target signal, was identified in the albino tissues.
Figure 7.
Accumulation Levels of Solubilized Proteins in GF-305 Peach Leaves.
(A) Protein profiles after SDS-PAGE on a 12% gel and staining with Coomassie blue. The external lanes contain molecular markers (M), with their sizes in kilodaltons indicated at left. The position of the large subunit of the ribulose-1,5-bisphosphate carboxylase/oxygenase (RBCL), the accumulation of which is very much reduced in the albino sector of a leaf infected by variant PC-C40, is indicated at right.
(B) and (C) Protein gel blot analyses with antibodies raised against the plastid-encoded D1 protein (B) and the nucleus-encoded and plastid-targeted FNR (C). Only the autoradiogram sections corresponding to each protein are shown.
PLMVd Replicates and Accumulates in Albino Leaf Sectors with Altered Plastids
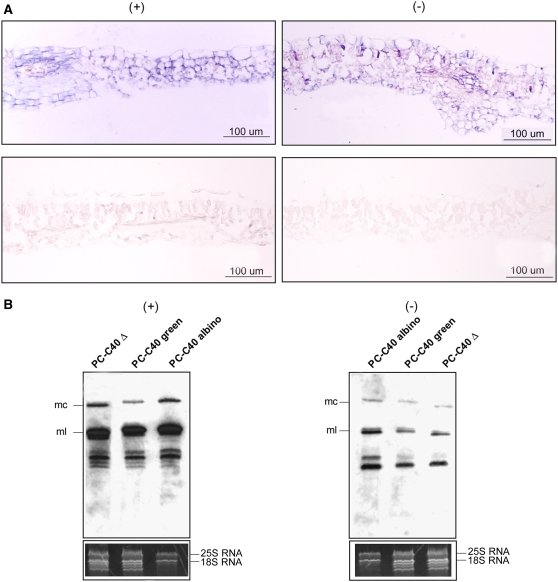
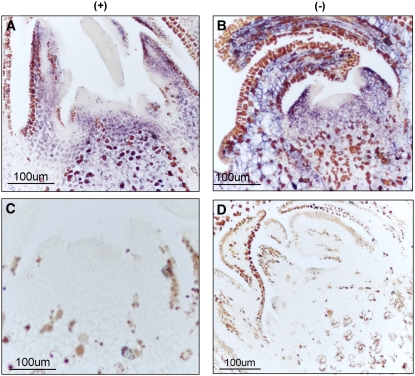
Because PLMVd replicates and accumulates in the chloroplast (Bussière et al., 1999), impairment of the plastid translation machinery observed in the albino leaf sectors raised the intriguing question of whether PLMVd was also able to replicate and accumulate in these sectors presenting altered plastids and essentially lacking plastid-encoded proteins. In situ hybridization with specific and equalized riboprobes detected (+) and (−) PLMVd RNAs in the albino leaf sectors from plants infected with variant PC-C40, but not in the noninoculated controls, thus showing that the hybridizations were specific (Figure 8A).
Figure 8.
Detection of PLMVd (+) and (−) Strands in GF-305 Peach Leaves Infected by Two Viroid Variants.
(A) In situ hybridization of transverse sections from the albino sector of a leaf infected by variant PC-C40 (top) and from a healthy leaf (bottom). The sections were hybridized with riboprobes for detecting (+) (left) and (−) (right) PLMVd strands. Hybridization signals appear with a blue-violet color.
(B) RNA gel blot hybridizations of different RNA preparations with riboprobes for detecting (+) (left) and (−) (right) PLMVd strands following fractionation by denaturing PAGE on a 5% gel. The positions of the monomeric circular (mc) and linear (ml) PLMVd RNAs are indicated at left. The bottom panels show the ethidium bromide pattern of the top section of the gels (for additional details, see the legend to Figure 5E). Results are representative of three independent experiments with similar outcome: the progeny of variant PC-C40 accumulated at a significantly higher level (50.5% ± 4.5%) in the albino sectors than in the green sectors of the same leaf. Loadings were equalized with respect to the nucleus-encoded rRNAs, which remain unaffected in symptomatic and asymptomatic tissues.
This result was confirmed by RNA gel blot analyses using the same riboprobes and RNA preparations from adjacent albino and green sectors of PC-expressing leaves (Figure 8B). PLMVd RNAs corresponding to the circular and linear forms of the two polarity strands were observed in both RNA preparations as well as in a preparation from a leaf infected by the latent variant PC-C40Δ, which was used as an additional control (Figure 8B). In agreement with previous data (Bussière et al., 1999; Delgado et al., 2005), PLMVd (+) RNAs were more abundant than their complementary (−) strands, as were the linear forms with respect to their circular counterparts (Figure 8B). Interestingly, the progeny of variant PC-C40 accumulated at higher levels in the albino sectors than in the green areas of the same leaf (Figure 8B). Equal loading of the RNA preparations was assessed by ethidium bromide staining of the polyacrylamide gels, which also showed that bands corresponding to plastid rRNAs were essentially undetectable in the preparation from the albino leaf sectors (Figure 8B, bottom panels).
As a consequence of the high self-complementarity of viroid RNAs, cross-hybridization between a strand-specific probe and viroid RNAs of the same polarity was reported previously (Owens and Diener, 1982; Branch and Robertson, 1984; Hutchins et al., 1985). This prompted us to use very stringent conditions: 70°C in the presence of 50% formamide or an equivalent commercial buffer for in situ and RNA gel blot hybridizations, respectively. Under these conditions, in vitro–synthesized (+) and (−) PLMVd RNAs failed to show detectable cross-hybridization in RNA gel blot analysis. Moreover, because the strand-specific riboprobes used in our experiments had been equalized previously, the intensity of the hybridization signals should approximately reflect the relative concentrations of (+) and (−) PLMVd RNAs in tissues and RNA preparations. Together, these data show that PLMVd replicates in the altered plastids of mesophyll cells from the albino leaf sectors of PC-expressing leaves and strongly suggest that the host proteins assisting PLMVd replication are nucleus-encoded.
In parallel RNA gel blot hybridizations, PLMVd sRNAs were detected in the RNA preparations from PLMVd-infected seedlings (see Supplemental Figure 4 online), and quantifications from three independent experiments showed that their levels followed the same trend as the genomic PLMVd RNA, irrespective of whether the tissue was asymptomatic or expressing PC. Therefore, generation of PLMVd sRNAs is not impaired in albino tissues.
