Abstract
The Arabidopsis thaliana MALE STERILITY1 (MS1) gene is critical for viable pollen formation and has homology to the PHD-finger class of transcription factors; however, its role in pollen development has not been fully defined. We show that MS1 transcription appears to be autoregulated by the wild-type MS1 transcript or protein. Using a functional green fluorescent protein (GFP) fusion to analyze the temporal and spatial expression of MS1, we demonstrate that the MS1:GFP protein is nuclear localized within the tapetum and is expressed in a developmentally regulated manner between late tetraspore and microspore release, then rapidly breaks down, probably by ubiquitin-dependent proteolysis. Absence of MS1 expression results in changes in tapetal secretion and exine structure. Microarray analysis has shown that 260 (228 downregulated and 32 upreglated) genes have altered expression in young ms1 buds. These genes are primarily associated with pollen wall and coat formation; however, a number of transcription factors and Cys proteases have also been identified as the putative primary regulatory targets of MS1. Ectopic expression of MS1 alters transcriptional regulation of vegetative gene expression, resulting in stunted plants with increased levels of branching, partially fertile flowers and an apparent increase in wall material on mature pollen. MS1 therefore plays a critical role in the induction of pollen wall and pollen coat materials in the tapetum and, ultimately, the production of viable pollen.
INTRODUCTION
Anther and pollen development is a critical phase in the plant life cycle that is vital for sexual reproduction and selective breeding, which involves a diverse range of gene interactions (Goldberg et al., 1993; McCormick, 2004; Scott et al., 2004; Ma, 2005). Many genes that impact male and female meiosis have now been characterized (Bhatt et al., 2001), but only a few genes that affect the later stages of development during tapetal maturation have been identified; these include MALE STERILITY2 (MS2) (Aarts et al., 1993, 1997), which has homology to a fatty acyl reductase involved in pollen wall formation, ABORTED MICROSPORES (AMS) (Sorensen et al., 2003), the rice (Oryza sativa) ortholog TAPETUM DEGENERATION RETARDATION (Li et al., 2006), which is involved in tapetal development and microspore development, TAPETAL DETERMINANT1 (TPD1) (Yang et al., 2003a), DYSFUNCTIONAL TAPETUM (DYT1) (Zhang et al., 2006), and the rice ortholog UNDEVELOPED TAPETUM1 (Jung et al., 2005), which are involved in tapetal/microsporocyte determination, and MS1 (Wilson et al., 2001).
One of the earliest genes required for cell division and differentiation in the anther is the SPOROCYTELESS (SPL)/NOZZLE (NZZ) gene (Schiefthaler et al., 1999; Yang et al., 1999). In the spl/nzz mutant, archesporial initiation occurs normally, but male and female sporocyte differentiation is halted and anther development fails to occur. Early tapetal initiation is also affected by the downstream genes EXTRA SPOROGENOUS CELLS (EXS)/EXCESS MICROSPOROCYTES1 (EMS1) (Canales et al., 2002; Zhao et al., 2002) and TPD1 (Yang et al., 2003a). Mutants in these genes have altered numbers of archesporial cells and an absence of tapetal and middle cell layers. Two other genes, Somatic Embyogenesis Receptor-Like Kinase1 (SERK1) and SERK2, also appear to have redundant functions during this early stage of tapetal initiation (Albrecht et al., 2005; Colcombet et al., 2005), and when mutated, result in more sporogenous cells and lack of a tapetal cell layer. Sporocyte formation occurs in these mutants, but a functional tapetum is required for completion of meiosis and the production of viable pollen. MYB33 and MYB65 also act redundantly to facilitate tapetal development around the stage of meiosis, and it has been shown that the expression of MYB33 is regulated by miRNAs; however, their effect on fertility is conditional based upon environmental conditions (Millar and Gubler, 2005). These genes, which are involved in tapetal initiation, are not affected in the dyt1 mutant, indicating that they are upstream of DYT1 (Zhang et al., 2006). In the dyt1 mutant, tapetum and meiosis initiation occurs, although tapetal development is abnormal with enlarged vacuoles and microspore degeneration. DYT1 has been proposed to be involved in the regulation of many tapetal genes, either directly or indirectly, including AMS and MS1 (Zhang et al., 2006). The ams mutant has a similar phenotype with premature microspore and tapetal degeneration and short stamen filaments, and the tapetum becomes abnormally enlarged and vacuolated (Sorensen et al., 2003).
The tapetum plays a major secretory role in sporogenesis and is critical in pollen wall and pollen coat formation. Although species-specific variation occurs, the pollen wall forms as two distinct layers: the exine, which is the outer sculptured part of the wall, containing sporopollenin, an aliphatic polymer; and the simple internal intine, which is composed of cellulose, pectin, and protein (Scott et al., 2004). In Arabidopsis thaliana, callose deposition has been shown to be critical for the early stages of exine formation, and in the absence of callose synthase (CalS5), the baculae and tectum form as globular structures rather than normal sculptured exine wall formation (Dong et al., 2005). The sporopollenin and the pollen coat are synthesized in the tapetum under the control of the sporophytic genome (Piffanelli et al., 1998). Correct deposition of sporopollenin is critical for viable pollen production, for example, in the Arabidopsis dex-1 mutant, primexine and sporopollenin deposition are disrupted and the pollen collapses and is inviable (Paxson-Sowders et al., 2001). Similar pollen degradation is seen in the Arabidopsis ms2 mutant, which carries a defect in tapetal fatty acyl transferase (Aarts et al., 1997). The tapetum has a highly regulated transient lifecycle; as pollen grain maturation occurs, the tapetal cells become increasingly vacuolated and accumulate elaioplasts and large cytoplasmic lipid bodies. Soon after the first pollen mitotic division, the tapetal cells undergo programmed cell death (PCD) and release their contents into the anther locule. This tapetal debris goes to form the pollen coat (tryphine and pollenkitt), which becomes embedded on or beneath the exine surface.
MS1 is a vital, sporophytic gene required for tapetal development and microspore maturation in higher plants. In ms1 mutants, the early stages of pollen mother cell (PMC) meiosis and microspore release occur normally, and the tapetum then becomes abnormally vacuolated and the microspores and tapetum degenerate (Wilson et al., 2001; Ito and Shinozaki, 2002; Ariizumi et al., 2005). The MS1 protein contains a PHD-finger motif that is found in a number of homeodomain proteins from a range of organisms from humans to yeast (Wilson et al., 2001; Ito and Shinozaki, 2002). The homologous proteins have diverse functions, although an Arabidopsis meiosis specific PHD-finger gene, MMD1 (Yang et al., 2003b)/DUET (Reddy et al., 2003), has been identified, but all appear to be involved in transcriptional regulation. It is speculated that the PHD-finger motif may be a site for combining a group of proteins into a large complex or for the recognition of specific targets associated with chromatin structure and regulation (Asaland et al., 1995), and recent work has shown that the PHD motif interacts with methylated histones to regulate chromatin conformation and gene expression (Shi et al., 2006; Wysocka et al., 2006).
We have shown using an MS1 translational green fluorescent protein (GFP) fusion that MS1 expression is spatially and temporally regulated to the nuclei of tapetal cells within the anther, from late tetraspore stage to the newly released microspores. Absence of MS1 expression results in changed secretion and development of the tapetum, resulting in abnormal pollen wall formation with aberrant deposition of the exine. Microarray analysis has shown that 260 (228 downregulated and 32 upregulated) genes have altered expression in young ms1 buds; of these, 94 also change in the spl and exs mutants (Wijeratne et al., 2007). These genes are primarily associated with pollen wall and coat formation; however, a number of transcription factors and Cys proteases have also been identified as the putative primary regulatory targets of MS1. Ectopic expression of MS1 alters transcriptional regulation of gene expression in vegetative tissues, resulting in stunted plants with increased levels of branching and partially fertile flowers and an apparent increase in wall material on mature pollen. MS1 therefore plays a critical role in the induction of pollen wall and pollen coat materials in the tapetum and, ultimately, the production of viable pollen.
RESULTS
MS1 Negatively Regulates Its Own Expression
Quantitative RT-PCR analysis of MS1 expression was conducted using pools of developmentally staged, closed buds consisting of (1) PMCs through to meiosis stage, (2) tetrads through to pollen mitosis I, and (3) mitosis II through to dehiscence (mitosis II). In the wild type, MS1 expression is maximal from PMCs through to meiosis. Low levels of expression (decreasing by a factor of 17 and 2.8 progressively) are also detected in young buds (mitosis I) and old buds (mitosis II) (Figure 1A), indicating that MS1 expression is from meiosis through to mitosis I stage. This agrees with previous promoter:β-glucuronidase reporter data (Wilson et al., 2001) but is slightly longer than was shown by in situ hybridization (Ito and Shinozaki, 2002), and this may be a reflection of the sensitivity of the detection methods. Expression of the ms1-1 mutant transcript was also analyzed; this mutant has a point mutation at the junction of the second intron and therefore fails to splice normally (Wilson et al., 2001). The mutation results in a truncated transcript that is predicted to produce a nonfunctional protein of 199 amino acids, which is missing the Leu zipper (LZ) and PHD domains of the full-length MS1 protein (672 amino acids) (Wilson et al., 2001).
Figure 1.
Expression Analysis of MS1.
(A) Quantitative RT-PCR of MS1 in wild-type Ler and ms1-1 buds.
(B) RT-PCR of MS1 gene in buds from Ler wild type and ms1-1. Lane 1, ms1-1 buds; lane 2, F1 heterozygote Ler/ms1-1; lane 3, wild-type Ler buds; lane 4, MS1-GFP–complemented line. The ms1-1 mutant line shows overexpression of the truncated MS1 gene compared with the wild type; however, there is no difference in the level of expression in the heterozygote, indicating that a single copy of the full-length wild-type MS1 transcript can downregulate its own and the mutant transcript.