Proplastids in the Apical Meristem of Albino Shoots Are Altered
The presence of plastids resembling proplastids in mesophyll cells from the albino sectors of mature leaves infected by PLMVd variants inducing PC suggested that this phenotype could result from impaired chloroplast development, a process that starts in the SAM (Kirk and Tilney-Bassett, 1978). To test this hypothesis, longitudinal sections of shoot apices from GF-305 seedlings infected by PLMVd variants differing in their pathogenicity were examined by transmission electron microscopy. The observations were extended to different apex sections, including the leaf primordia and the meristematic cells containing proplastids, at various developmental stages. Most proplastids from the albino shoot apices showed structural alterations already evident in the meristematic cells (Figure 9A), namely rudimentary thylakoids with enlarged interspaces forming vesicles (Figure 9E). Similar malformations were not detected in noninoculated controls or in tissues infected with latent variants PC-C40Δ and PC-P1.159 (Figures 9B to 9D), the proplastids of which had stacked thylakoids (Figures 9F to 9H). These data support the notion that impairment of an early step of chloroplast development is specific to PLMVd variants inducing PC, and they also suggest that the other alterations found in chloroplasts from green leaf tissues infected by symptomatic and latent variants (Figures 4C and 4D; see Supplemental Figure 3 online) most likely result from interference with a later stage of chloroplast differentiation. Consistent with this view, no specific alterations were detected in SAM proplastids from tissues infected with the mosaic-inducing variant GDS6. A corollary from these data is that at least some PLMVd variants should be able to elude the surveillance system limiting RNA invasion into the SAM (Foster et al., 2002), although at this point we could not rule out the possibility that SAM invasion was a general feature of all PLMVd variants irrespective of their pathogenicity.
Figure 9.
Transmission Electron Microscopy Images of Cells and Proplastids from the SAM of GF-305 Peach Seedlings.
(A) SAM cell from an albino shoot infected by variant PC-C40.
(B) SAM cell from a healthy control.
(C) and (D) SAM cells from shoots infected by the latent variants PC-C40Δ (C) and PC-P1.159 (D).
(E) to (H) Closer views of proplastids from the same tissue sections presented in (A) to (D), respectively. Proplastids from shoots infected by latent variants ([G] and [H]) do not show thylakoid alterations with respect to the healthy control (F), in contrast with thylakoids from the albino shoot infected by variant PC-C40, in which enlarged interspaces are clearly visible (E).
cw, cell wall; m, mitochondrion; n, nucleus; pp, proplastid; v, vacuole.
PLMVd Invades the SAM
To address the possibility of PLMVd invasion of the apical meristem, longitudinal sections of shoot apices from GF-305 seedlings infected with variants PC-C40, GDS6, and PC-P1.159, thus expressing PC, mosaic, and no symptoms, respectively, were examined by in situ hybridization with strand-specific PLMVd riboprobes. This experiment allowed us to investigate both the replication and accumulation of the invading viroid RNA. PLMVd RNAs of both polarities were detected in the apices of all of the PLMVd-infected shoots analyzed, regardless of the variant and its pathogenicity (Figures 10A and 10B; see Supplemental Figure 5 online). Hybridization signals were uniformly distributed in the shoot apex, including the SAM and the first leaf primordia (Figures 10A and 10B; see Supplemental Figure 5 online), except in a few cell layers at the very tip of the SAM. However, we cannot exclude the possibility of viroid accumulation in these cell layers below the detection limit of the technique. No hybridization signal was observed in noninfected controls (Figures 10C and 10D), in which only unspecific precipitates of unknown origin, also present in infected shoots, were visible. Detection of PLMVd (+) and (−) RNAs under the stringent hybridization conditions used shows that PLMVd invades the SAM and most likely the proplastids, which therefore appear competent for viroid replication at a very early developmental stage. Moreover, the ability to invade the SAM is a general property of PLMVd variants, because no correlation was found with their pathogenicity.
Figure 10.
Detection of PLMVd (+) and (−) Strands in the SAM of GF-305 Seedlings.
In situ hybridizations with riboprobes for detecting (+) (left) and (−) (right) PLMVd strands in longitudinal SAM sections from albino shoots infected by variant PC-C40 ([A] and [B]) and from a healthy shoot ([C] and [D]). Hybridization signals appear with a blue-violet color different from the brown precipitates of unknown origin also visible in the healthy control.
DISCUSSION
PC PLMVd Variants and Certain Variegated Plant Mutants Induce Similar Leaf and Cell Alterations
Previous findings that PC symptoms are triggered by PLMVd variants with specific hairpin insertion in the genomic RNA (Malfitano et al., 2003; Rodio et al., 2006) have been complemented here with light microscopy and transmission electron microscopy analyses of symptomatic tissues. These observations established a close parallelism with defects produced by certain variegated mutants of dicotyledonous plants (Chatterjee et al., 1996; Wang et al., 2000; Bellaoui et al., 2003; Bisanz et al., 2003; Bellaoui and Gruissem, 2004). In such mutants, assembly of the chloroplast thylakoid membrane—one of the early events in chloroplast development (Vothknecht and Westhoff, 2001)—is impaired. This alteration is generally accompanied by defects in the organization of the palisade cell layer, which likely result from malfunctioning of the plastid-to-nucleus signaling pathways involved in normal leaf differentiation. Because the absence of a completely differentiated palisade cell layer in the albino leaf sectors induced by the PC-C40 variant (Figure 3) is associated with the presence of plastids lacking thylakoid membranes (Figure 4), these results strongly suggest that the cellular events leading to PC symptom expression also involve the inhibition of chloroplast development at an early stage. We also observed milder alterations in chloroplasts of green leaf sectors from plants infected by PC-inducing or latent variants. Therefore, PLMVd induces plastid ultrastructural defects even when macroscopic leaf symptoms are not visible, which might account for the lower productivity and premature senescence observed in cultivated peach plants infected by latent PLMVd strains (Desvignes, 1986; Flores et al., 2006).
The Ability of a Non-Protein-Coding RNA to Perturb Cellular Functions Resides in a Specific Motif
The observation that the processing of plastid rRNAs is affected (Figure 5) and that typical plastid-encoded proteins like D1 and RBCL (Figure 7) accumulate at very low levels in the albino sectors of PC-expressing leaves reinforced the link with certain plant mutants inducing variegated phenotypes in which similar biochemical defects have been reported (Bellaoui et al., 2003; Bisanz et al., 2003; Bellaoui and Gruissem, 2004; Bollenbach et al., 2005). Impairment of the p23S-4.5S rRNA processing was associated in our system with reduced accumulation of the mature plastid rRNAs, a situation also reported in some of these mutants (Bollenbach et al., 2005), suggesting the existence of additional deleterious effects on the transcription of plastid rRNAs. Since impairment of plastid transcription and translation (Figures 6 and 7) was not detected in tissues infected by latent and mosaic-inducing PLMVd variants, which lack the PC determinant, as well as by PC-C40Δ, in which the determinant was removed by site-directed mutagenesis, these results showed that a motif of only 12 to 13 nucleotides makes a non-protein-coding RNA competent for interfering with chloroplast development. These observations pose interesting questions on the primary molecular mechanism, which might involve direct interaction of the mature viroid RNA containing the PC determinant with a host factor needed for plastid rRNA maturation or viroid-induced silencing of a specific host gene product also mediating this process.