Expression of the ms1-1 mutant transcript is at a significantly higher level and for a longer duration than the wild-type MS1 transcript (Figure 1). Extremely high levels of expression are seen in both PMC to meiosis (27-fold increase) and tetrad to mitosis I stages (600-fold increase) in ms1 compared with the wild type. During mitosis I, a reduction (17-fold) in expression is seen in the wild type; however, this does not occur in the ms1-1 mutant, further increasing the difference in expression at this stage. In ms1-1, expression is also maintained during mitosis II stage, albeit at a slightly lower level than in the young mutant buds; however, this is still 11-fold increased compared with the wild type. RT-PCR was also conducted on our other ms1 alleles; all of these mutants result from single-base mutations that are predicted to form partial transcripts and lose the PHD-finger motif (Wilson et al., 2001). Increased expression of the mutant transcripts is seen in all of the ms1 alleles compared with the wild-type buds (see Supplemental Figure 1 online); however, this increase is greatest in the ms1-1/ms1-6 alleles, which show a failure of splicing of the second intron. However, equal expression of the wild type and mutant transcript is seen in the F1 Landsberg erecta (Ler) × ms1-1 heterozygote (Figure 1B), indicating that wild-type MS1 expression is not affected by the ms1-1 mutant transcript or mutant protein and that a single copy of the full-length wild-type MS1 transcript can downregulate both its own expression and that of the mutant transcript.
The MS1:GFP Translational Fusion Complements the ms1 Mutation
MS1 protein expression was localized using translational GFP fusions (see Supplemental Figure 2 online) under the control of the native MS1 promoter, which were transformed into heterozygous ms1 ttg plants and the subsequent generations screened to identify homozygous ms1 complemented lines. Both the N- and the C-terminal MS1:GFP fusions rescued the male-sterile phenotype of the ms1 ttg lines to full fertility; however, the internal GFP construct in which the fluorescent tag was inserted upstream of the LZ motif was not functional. RT-PCR analysis of the complemented lines showed the presence of the ms1-1 mutant transcript and the MS1 and NPTII transgene (Figure 1B). Stronger GFP signal was seen in the lines carrying the C-terminal construct compared with the N-terminal construct, and these were used for the subsequent expression analysis. Hybridization of GFP polyclonal antibodies to total protein isolated from MS1:GFP C-terminal fusion lines showed a 107-kD band (77 kD for MS1; 30.6 kD for GFP protein) corresponding to the MS1:GFP fusion protein in buds as well as from excised anthers in the transgenic lines (see Supplemental Figure 3 online), which was absent from the wild-type buds.
MS1 Is Transiently Expressed in the Tapetum and Is Nuclear Localized
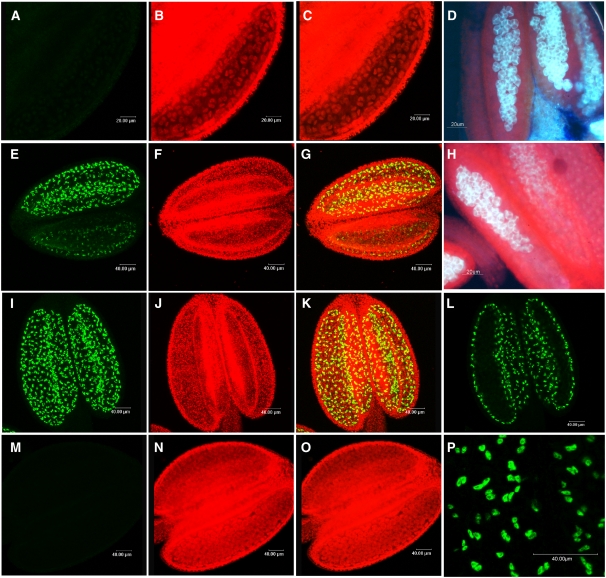
Expression of the MS1:GFP fusion protein is only detected in the tapetal tissues of the anther, and no signal is seen in the microspores or in the vegetative tissues. Expression starts in the tapetum as the callose wall separating the tetraspores begins to degrade (Figures 2E to 2H); no MS1 protein expression is detected before this stage (Figures 2A to 2D; early tetraspore stage). The timing of callose breakdown is developmentally regulated and is critical for pollen viability (Chasan, 1992; Dong et al., 2005) and occurs sequentially across the anther locules (Figure 2H); a similar progression of MS1:GFP expression is observed. Once the entire callose wall has broken down and the microspores are released, maximal MS1 expression is seen within the tapetum across the whole locule (Figures 2I, 2K, and 2L). Immediately after microspore release, pollen wall development and microspore mitosis occur, and the tapetum is clearly discernable at this stage (Figure 2N), but the MS1:GFP fusion protein is no longer detected (Figures 2M and 2O). The MS1:GFP fusion protein is therefore only detectable for a short developmental period of ∼12 h from late tetraspore until the free microspore stage. This is similar to the MS1 RNA expression pattern, with translation occurring soon after detection of the transcript; however, the MS1 transcript can be detected for an extended period beyond this, suggesting a process of developmentally regulated breakdown. However, if anthers are excised from the plant and maintained at 4°C in a humid environment, the developmental process is halted and the microspores fail to go through mitosis I. Under these conditions, the MS1:GFP fusion protein persists and does not go through normal breakdown. MS1:GFP signal is detected for at least 4 d, after which point the anthers undergo general degeneration and analysis was discontinued (data not shown).
Figure 2.
MS1 Expression in Transgenic Lines Carrying a Functional C-terminal MS1:GFP Fusion Protein.
(A), (E), (I), (L), (M), and (P) The 488-nm excitation filter showing GFP expression.
(B), (F), (J), and (N) The 560-nm excitation filter showing chlorophyll autofluorescence.
(C), (G), (K), and (O) Superimposed GFP and chlorophyll autofluorescence.
(D) and (H) Aniline blue staining, visualized using a 490-nm excitation filter.
No expression of the MS1:GFP fusion protein is seen during the early tetraspore stage ([A] to [D]). During this stage, breakdown of the callose has not started, as shown by aniline blue staining (D). During late tetraspore stage ([E] to [H]), when callose breakdown has started (H), expression of the MS1:GFP fusion protein is seen in different regions of the anther where development is most advanced. Maximal expression of the MS1:GFP fusion protein is seen during microspore release ([I] and [K]), and expression is confined to the tapetal tissue within the anthers (L) and is nuclear localized (P). No MS1:GFP expression is seen by microspore mitosis I ([M] to [O]).
The deduced MS1 protein sequence contains a predicted N-terminal nuclear localizing sequence PKKR (http://psort.hgc.jp), and previous transient expression studies using the truncated N-terminal region of the MS1 polypeptide suggested that the MS1 protein might be nuclear localized (Ito and Shinozaki, 2002). The functional MS1:GFP fusion protein was detected within the nuclei of the tapetal cells (Figures 2L and 2P); no cytoplasmic expression was seen, thus confirming that the predicted nuclear localizing sequence is effective for nuclear targeting of the MS1 protein within the tapetum.
MS1 Affects Pollen Wall Development and Anther Gene Expression
Transcript profiling of the ms1 ttg mutant was performed on closed buds that had been staged by microscopy and RT-PCR into two developmental groups: (1) young, from formation of the sporogeneous tissues to pollen mitosis I; (2) old, pollen mitosis II through to dehiscence. Statistical analysis of microarrays (SAM) (Tusher et al., 2001) was used to analyze the data, a false discovery cutoff of 5% was used for the initial selection of candidate genes, and a secondary filtering of >log2 fold difference was then applied.
Comparisons between the two developmental stages showed that in the wild type, more genes are upregulated in the young buds (up to mitosis I) compared with the later stages of mitosis II through to dehiscence; this agrees with previous reports in the literature of the pollen transcriptome (Honys and Twell, 2004). These changes in gene expression correlate with the stages of cell differentiation and biosynthesis that occur during the early stages of pollen maturation. In the ms1 mutant, a general reduction in gene expression was seen. The MS1 gene is functional at mid stages of development; therefore, changes in gene expression were expected during the later stages of development after MS1 expression; however, no significant ms1-associated expression changes were detected in this older material. However, 966 genes (SAM analysis, false discovery rate [FDR] <5%; see Supplemental Data Set 1 online) showed differential expression during the early stages of development in the ms1 mutant compared with the wild-type buds; however, when filtered for changes of >log2 fold, the number of genes reduced to 314, and of these, only 260 (228 downregulated; 32 upregulated; see Supplemental Data Set 1 online) were identified as expressed in the anther (J. Song and Z.A. Wilson, unpublished data). Of the 260 genes, 161 were found to be also expressed in the pollen (Honys and Twell, 2004), while 99 were expressed in the anther but not found within the pollen transcriptome set (see Supplemental Figure 4 online). Comparisons were also made to array data sets for the spl and ems mutants, and 1954 and 525 genes were seen to change in the spl and ems1 mutants, respectively (Wijeratne et al., 2007). The differences in numbers of genes changing reflect the relative position of the mutants in the network of pollen development, with SPL active very early, followed by EMS1, and then at a later stage of development MS1. The 94 genes that change in all three mutants reflect genes that are downstream of MS1 as either direct or indirect targets (see Supplemental Data Set 2 online). The other remaining 166 are likely to act in parts of the pollen development pathway that are independent of SPL and EMS1.
Among the genes that are very highly upregulated in the young ms1 tissue and are expressed in the anther tissues are At1g23760 (putative polyygalacuronase isoenzyme; +10.6), two unknown proteins (At2g34870 [+9.3] and At5g46230 [+5.2]), At2g42990 (putative APG isolog; +5.6), At5g15140 (putative aldose 1-epimerase-like; +5.2), and At1g01280 (cytochrome P450; +3.8). The downregulated genes showing the greatest changes in ms1 young buds are genes associated with pollen wall development, including At5g07550 (Gly-rich protein PUTG1; −94.2), At1g67990 (putative S-adenosyl-l-Met; −73.4), At1g68875 (expressed protein; −109.9), At3g51590 (lipid transfer protein-like; −51.9), At5g07560 (oleosin-like; −61.5), and At1g75910 (anther-specific Pro- rich-like protein; −43.1).
Expression data obtained from the array hybridization were verified by RT-PCR (see Supplemental Figure 5 online) using a group of key genes selected because they had high fold change and/or functional relevance that included anther-specific genes (At1g20120, At1g75910, and At1g75930), transcription factors (At2g16910 and At2g46130), pollen wall (At4g37900, At3g51590, At5g07560, At5g09520, and At5g07550), lignin biosynthesis (At1g67990), and high fold changes in genes related to the cytochrome P450 (At4g31970) and hypothetical proteins (At1g23570 and At1g68875) with no homology with any other Arabidopsis protein. All data confirmed the expression changes seen in the arrays.