Direct interactions between invading RNAs and host factors have been proposed to regulate symptom expression not only in nuclear viroids (Diener et al., 1993) but also in viral satellite RNAs (Taliansky and Robinson, 1997) and viruses (Rodríguez-Cerezo et al., 1991). This scenario is particularly attractive in this instance because the PC determinant maps at a hairpin, a structural element frequently involved in RNA–RNA and RNA–protein interactions. Moreover, comparisons among the progeny resulting from natural and artificial variants inducing PC have revealed that the sequence variability preserves the hairpin structure and the U-rich capping loop (Malfitano et al., 2003; Rodio et al., 2006).
Speculations on the nature of the host factor interacting with the PC-inducing PLMVd variants seem premature at this stage. Intriguingly, however, one of the variegated mutants with a phenotype resembling PC, the recessive variegated and distorted leaf mutant of tobacco (Nicotiana tabacum), has been mapped at a nuclear gene potentially encoding a plastid DEAD box RNA helicase (Wang et al., 2000). An enzyme of this class with affinity for structured RNA motifs might interact with PC-inducing PLMVd variants and, as a consequence, become compromised in its role in chloroplast differentiation and leaf morphogenesis.
The alternative mechanism involving vd-sRNAs as the effectors of PC symptoms appears less likely, because vd-sRNAs are present in albino and green sectors of symptomatic leaves (see Supplemental Figure 4 online). However, at this stage, we cannot exclude the involvement in pathogenesis of vd-sRNAs resulting from the PC determinant. Cloning and sequencing of the population of PLMVd sRNAs from tissues infected by PC-inducing and latent variants may provide hints in this direction: the presence of vd-sRNAs from the PC determinant would be compatible with an RNA silencing–based hypothesis, whereas their absence would make a case against it.
Altered Plastids Support PLMVd Replication, the Import of Nucleus-Encoded Proteins, and NEP-Dependent Transcription
The involvement of the nucleoplasmic RNA polymerase II in the replication of PSTVd and other members of the family Pospiviroidae is well documented based on the inhibitory effects of nanomolar concentrations of α-amanitin and on the immunoprecipitation of (+) and (−) viroid strands with a monoclonal antibody against the C-terminal domain of the largest subunit of RNA polymerase II (for review, see Flores et al., 2005a). However, while the participation of a chloroplastic RNA polymerase is generally accepted in the family Avsunviroidae, the specific nature of this enzyme remains controversial. From the lack of inhibitory effects of tagetitoxin on the elongation of ASBVd (+) and (−) strands in a chloroplastic transcription assay from infected avocado (Persea americana), the involvement of a NEP has been proposed (Navarro et al., 2000). On the other hand, the contribution of PEP to the replication of PLMVd was inferred from in vitro transcription studies with the RNA polymerase of Escherichia coli, leading to the hypothesis that different chloroplastic RNA polymerases could mediate the replication of different members of the family Avsunviroidae (Pelchat et al., 2001).
Our present results argue against this latter hypothesis and, instead, support a role for NEP in the replication of ASBVd and PLMVd. Indeed, RNA gel blot hybridizations showed that in the albino leaf sectors induced by the PC-C40 variant, PLMVd (+) and (−) circular and linear forms accumulate to levels even higher than those found in the green adjacent areas (Figure 8B). The possibility that viroid RNAs are synthesized in the green sectors and then translocated to the adjacent albino sectors appears unlikely because PLMVd RNAs, which included the (−) circular form (a key replicative intermediate), were detected in leaves displaying the albino phenotype in the complete foliar area since their emergence. These results indicate active viroid replication in altered plastids in which rRNA processing and accumulation are impaired, as is the translation of plastid-encoded proteins like those forming the PEP core. Moreover, the altered plastids can import nucleus-encoded proteins (Figure 7) and sustain active NEP-dependent but not PEP-dependent transcription (Figure 6), thus showing that they retain key functional properties. PLMVd replication was also detected by in situ hybridization in cells of the SAM containing proplastids at a very early developmental stage (Figure 10), at which PEP activity is presumably very low. Since at least two different NEP activities have been reported in chloroplasts (Allison et al., 1996; Bligny et al., 2000), which one is actually involved in viroid replication remains to be determined. Furthermore, replication of PLMVd in plastids in which protein translation is impaired suggests that any other host protein assisting viroid replication should be nucleus-encoded, including an RNA ligase, if circularization of the monomeric forms that result from hammerhead-mediated self-cleavage is an enzyme-catalyzed reaction. In support of this view, a chloroplast protein that binds ASBVd in vivo and facilitates the hammerhead-mediated self-cleavage of a dimeric ASBVd RNA in vitro is nucleus-encoded (Daròs and Flores, 2002).
Viroids as Models for Studying RNA Trafficking into the SAM
The existence of a surveillance system regulating the selective entry of RNAs into the SAM has been documented (Foster et al., 2002). In this context, recent data support the notion that an RNA-silencing mechanism (Schwach et al., 2005), most likely temperature-dependent (Qu et al., 2005), operates in the exclusion of certain viral RNAs from the SAM. The same mechanism could explain the exclusion of PSTVd, the type species of nucleus-replicating viroids, from the vegetative and floral apical meristems (Qi and Ding, 2003a). However, the identification of PLMVd (+) and (−) RNAs in the SAM of infected peach (Figure 10) shows that a chloroplast-replicating viroid overcomes this surveillance system. PLMVd trafficking into the SAM may depend on the plastid localization of this viroid, in which no RNA silencing has been reported to date. In support of this view, we have observed that ASBVd, which also replicates and accumulates in the chloroplast and can induce symptoms similar to PC (Semancik and Szychowski, 1994), also invades the SAM (data not shown). Our results highlight the potential of viroids for further dissection of RNA trafficking in plants, establish an important additional difference between representative members of the two viroid families, and also explain why obtaining PLMVd-free peach plants by in vitro shoot-tip grafting is very difficult (Barba et al., 1995).
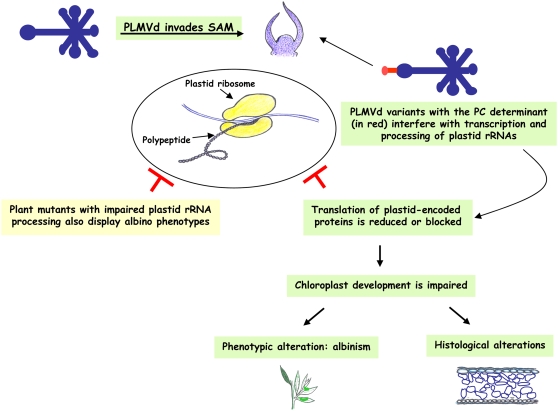
SAM invasion by PLMVd, together with the other results reported here, support a model that traces a link between the macroscopic PC symptoms, the associated cellular and biochemical alterations, and the presence of a specific structural motif in the viroid RNA (Figure 11). Invasion by PC-inducing variants of the SAM, the site where chloroplast development from proplastids starts, would interfere with plastid rRNA maturation, blocking plastid protein translation and inhibiting chloroplast differentiation, ultimately leading to an albino phenotype resembling that of certain variegated mutants in which plastid rRNA maturation is impaired. These results suggest that common developmental pathways are altered by an alien non-protein-coding RNA and by mutations affecting endogenous genes.