The MS1 gene (At5g22260) is not represented on the ATH1 chip; however, the expression of other genes known to be associated with pollen development, including: CER6/CUT1 (At1g68530), MS2 (At3g11980), ATA20 (At3g15400), ATA7 (At4g28395), SUP (At3g23130), TES (At3g43210), POLLENLESS/MS5 (At4g20900), and NZZ (At4g27330) were also analyzed by RT-PCR to verify the microarray expression data (see Supplemental Figure 6 online). Early genes associated with anther development, meiosis, and tapetal initiation (SUP, MS5, TES, and NZZ), ATA7, or CER6, which is associated with wax biosynthesis (Hooker et al., 2002), do not show significant changes in expression in the ms1 mutant. However ATA20, which has homology to lipid transfer proteins (Rubinelli et al., 1998) and is expressed later during pollen wall development, showed significant downregulation. MS2 (Aarts et al., 1993, 1997), which has homology to a fatty acyl reductase involved in pollen wall formation, showed a slightly prolonged expression in older ms1 buds (see Supplemental Figure 6 online); however, SAM analysis did not show this as significant, and since this occurs after most of the ms1-related changes, it seems unlikely that there is a direct link between the regulation of MS2 and MS1.
Microarray data from the ems1 mutant, which has impaired tapetal development and is active at an earlier stage than MS1, and DYT1 indicated a number of genes in the pollen development pathway (Zhang et al., 2006). Many of these genes do not show altered expression in the ms1 mutant, although the genes encoding for a lipid transfer protein (At3g51590), a hexose transport protein (At1g07340), polyadenylated binding protein 5 (At1g71770), and β-1,3-glucanase (AT3g23770) show changed expression in the ms1 mutant and therefore can be placed downstream of EMS1 and DYT1 (Table 1).
Table 1.
Expression in the ms1 Mutant of Genes Previously Reported as Altered in Expression by EMS/SPL and Involved in Pollen Development
| Gene Name | Locus | Description | Fold Change in the ems1 Mutanta | Change of Expression in dyt1a | Log2 Fold Change in ms1 Young Buds |
|---|---|---|---|---|---|
| MS5 | At4g20900 | Male sterility MS5; pollenless3 | −3 | 0 | 1 |
| SDS | At1g14750 | SOLO DANCER, a putative cyclin | −3 | 0 | 1 |
| SPL | At4g27330 | SPL/NZZ | 1 | 0 | 1 |
| EMS1 | At5g07280 | EMS1/EXS | −7 | 0 | 1 |
| AtMYB32 | At4g34990 | MYB-like protein myb-related protein Y49 | −5 | 0 | 1 |
| At1G06170 | Basic helix-loop-helix family protein | −33 | 0 | 1 | |
| MYB65 | At3g11440 | MYB family transcription factor (MYB65) | −2 | 0 | 1 |
| AtMYB4 | At4g38620 | Putative transcription factor (MYB4) | 1 | 0 | 1 |
| MYB33 | At5g06100 | MYB family transcription factor (MYB33) | 1 | 0 | 1 |
| MS5 like | At5g44330 | MS5 family protein | −60 | Down | 1 |
| A9 | At5g07230 | Protease inhibitor/seed storage/lipid transfer protein, tapetum-specific protein A9 | −63 | Down | 1 |
| FLF | At5g10140 | MADS box protein FLOWERING LOCUS F (FLF) | −12 | Down | 1 |
| RCK/AtMER3 | At3g27730 | ROCK-N-ROLLERS/At MER3, an ATP-dependent DNA helicase | −2 | Down | 1 |
| MS2 | At3g11980 | MS2 | −56 | Down | 1 |
| At3g60580 | Zinc-finger protein-like ZPT3-3, Petunia hybrida | −11 | — | ||
| At1g69500 | Cytochrome P450 | −97 | — | 3 | |
| A6 | At4g14080 | Glycosyl hydrolase family 17 protein/anther-specific protein (A6) | −90 | 3 | |
| ATA1 | At3g42960 | Alcohol dehydrogenase (ATA1) | −70 | Down | 2 |
| ATMYB103 | At5g56110 | Regulate the tapetum and trichome development, anther-specific gene | −3 | Down | 2 |
| At3g13220 | ABC transporter | −55 | Down | 2 | |
| AMS | At2g16910 | AMS | −37 | Down | 2 |
| At3g28470 | MYB transcription factor | −33 | Down | 4 | |
| LPT12 | At3g51590 | Lipid transfer protein | −11 | Down | −52 |
| At3g23770 | β-1,3-glucanase | −29 | Down | −6 | |
| ATSTP2 | At1g07340 | Hexose transporter | −3 | Down | −3 |
| PAB5 | At1g71770 | Polyadenylate binding protein 5 | −8 | Down | −2b |
| UBQ1 | At3g52590 | UBIQUITIN EXTENSION PROTEIN1 (UBQ1) | 1 | 1 | 1 |
Data are taken from Zhang et al. (2006); –, not analyzed.
Not identified by SAM analysis at 5% FDR cutoff.