Figure 11.
Model for Pathogenesis Based on the Ability of PLMVd RNA to Invade the SAM and Alter the Proplastid Developmental Program.
METHODS
Plant Material and Growing Conditions
Leaves and shoot apices analyzed in this study were from GF-305 peach (Prunus persica) seedlings healthy or slash-inoculated with in vitro transcripts of natural and artificial PLMVd variants inducing different leaf symptoms or no symptoms (see below). To stimulate symptom expression, 6 weeks after inoculation the seedlings were chilled at 4°C in darkness for 6 to 8 weeks and then transferred back to the greenhouse, and new flushes emerged soon after. When indicated, expanded leaves from the PC-inducing PLMVd field isolate PC-P1, characterized previously (Rodio et al., 2006), were also examined.
PLMVd Variants, in Vitro Transcription, and Plant Inoculation
Previous studies showed that the PLMVd variants PC-C40 (Malfitano et al., 2003), GDS6 (Ambrós et al., 1999), and PC-P1.159 (Rodio et al., 2006) induce in GF-305 peach seedlings severe chlorosis (albinism-variegation), mosaics, and no symptoms, respectively. Variant PC-C40Δ, generated by site-directed deletion of the 12-nucleotide fragment containing the PC determinant in variant PC-C40, infects peach seedlings symptomlessly (Malfitano et al., 2003). Recombinant plasmids containing head-to-tail PLMVd-cDNA dimeric inserts corresponding to these variants were linearized with appropriate restriction enzymes and then transcribed in vitro with T7 or SP6 RNA polymerase. Transcription products were separated by PAGE on 5% gels containing 8 M urea and 40% formamide, and those of monomeric length were eluted, recovered by ethanol precipitation, and slash-inoculated into GF-305 peach seedlings that were subsequently treated as indicated above.
Electron and Light Microscopy
For thin sectioning, shoot apical tips or excised leaf pieces were processed according to standard procedures (Martelli and Russo, 1984): fixation at 4°C for 2 h in 4% glutaraldehyde in 0.05 M potassium phosphate, pH 7.2, postfixation for 2 h at 4°C in 1% osmium tetroxide in the same buffer, staining overnight in 0.5% aqueous uranyl acetate at 4°C, dehydratation in ethanol, and embedding in Spurr's resin. Thin sections were stained with lead citrate before observation with a Philip Morgagni 282D electron microscope. Transverse sections of leaf tissues embedded in Spurr's resin as indicated above were attached to glass slides using poly-l-Lys, stained with 1% (w/v) toluidine blue, and examined with an Axioplan 2 Axiophot 2 (Zeiss) light microscope.
Hybridization Probes
The full-length PLMVd cDNA of variant PC-P1.159 (Rodio et al., 2006) was cloned in both orientations in the vector pGEM-T-Easy (Promega), generating the recombinant plasmids pPL159.4m and pPL159.5m. These plasmids, linearized with SpeI, served to synthesize PLMVd riboprobes of (−) and (+) polarity, respectively, using the digoxigenin (DIG) RNA labeling kit (Roche Diagnostics) and T7 RNA polymerase. The DIG-labeled transcripts were quantified following the manufacturer's instructions, and similar amounts of each polarity probe were used for the RNA gel blot and the in situ hybridizations to estimate the relative proportions of both PLMVd strands. DIG-labeled cDNA probes for detecting plastid rRNAs were prepared using the PCR DIG probe synthesis kit (Roche Diagnostics) and the genomic DNA from GF-305 peach seedlings obtained with the DNeasy plant mini kit (Qiagen). In the absence of any previous sequence information on the peach plastid rRNAs, amplifications were performed with the following primers designed to specifically amplify cDNA fragments of plastid rRNAs from Arabidopsis thaliana (Bellaoui et al., 2003): 5′-ACCCTCGTGTTTAGTTGC-3′ and 5′-CGCACCTTCCAGTACGGC-3′ for 16S rRNA; 5′-TGAACTGCTGCTGAATCC-3′ and 5′-CGCTTTCGCTACGGCTCC-3′ for 23S rRNA; 5′-GCACAGCCGAGACAGCAAC-3′ and 5′-GAACAATGGTTTTTTCGTG-3′ for 4.5S rRNA; and 5′-CACAATCACTAGCCAATATGC-3′ and 5′-GGGATGATTTCATAATTGGGG-3′ for 5S rRNA. Restriction analysis or sequencing of the PCR-amplified peach cDNAs confirmed that they had the expected length and sequence.
RNA Extraction and RNA Gel Blot Hybridization
Nucleic acids were extracted from 50 mg of leaf pieces with phenol-chloroform and recovered by ethanol precipitation (Dalmay et al., 1993). Following digestion with RQ1 DNase (Promega) and phenol extraction, RNAs were recovered by ethanol precipitation, fractionated by electrophoresis on 1.2% agarose gels containing formaldehyde or by PAGE on 5% gels containing 8 M urea, stained with ethidium bromide, and transferred to Hybond-N+ (Roche Diagnostics) membranes. To confirm equal loading, the fluorescence emitted after UV irradiation by nuclear rRNAs stained with ethidium bromide was quantified using Bio-Rad Quantity One software. Prehybridization and hybridization with RNA or DNA probes were performed in DIG-Easy Hyb Granules solution (Roche Diagnostics) according to the manufacturer's instructions with the only difference that, when indicated, the incubation temperature with riboprobes was increased to 70°C. Membranes were washed and incubated with an anti-DIG antibody conjugated to alkaline phosphatase before adding the chemiluminescent substrate CDP-Star (Roche Diagnostics). Signals were detected by a ChemiDoc analyzer and quantified with Quantity One software (Bio-Rad).
In Situ Hybridization
Small tissue pieces from leaves and shoots apices were fixed at 4°C overnight with 4% p-formaldehyde in 0.1 M sodium phosphate, pH 7.2, containing 0.1% Tween 20, dehydrated, embedded in paraffin, and sectioned to 7 μm. In situ hybridization was performed according to Jackson (1992) with some modifications. DIG-labeled riboprobes were added to the hybridization solution containing 50% deionized formamide, 1% Denhardt's solution (1× Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), 1 mg/mL tRNA, 10% dextran sulfate, 300 mM NaCl, 10 mM Tris-HCl, pH 6.8, 10 mM saline phosphate buffer, and 5 mM EDTA. To increase the stringency, the hybridization and the three subsequent washes (for 30, 90, and 60 min in a buffer consisting of 2× SSC [1× SSC is 0.15 M NaCl and 0.015 M sodium citrate] and 50% formamide) were performed at 70°C. The washes were followed by treatment for 1 h with blocking solution (Blocking reagent; Roche Diagnostics). The sections were then incubated for 2 h at room temperature with alkaline phosphatase–conjugated anti-DIG antibody (1:2000 dilution) in TBS buffer (100 mM Tris-HCl, pH 7.5, and 400 mM NaCl) containing 0.3% Triton X-100 and 1% BSA. Following three final washes for 20 min in the same buffer, alkaline phosphatase was detected by the nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate procedure according to the supplier (Roche Diagnostics). A bright-field microscope (E600; Nikon) was used for sample visualization and photography.
Transcript Analysis
Transcript levels were estimated by RT-PCR. RNA preparations equalized as indicated previously were used for reverse transcription with random octamers and avian myeloblastosis virus reverse transcriptase following the protocol provided by the supplier (Promega). Aliquots (2 μl) of serial dilutions (1:2, 1:20, and 1:200) of the resulting cDNAs were added to each amplification reaction, which was performed in a final volume of 25 μL with the Go-Taq DNA polymerase, also following the protocol provided by the supplier (Promega). The cycling program (initial denaturation for 2 min at 95°C; followed by 25 cycles of 15 s at 94°C, 30 s at 50°C, and 30 s at 72°C; with a final extension of 5 min at 72°C) was adopted because preliminary experiments showed that amplification of the cDNAs being analyzed was in the logarithmic phase. To amplify the cDNAs of the 18S nucleus-encoded rRNA and the transcripts of the plastid psbA and rbcL genes, pairs of specific primers were designed from the corresponding peach gene sequences: 18S-FW (5′-CATTCAAATATCTGCCCTATCAACTTTCG-3′) and 18S-RV (5′-TGCCTTCCTTGGATGTGGTAGC-3′) leading to a cDNA of 132 bp of the 18S rRNA; psbA-FW (5′-ACTGGATAACCAGCACTGAAAACCGTC-3′) and psbA-RV (5′-ACAGCAGTAGCAGCTGCAACAGGAGCTG-3′) leading to a cDNA of 415 bp of the plastid psbA transcript; and rbcL-FW (5′-ACTGACGGGCTTACTTACTAGTCTTGAC-3′) and rbcL-RV (5′-ATAAACTGCTCTACCGTAATTCTTAGCG-3′) leading to a cDNA of 360 bp of the plastid rbcL transcript. In the absence of sequence data for the peach gene rpoB, primers rpoB-FW (5′-ACCCAAGCGAGCATTCAAATATCTG-3′) and rpoB-RV (5′-TAGGTGGTAGAGGTCGAGTTATTGATG-3′) were derived from the gene rpoB of Arabidopsis; the PCR-amplified peach product obtained with this primer pair had the expected size (282 bp). Sequencing of all of the PCR-amplified products confirmed that they corresponded to the expected cDNAs.
Protein Analysis
Pieces (100 mg) of GF-305 peach leaves were homogenized in 10 volumes of extraction buffer (Laemmli, 1970). After boiling the samples for 3 min, the solubilized proteins were resolved by 12% SDS-PAGE and stained with Coomassie Brilliant Blue or electrotransferred onto polyvinylidene difluoride membranes (Hybond-P; Amersham Biosciences) for protein gel blot analyses. The membranes were probed with anti-rabbit polyclonal antisera raised against the plastid-encoded psbA/D1 protein (Agrisera) and the FNR protein coded by the petH nuclear gene (generously supplied by R. Herrmann). After incubating with a peroxidase-linked goat anti-rabbit IgG (Pierce), the complexes were detected by an enhanced chemiluminescence assay (ECL; Amersham Biosciences).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AJ550912 (PLMVd variant PC-C40), DQ222062 (PLMVd variant PC-P1.159), AJ005303 (PLMVd variant GDS6), L28749 (peach 18S nucleus-encoded rRNA), AF410189 (peach plastid psbA gene), AF411493 (peach plastid rbcL gene), and AP000423 (plastid rpoB gene of Arabidopsis).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Primary and Secondary Structures of the PC-Inducing and Latent PLMVd Variants Used in This Study.
Supplemental Figure 2. Close View of a Plastid, Resembling a Proplastid, in the Albino Sector of a Leaf Infected by PLMVd Variant PC-C40.
Supplemental Figure 3. Transmission Electron Microscopy Images of Cells and Plastids of Mature GF-305 Peach Leaves.
Supplemental Figure 4. Detection of PLMVd sRNAs in GF-305 Peach Leaves Infected by Three Viroid Variants.
Supplemental Figure 5. Detection of PLMVd (+) and (−) Strands in the SAM of GF-305 Seedlings Infected by Different Viroid Variants.
Supplementary Material
Acknowledgments
We are grateful to R. Bassi, R. Herrmann, and T. Cardi for suggestions (and in the case of R. Herrmann for supplying the anti-FNR antibody) and to G.P. Martelli for critical reading of the manuscript. We also thank A. Ahuir for technical assistance. Work in R.F.'s laboratory was partially supported by the Ministerio de Educación y Ciencia (MEC; Grant BFU2005-06808/BMC) and the Generalidad Valenciana (GV; Grants ACOMP06/141 and ACOMP07/268) of Spain. F.D.S.'s and R.F.'s laboratories were jointly supported by the Consiglio Nazionale delle Ricerche–Consejo Superior de Investigaciones Científicas project 2004IT0028. During this work, M.-E.R. received a predoctoral fellowship from the Università degli Studi di Bari and the Antonio Ciccarone Prize from the Mediterranean Phytopathological Union and S.D. received postdoctoral fellowships from MEC and GV and an I3P contract from the Consejo Superior de Investigaciones Científicas.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Francesco Di Serio (f.diserio@ba.ivv.cnr.it).
Online version contains Web-only data.