Italics, upregulation in ms1; bold, downregulation in ms1. UBQ1 was used as a control.
The 228 downregulated genes showing differential expression in ms1 fall into a number of functional groups (see Supplemental Figure 4 online); a large number of them (75) are associated with catalytic activities, principally of hydrolase activity, which are associated with cell wall biosynthesis and lipid-associated genes involved in the production of pollen wall materials. Cluster analysis of gene expression identified a number of groups that showed coordinated expression profiles in ms1, for example, the tapetal oleosin and APG-like genes (Figure 3). The oleosin genes comprise a family of ∼20 members, many of which are associated with seed oil body formation, but some have been linked to pollen wall development. Seed-related oleosin genes do not show alterations in expression in ms1 (see Supplemental Table 1 online), indicating the specificity of the MS1-regulated pathway.
Figure 3.
Clustered Expression of Oleosin and APG Genes in Wild-Type and ms1 Buds.
Affymetrix array data from closed buds from Ler, Ler ttg, and ms1 ttg show coordinated regulation of expression. Three replicates were used for Ler and ms1 ttg and two for ttg samples.
(A) Expression and gene information.
(B) Gene clustering of expression profiles.
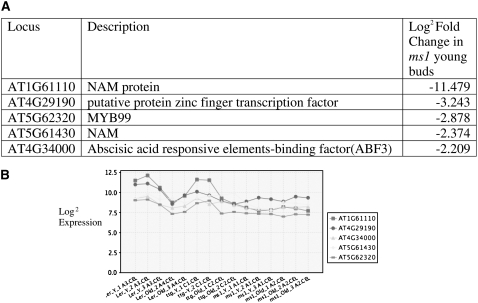
A number of transcription factors show downregulated expression in ms1 (see Supplemental Table 2 online), and four of these show clustering of expression, suggesting coordinate regulation (Figure 4), indicating that they are likely direct targets for MS1. Another group of genes that are overrepresented in the data set are Cys proteases. These show varying levels of downregulation ranging from 33- to 2-fold (Figure 5).
Figure 4.
Clustered Expression of Transcription Factors Showing Altered Expression in ms1 Mutant Buds.
Affymetrix array data from closed buds from Ler, Ler ttg, and ms1 ttg show coordinated regulation of expression. Three replicates were used for Ler and ms ttg and two for ttg samples.
(A) Expression and gene information.
(B) Gene clustering of expression profiles.
Figure 5.
Clustered Expression of Cys Protease Genes in ms1 Mutant Buds.
Affymetrix array data from closed buds from Ler, Ler ttg, and ms1 ttg show coordinated regulation of expression. Three replicates were used for Ler and ms ttg and two for ttg samples.
(A) Expression and gene information.
(B) Gene clustering of expression profiles.
Abnormal Secretion of Pollen Wall Materials Occurs in ms1
The pollen wall can be divided into two distinct regions: the inner intine and outer exine. In the ms1 mutant, exine formation is abnormal, with initial deposition of prime-exine occurring but with only limited, sporopollenin deposition (Ito and Shinozaki, 2002; Ariizumi et al., 2005; Vizcay-Barrena and Wilson, 2006). We conducted further ultrastructural analysis on wild-type Ler and ms1 buds to determine the effect of MS1 on tapetal development and specifically secretion of pollen wall material. Once the microspores are released from the callose wall, the tapetal cells start to appear abnormal and secretion into the locule is affected. After the first mitotic division, the wild-type tapetal cells contain a large number of inclusions, lipid deposits in the plastids become larger than in previous stages, and cytoplasmic lipid droplets fuse to form large drops. High numbers of vesicles containing fibrillar material are present in the cytoplasm (Figures 6A, 6C, and 6E). In the wild type, vesicles can be seen to migrate to the inner tapetal membrane, which later breaks down to aid release of the pollen wall material (Figures 6C and 6E). Whereas in ms1 the materials accumulate in the tapetum, these small vesicles form an extensive network of vesicles that progressively fuse and effectively engulf the entirety of the tapetum cells, coalescing to form large vacuoles. Surprisingly, these vesicles do not seem to fuse with the cell membrane but with each other (Figures 6B, 6D, and 6F). Therefore, the normal passage of pollen wall materials in vesicles through the tapetum for secretion into the locule fails to occur. Lipid droplets can be identified in the cytoplasm and inside the vacuoles; also, tryphine inclusions similar to those observed in the wild type are found in the mutant tapetal cells. Decreases in elaioplast numbers are also seen in the mutant, whereas in the wild type, these are numerous and contain large amounts of lipid materials. This correlates with the decrease in expression of genes linked to fatty acid and lipid biosynthesis seen in the ms1 transcriptomic data. The tapetal cells then go through autophagic breakdown of the vacuoles and necrotic-based cell death as opposed to the usual process of tapetal PCD (Vizcay-Barrena and Wilson, 2006).
Figure 6.
Transmission Electron Micrographs of Cross Sections through Wild-Type and ms1 Mutant Anthers during Ring-Vacuolate Stage/Microspore Mitosis.
(A), (C), and (E) The wild type.
(B), (D), and (F) ms1.