References
- Allison, L.A., Simon, L.D., and Maliga, P. (1996). Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 15 2802–2809. [PMC free article] [PubMed] [Google Scholar]
- Ambrós, S., Hernández, C., Desvignes, J.C., and Flores, R. (1998). Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: Implications of the existence of constraints limiting the heterogeneity of viroid quasi-species. J. Virol. 72 7397–7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrós, S., Hernández, C., and Flores, R. (1999). Rapid generation of genetic heterogeneity in progenies from individual cDNA clones of peach latent mosaic viroid in its natural host. J. Gen. Virol. 80 2239–2252. [DOI] [PubMed] [Google Scholar]
- Barba, M., Cupidi, A., Loreti, S., Faggioli, F., and Martino, L. (1995). In vitro micrografting: A technique to eliminate peach latent mosaic viroid from peach. Acta Hortic. 386 531–535. [Google Scholar]
- Barkan, A. (1993). Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D. (2004). RNA silencing in plants. Nature 431 356–363. [DOI] [PubMed] [Google Scholar]
- Bellaoui, M., and Gruissem, W. (2004). Altered expression of the Arabidopsis ortholog of DCL affects normal plant development. Planta 219 819–826. [DOI] [PubMed] [Google Scholar]
- Bellaoui, M., Keddie, J.S., and Gruissem, W. (2003). DCL is a plant-specific protein required for plastid ribosomal RNA processing and embryo development. Plant Mol. Biol. 53 531–543. [DOI] [PubMed] [Google Scholar]
- Bisanz, C., Begot, L., Carol, P., Perez, P., Bligny, M., Pesey, H., Gallois, J.L., Lerbs-Mache, S., and Mache, R. (2003). The Arabidopsis nuclear DAL gene encodes a chloroplast protein which is required for the maturation of the plastid ribosomal RNAs and is essential for chloroplast differentiation. Plant Mol. Biol. 51 651–663. [DOI] [PubMed] [Google Scholar]
- Bligny, M., Courtois, F., Thaminy, S., Chang, C.C., Lagrange, T., Baruah-Wolff, J., Stern, D., and Lerbs-Mache, S. (2000). Regulation of plastid rDNA transcription by interaction of CDF2 with two different RNA polymerases. EMBO J. 19 1851–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach, T.J., Lange, H., Gutierrez, R., Erhardt, M., Stern, D.B., and Gagliardi, D. (2005). RNR1, a 30-50 exoribonuclease belonging to the RNR superfamily, catalyzes maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res. 33 2751–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfiglioli, R.G., McFadden, G.I., and Symons, R.H. (1994). In situ hybridization localizes avocado sunblotch viroid on chloroplast thylakoid membranes and coconut cadang cadang viroid in the nucleus. Plant J. 6 99–103. [Google Scholar]
- Bonfiglioli, R.G., Webb, D.R., and Symons, R.H. (1996). Tissue and intra-cellular distribution of coconut cadang cadang viroid and citrus exocortis viroid determined by in situ hybridization and confocal laser scanning and transmission electron microscopy. Plant J. 9 457–465. [Google Scholar]
- Branch, A.D., Benenfeld, B.J., and Robertson, H.D. (1988). Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc. Natl. Acad. Sci. USA 85 9128–9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch, A.D., and Robertson, H.D. (1984). A replication cycle for viroids and other small infectious RNAs. Science 223 450–454. [DOI] [PubMed] [Google Scholar]
- Brodersen, P., and Voinnet, O. (2006). The diversity of RNA silencing pathways in plants. Trends Genet. 22 268–280. [DOI] [PubMed] [Google Scholar]
- Bussière, F., Lehoux, J., Thompson, D.A., Skrzeczkowski, L.J., and Perreault, J.P. (1999). Subcellular localization and rolling circle replication of peach latent mosaic viroid: Hallmarks of group A viroids. J. Virol. 73 6353–6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussière, F., Ouellet, J., Côté, F., Lévesque, D., and Perreault, J.P. (2000). Mapping in solution shows the peach latent mosaic viroid to possess a new pseudoknot in a complex, branched secondary structure. J. Virol. 74 2647–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, M., Sparvoli, S., Edmunds, C., Garosi, P., Findlay, K., and Martin, C. (1996). DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J. 15 4194–4207. [PMC free article] [PubMed] [Google Scholar]
- Dalmay, T., Rubino, L., Burgyán, J., Kollár, A., and Russo, M. (1993). Functional analysis of cymbidium ringspot virus genome. Virology 194 697–704. [DOI] [PubMed] [Google Scholar]
- Daròs, J.A., and Flores, R. (2002). A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 21 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daròs, J.A., Marcos, J.F., Hernández, C., and Flores, R. (1994). Replication of avocado sunblotch viroid: Evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl. Acad. Sci. USA 91 12813–12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Peña, M., and Flores, R. (2002). Chrysanthemum chlorotic mottle viroid RNA: Dissection of the pathogenicity determinant and comparative fitness of symptomatic and non-symptomatic variants. J. Mol. Biol. 321 411–421. [DOI] [PubMed] [Google Scholar]
- De la Peña, M., Navarro, B., and Flores, R. (1999). Mapping the molecular determinant of pathogenicity in a hammerhead viroid: A tetraloop within the in vivo branched RNA conformation. Proc. Natl. Acad. Sci. USA 96 9960–9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado, S., Martínez de Alba, A.E., Hernández, C., and Flores, R. (2005). A short double-stranded RNA motif of peach latent mosaic viroid contains the initiation and the self-cleavage sites of both polarity strands. J. Virol. 79 12934–12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvignes, J.C. (1986). Peach latent mosaic and its relation to peach mosaic and peach yellow mosaic virus diseases. Acta Hortic. 193 51–57. [Google Scholar]
- Diener, T.O. (1972). Potato spindle tuber viroid. VIII. Correlation of infectivity with a UV-absorbing component and thermal denaturation properties of the RNA. Virology 50 606–609. [DOI] [PubMed] [Google Scholar]
- Diener, T.O. (2003). Discovering viroids—A personal perspective. Nat. Rev. Microbiol. 1 75–80. [DOI] [PubMed] [Google Scholar]
- Diener, T.O., Hammond, R.W., Black, T., and Katze, M.G. (1993). Mechanisms of viroid pathogenesis: Differential activation of the interferon-induced, double-stranded RNA-activated, Mr 68 000 protein kinase by viroid strains of varying pathogenicity. Biochimie 75 533–538. [DOI] [PubMed] [Google Scholar]
- Ding, B., and Itaya, A. (2007). Viroid: A useful model for studying the basic principles of infection and RNA biology. Mol. Plant Microbe Interact. 20 7–20. [DOI] [PubMed] [Google Scholar]
- Ding, B., Itaya, A., and Zhong, X. (2005). Viroid trafficking: A small RNA makes a big move. Curr. Opin. Plant Biol. 8 606–612. [DOI] [PubMed] [Google Scholar]
- Ding, B., Kwon, M.-O., Hammond, R., and Owens, R.A. (1997). Cell-to-cell movement of potato spindle tuber viroid. Plant J. 12 931–936. [DOI] [PubMed] [Google Scholar]
- Fadda, Z., Daròs, J.A., Fagoaga, C., Flores, R., and Duran-Vila, N. (2003). Eggplant latent viroid, the candidate type species for a new genus within the family Avsunviroidae (hammerhead viroids). J. Virol. 77 6528–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores, R., Daròs, J.A., and Hernández, C. (2000). The Avsunviroidae family: Viroids with hammerhead ribozymes. Adv. Virus Res. 55 271–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores, R., Delgado, S., Rodio, M.E., Ambrós, S., Hernández, C., and Di Serio, F. (2006). Peach latent mosaic viroid: Not so latent. Mol. Plant Pathol. 7 209–221. [DOI] [PubMed] [Google Scholar]
- Flores, R., Hernández, C., Martínez de Alba, E., Daròs, J.A., and Di Serio, F. (2005. a). Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43 117–139. [DOI] [PubMed] [Google Scholar]
- Flores, R., Randles, J.W., Owens, R.A., Bar-Joseph, M., and Diener, T.O. (2005. b). Viroids. In Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses, C.M. Fauquet, M.A. Mayo, J. Maniloff, U. Desselberger, and A.L. Ball, eds (London: Elsevier/Academic Press), pp. 1145–1159.