(A) The wild-type tapetal cells show a compact cytoplasm with some vacuoles containing fibrillar material and plastids with large inclusions. Big lipid droplets can be seen in the cytoplasm. Bar = 5 μm.
(B) By contrast, in the mutant, the cytoplasm is filled with big vacuoles, and fewer plastids are visible. Bar = 10 μm.
(C) and (D) Tapetal cells at the ring-vacuolate stage. In the wild type (C), plastid inclusions (asterisks) have become more electron transparent. Some small vacuoles are present in the cytoplasm. In the mutant (D), the vacuoles are increased in number and larger in size. They keep at a distance of the outer tangential membrane, and they seem to contain portions of cytoplasm and organelles, such as mitochondria (arrowhead). Bars = 2 μm.
(E) and (F) Enlarged region of tapetum in the wild type and the mutant, respectively.
(E) In the wild type, the tapetal inner membrane has degraded, while the outer membrane is still present. Plastid inclusions and vesicles containing fibrillar material (arrows) fuse to the membrane releasing material into the anther locule.
(F) However, in the mutant, the inner membrane is still present, and larger vacuoles are present due to the fusing of the vesicles. Bars in (E) and (F) = 1 μm.
Ms, microspore; En, endothecium; ML, middle cell layer; L, lipid globules; p, plastid; V, vacuole; N, nucleus.
Overexpression of MS1 Results in Stunted Plants and Altered Vegetative Expression
The MS1 gene was overexpressed under the control of the cauliflower mosaic virus 35S promoter (CaMV35S). High levels of ectopic MS1 expression were detected in leaves of the transgenic lines, while no expression of MS1 was seen in wild-type leaf material (Figure 7B). The 35S:MS1 lines were stunted, often forming flowering stems with only a minimal rosette (Figures 7A and 7C). These lines showed a range of severity of phenotypes linked to the level of MS1 expression. In the most severe cases, they showed slow development with a very small rosette, densely packed and short flowering stem (Figure 7C), and the flowers were often abnormal with shorter petals and sepals, which means that the anthers protruded at an earlier developmental stage than normal (Figure 7E). However, these anthers were also actually shorter than their wild-type equivalent (Figures 7D and 7E). As seen with many of the male-sterile mutants, the longevity of the plants was enhanced, with flowering continuing for much longer than in the wild type. In other less severe lines, flower initiation occurred normally; however, increased levels of branching were observed. In some cases the flowers were semi- or completely sterile; however, this sterility partially broke down as the plants aged. The phenotypes of these sterile flowers were distinct from that of the ms1 mutant, with mature pollen degeneration occurring rather than the earlier degeneration of microspores seen in ms1; therefore, this is not due to silencing of the MS1 gene. The leaves and tissues of the 35S:MS1 overexpression lines were often distorted and twisted; this was particularly evident in the anthers that showed increased indentations on the epidermal surface. Analysis of pollen from these lines showed an excess deposition of wall material and a loss of the regular structure of the pollen wall (Figures 7H and 7I).
Figure 7.
Overexpression of MS1 cDNA Using the CaMV35S Promoter.
(A) Wild-type Ler plant and two separate MS1-overexpressing lines.
(B) RT-PCR of the MS1 gene and actin control in wild-type and MS1 overexpression lines. Lanes 1 and 2, Ler wild type; lanes 3 to 6, 35S:MS1 overexpression lines. L, leaf tissue; B, closed buds.
(C) Extreme phenotype of minimal rosette, small size, and increased branching.
(D) Wild-type inflorescence and fully developed siliques indicating that they are fully self-fertile.
(E) Inflorescence of a severe overexpression line, showing increased numbers of flowers and aberrant flowers with small petals and sepals and protruding anthers (arrow). Anthers are short and flowers are semifertile.
(F) and (G) Mature pollen from the wild type.
(H) and (I) Mature pollen from MS1 overexpression line ([F] and [H]) scanning electron microscopy analysis.
(G) and (I) Alexander staining (Alexander, 1969). The pollen from the MS1 overexpression lines shows uneven surface indicative of increased deposition of pollen wall material (arrows).
Bars = 10 μm in (F) and (H) and 20 μm in (G) and (I).
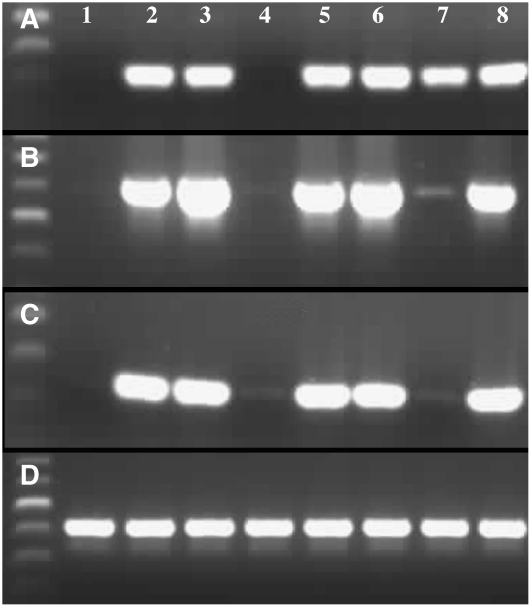
Microarray analysis has identified a number of genes with altered expression in the ms1 mutant buds, and three of these, which are associated with lipid biosynthesis and pollen wall development (At5g07550, At5g07410, and At5g07560) and show reproducible downregulation in ms1 buds, were analyzed in the 35S:MS1 overexpression lines by RT-PCR. Expression is observed in wild-type buds, but no wild-type vegetative expression is seen (Figures 8A to 8C). However, in some of the MS1 overexpression material, expression is seen in both bud and leaf material, suggesting that expression of these floral-specific transcripts is being activated by the ectopically expressed MS1 (Figures 8A to 8C).