- Foster, T.M., Lough, T.J., Emerson, S.J., Lee, R.H., Bowman, J.L., Forster, R.L., and Lucas, W.J. (2002). A surveillance system regulates selective entry of RNA into the shoot apex. Plant Cell 14 1497–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, G., and Pallás, V. (2001). Identification of an in vitro ribonucleoprotein complex between a viroid RNA and a phloem protein from cucumber plants. Mol. Plant Microbe Interact. 14 910–913. [DOI] [PubMed] [Google Scholar]
- Gómez, G., and Pallás, V. (2004). A long-distance translocatable phloem protein from cucumber forms a ribonucleoprotein complex in vivo with hop stunt viroid RNA. J. Virol. 78 10104–10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, H.J., Domdey, H., Lossow, C., Jank, P., Raba, M., Alberty, H., and Sänger, H.L. (1978). Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 273 203–208. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P.T., Allison, L.A., and Maliga, P. (1997). The two RNA polymerases encoded by nuclear and plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16 4041–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harders, J., Lukacs, N., Robert-Nicoud, M., Jovin, J.M., and Riesner, D. (1989). Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J. 8 3941–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández, C., and Flores, R. (1992). Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro through hammerhead structures. Proc. Natl. Acad. Sci. USA 89 3711–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, W.R., Prombona, A., Fieder, B., Subramanian, A.R., and Borner, T. (1993). Chloroplast rps 15 and the rpo B/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: Evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J. 12 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins, C., Rathjen, P.D., Forster, A.C., and Symons, R.H. (1986). Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 14 3627–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins, C.J., Keese, P., Visvader, J.E., Rathjen, P.D., McInnes, J.L., and Symons, R.H. (1985). Comparison of multimeric plus and minus forms of viroids and virusoids. Plant Mol. Biol. 4 293–304. [DOI] [PubMed] [Google Scholar]
- Itaya, A., Folimonov, A., Matsuda, Y., Nelson, R.S., and Ding, B. (2001). Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol. Plant Microbe Interact. 14 1332–1334. [DOI] [PubMed] [Google Scholar]
- Itaya, A., Matsuda, Y., Gonzales, R.A., Nelson, R.S., and Ding, B. (2002). Potato spindle tuber viroid strains of different pathogenicity induces and suppresses expression of common and unique genes in infected tomato. Mol. Plant Microbe Interact. 15 990–999. [DOI] [PubMed] [Google Scholar]
- Itaya, A., Zhong, X.H., Bundschuh, R., Qi, Y.J., Wang, Y., Takeda, R., Harris, A.R., Molina, C., Nelson, R.S., and Ding, B. (2007). A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J. Virol. 81 2980–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D.P. (1992). In situ hybridization in plants. In Molecular Plant Pathology: A Practical Approach, D.J. Bowles, S.J. Gurr, and M.J. McPherson, eds (Oxford, UK: Oxford University Press), pp. 163–174.
- Kidner, C.A., and Martienssen, R.A. (2005). The developmental role of microRNA in plants. Curr. Opin. Plant Biol. 8 38–44. [DOI] [PubMed] [Google Scholar]
- Kirk, J.T.O., and Tilney-Bassett, R.A.E. (1978). The Plastids: Their Chemistry, Structure, Growth and Inheritance, 2nd ed. (Amsterdam: Elsevier-North Holland).
- Krause, K., Maier, R.M., Kofer, W., Krupinska, K., and Herrmann, R.G. (2000). Disruption of plastid-encoded RNA polymerase genes in tobacco: Expression of only a distinct set of genes is not based on selective transcription of the plastid chromosome. Mol. Gen. Genet. 263 1022–1030. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Legen, J., Kemp, S., Krause, K., Profanter, B., Herrmann, R.G., and Maier, R.M. (2002). Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 31 171–188. [DOI] [PubMed] [Google Scholar]
- Lima, M.I., Fonseca, M.E.N., Flores, R., and Kitajima, E.W. (1994). Detection of avocado sunblotch viroid in chloroplasts of avocado leaves by in situ hybridization. Arch. Virol. 138 385–390. [DOI] [PubMed] [Google Scholar]
- Malfitano, M. (2000). Distribuzione di Varianti di Sequenza del Viroide del Mosaico Latente Del Pesco in Isolati di Campo Italiani. PhD dissertation (Naples, Italy: Università degli Studi di Napoli Federico II).
- Malfitano, M., Di Serio, F., Covelli, L., Ragozzino, A., Hernández, C., and Flores, R. (2003). Peach latent mosaic viroid variants inducing peach calico contain a characteristic insertion that is responsible for this symptomatology. Virology 313 492–501. [DOI] [PubMed] [Google Scholar]
- Markarian, N., Li, H.W., Ding, S.W., and Semancik, J.S. (2004). RNA silencing as related to viroid induced symptom expression. Arch. Virol. 149 397–406. [DOI] [PubMed] [Google Scholar]
- Martelli, G.P., and Russo, M. (1984). Use of thin sectioning for visualization and identification of plant viruses. Methods Virol. 8 143–224. [Google Scholar]
- Martín, R., Arenas, C., Daròs, J.A., Covarrubias, A., Reyes, J.L., and Chua, N.H. (2007). Characterization of small RNAs derived from citrus exocortis viroid (CEVd) in infected tomato plants. Virology 367 135–146. [DOI] [PubMed] [Google Scholar]
- Martínez de Alba, A.E., Flores, R., and Hernández, C. (2002). Two chloroplastic viroids induce the accumulation of the small RNAs associated with post-transcriptional gene silencing. J. Virol. 76 13094–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez de Alba, A.E., Sägesser, R., Tabler, M., and Tsagris, M. (2003). A bromodomain-containing protein from tomato specifically binds potato spindle tuber viroid RNA in vitro and in vivo. J. Virol. 77 9685–9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima, A., Hanaoka, M., Motohashi, R., Seki, M., Shinozaki, K., Kanamaru, K., Takahashi, H., and Tanaka, K. (2004). DNA microarray analysis of plastid gene expression in an Arabidopsis mutant deficient in a plastid transcription factor sigma, SIG2. Biosci. Biotechnol. Biochem. 68 694–704. [DOI] [PubMed] [Google Scholar]
- Navarro, B., and Flores, R. (1997). Chrysanthemum chlorotic mottle viroid: Unusual structural properties of a subgroup of self-cleaving viroids with hammerhead ribozymes. Proc. Natl. Acad. Sci. USA 94 11262–11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, J.A., Daròs, J.A., and Flores, R. (1999). Complexes containing both polarity strands of avocado sunblotch viroid: Identification in chloroplasts and characterization. Virology 253 77–85. [DOI] [PubMed] [Google Scholar]
- Navarro, J.A., Vera, A., and Flores, R. (2000). A chloroplastic RNA polymerase resistant to tagetitoxin is involved in replication of avocado sunblotch viroid. Virology 268 218–225. [DOI] [PubMed] [Google Scholar]
- Owens, R.A., Blackburn, M., and Ding, B. (2001). Possible involvement of the phloem lectin in long-distance viroid movement. Mol. Plant Microbe Interact. 14 905–909. [DOI] [PubMed] [Google Scholar]
- Owens, R.A., and Diener, T.O. (1982). RNA intermediates in potato spindle tuber viroid replication. Proc. Natl. Acad. Sci. USA 79 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens, R.A., Steger, G., Hu, Y., Fels, A., Hammond, R.W., and Riesner, D. (1996). RNA structural features responsible for potato spindle tuber viroid pathogenicity. Virology 222 144–158. [DOI] [PubMed] [Google Scholar]
- Palukaitis, P. (1987). Potato spindle tuber viroid: Investigation of the long-distance, intra-plant transport route. Virology 158 239–241. [DOI] [PubMed] [Google Scholar]
- Papaefthimiou, I., Hamilton, A.J., Denti, M.A., Baulcombe, D.C., Tsagris, M., and Tabler, M. (2001). Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res. 29 2395–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelchat, M., Côté, F., and Perreault, J.P. (2001). Study of the polymerization step of the rolling circle replication of peach latent mosaic viroid. Arch. Virol. 146 1753–1763. [DOI] [PubMed] [Google Scholar]
- Pelissier, T., and Wassenegger, M. (2000). A DNA target of 30 bp is sufficient for RNA-directed DNA methylation. RNA 6 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prody, G.A., Bakos, J.T., Buzayan, J.M., Schneider, I.R., and Bruening, G. (1986). Autolytic processing of dimeric plant virus satellite RNA. Science 231 1577–1580. [DOI] [PubMed] [Google Scholar]
- Qi, Y., and Ding, B. (2003. a). Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 15 1360–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Y., and Ding, B. (2003. b). Differential subnuclear localization of RNA strands of opposite polarity derived from an autonomously replicating viroid. Plant Cell 15 2566–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, F., Ye, X., Hou, G., Sato, S., Clemente, T.E., and Morris, T.J. (2005). RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 79 15209–15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reanwarakorn, K., and Semancik, J.S. (1998). Regulation of pathogenicity in hop stunt viroid-related group II citrus viroids. J. Gen. Virol. 79 3163–3171. [DOI] [PubMed] [Google Scholar]
- Rodio, M.E., Delgado, S., Flores, R., and Di Serio, F. (2006). Variants of peach latent mosaic viroid inducing peach calico: Uneven distribution in infected plants and requirements of the insertion containing the pathogenicity determinant. J. Gen. Virol. 87 231–240. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Cerezo, E., Klein, P.G., and Shaw, J.G. (1991). A determinant of disease symptom severity is located in the 3′-terminal noncoding region of the RNA of a plant virus. Proc. Natl. Acad. Sci. USA 88 9863–9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnölzer, M., Haas, B., Ramm, K., Hofmann, H., and Sänger, H.L. (1985). Correlation between structure and pathogenicity of potato spindle tuber viroid (PSTV). EMBO J. 4 2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach, F., Vaistij, F.E., Jones, L., and Baulcombe, D.C. (2005). An RNA-dependent RNA polymerase prevents meristem invasion by Potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semancik, J.S., and Szychowski, J.A. (1994). Avocado sunblotch disease: A persistent viroid infection in which variants are associated with differential symptoms. J. Gen. Virol. 75 1543–1549. [DOI] [PubMed] [Google Scholar]
- Symons, R.H. (1981). Avocado sunblotch viroid: Primary sequence and proposed secondary structure. Nucleic Acids Res. 9 6527–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabler, M., and Tsagris, M. (2004). Viroids: Petite RNA pathogens with distinguished talents. Trends Plant Sci. 9 339–348. [DOI] [PubMed] [Google Scholar]
- Taliansky, M.E., and Robinson, D.J. (1997). Trans-acting untranslated elements of groundnut rosette virus satellite RNA are involved in symptom production. J. Gen. Virol. 78 1277–1285. [DOI] [PubMed] [Google Scholar]
- Visvader, J.E., and Symons, R.H. (1986). Replication of in vitro constructed viroid mutants: Location of the pathogenicity-modulating domain in citrus exocortis viroid. EMBO J. 13 2051–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vothknecht, U.C., and Westhoff, P. (2001). Biogenesis and origin of thylakoid membranes. Biochim. Biophys. Acta 1541 91–101. [DOI] [PubMed] [Google Scholar]
- Wang, M.B., et al. (2004). On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. USA 101 3275–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.C., Duby, G., Purnelle, B., and Boutry, M. (2000). Tobacco VDL gene encodes a plastid DEAD box RNA helicase and is involved in chloroplast differentiation and plant morphogenesis. Plant Cell 12 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger, M., Heimes, S., Riedel, L., and Sänger, H.L. (1994). RNA-directed de novo methylation of genomic sequences in plants. Cell 76 567–576. [DOI] [PubMed] [Google Scholar]
- Woo, Y.M., Itaya, A., Owens, R.A., Tang, L., Hammond, R.W., Chou, H.C., Lai, M.M.C., and Ding, B. (1999). Characterization of nuclear import of potato spindle tuber viroid RNA in permeabilized protoplasts. Plant J. 17 627–635. [Google Scholar]
- Zhao, Y., Owens, R.A., and Hammond, R.W. (2001). Use of a vector based on potato virus X in a whole plant assay to demonstrate nuclear targeting of potato spindle tuber viroid. J. Gen. Virol. 82 1491–1497. [DOI] [PubMed] [Google Scholar]
- Zhong, X., Tao, X., Stombaugh, J., Leontis, N., and Ding, B. (2007). Tertiary structure and function of an RNA motif required for plant vascular entry to initiate systemic trafficking. EMBO J. 26 3836–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Green, L., Woo, Y.M., Owens, R.A., and Ding, B. (2001). Cellular basis of potato spindle tuber viroid systemic movement. Virology 279 69–77. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Qi, Y., Xun, Y., Owens, R., and Ding, B. (2002). Movement of potato spindle tuber viroid reveals regulatory points of phloem-mediated RNA traffic. Plant Physiol. 130 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko, M.K., and Day, A. (2002). Differential regulation of genes transcribed by nucleus-encoded plastid RNA polymerase, and DNA amplification, within ribosome-deficient plastids in stable phenocopies of cereal albino mutants. Mol. Genet. Genomics 267 27–37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.