Figure 8.
RT-PCR Analysis in the Overexpressing Lines of Genes That Are Floral Specific in Wild-Type Tissue but Show Altered Expression during Anther Development in the ms1 Mutant.
At5g07550 (A), At5g07410 (B), At5g07560 (C), and control (D) RT-PCR using actin-2. Lane 1, Ler leaf; lane 2, Ler bud; lane 3, B5-MS1-4 OEx bud; lane 4, B5- MS1-4 overexpression leaf; lane 5, B5-MS1-5-6 overexpression leaf; lane 6, B5-MS1-12 overexpression bud; lane 7, B5-MS11-23 overexpression leaf; lane 8, B5-MS1-37 overexpression bud. Ectopic induction of expression in the leaves is observed in the MS1 overexpression lines.
DISCUSSION
The MS1:GFP fusion protein is expressed in a developmentally regulated manner, associated with the late tetraspore to newly released microspore stage. An increase of ms1 transcript is detected in the ms1 mutant, implying that self-regulatory feedback, based upon a functional MS1 transcript, is limiting wild-type MS1 expression. The MS1 protein also appears to be actively degraded prior to microspore mitosis I, suggesting that its removal is required for pollen development progression. Similar classes of proteins containing PHD-finger motifs function by alteration of the chromatin, via protein complexes, to modify transcription. The putative function of MS1 as a transcriptional regulator is supported by the nuclear localization of the protein, downregulation of high numbers of genes linked to floral development, and the altered gene expression in the MS1-overexpressing lines and in the ms1 mutant.
MS1 Shows Posttranscriptional Regulation
The ms1 mutant alleles show an upregulation and prolonged expression of the mutant ms1 transcript compared with wild-type MS1 and the heterozygous ms1MS1 plants, indicating that self-regulation of MS1 expression is due to the full-length functional MS1 transcript or protein. Downregulation occurs around the stage of MS1 protein expression (Figures 1A and 2), suggesting that translation of the functional MS1 protein is required and, since downregulation is not observed in the 35S:MS1 lines, that the native MS1 promoter is critical for this regulation. In the ms1-1/ms1-6 mutant, both the LZ and PHD-finger domain are deleted from the MS1 transcript. Our other ms1 alleles result in variously sized partial transcripts; in ms1-3 and ms1-2, the LZ is present. However, in all of the mutant alleles, the PHD-finger motif is absent (Wilson et al., 2001). All of the ms1 alleles show upregulation of the ms1 mutant transcripts regardless of whether the LZ is present, suggesting that there may be a conformational problem with the truncated mutant polypeptides and/or that the PHD motif is critical in binding to the MS1 promoter and downregulating its expression. Overexpression of MS1 was found to have a detrimental effect on plant development; one could therefore imagine that a tightly regulated system of expression is required to moderate the effects of MS1 expression. Autoregulation of gene expression has been seen in other systems in Arabidopsis, for example, expression of the RNA binding protein FCA is negatively regulated by its own expression via alternative splicing within intron 3, which is mediated by the FCA WW protein interaction domain (Quesada et al., 2003). The ms1-1/ms1-6 allele shows a greater expression compared with the other alleles, suggesting that the lack of splicing at the second intron effects downregulation of the transcript; this may indicate that the regulation is linked to splicing, as seen with FCA (Quesada et al., 2003), or that there are regulatory sequences contained in the second intron. However, no evidence of alternative splicing of MS1 has been seen in wild-type anthers (Z.A. Wilson and C. Yang, unpublished data).
MS1 Protein Breakdown Is Developmentally Regulated
MS1:GFP expression is limited to the nuclei of the anther tapetum from late-tetraspore, during callose breakdown, to newly released microspores (Figure 2). No expression is observed in the microspores, which is supported by the lack of a gametophytic-associated phenotype (Wilson et al., 2001). MS1:GFP expression is short-lived; however, the protein appears to be stable in the absence of developmental progression. Based upon the developmental staging of Smyth et al. (1990), expression occurs for ∼12 h, representing a single bud within the inflorescence. This contrasts with other tapetal-specific transcription factors, for example, AMS, which is expressed throughout tapetal development (Sorensen et al., 2003). The spatial and temporal expression of the MS1:GFP fusion protein corresponds to the start of phenotypic abnormalities observed in the ms1 mutant (Dawson et al., 1993). However, this contrasts with the quantitative RT-PCR data (Figure 1) and the observations from MS1-promoter:β-glucuronidase fusions (Wilson et al., 2001), which indicate that MS1 transcription is for a significantly longer period, from late meiosis through to pollen mitosis II, suggesting that posttranscriptional regulation of the MS1 transcript may also be occurring.
The transient nature of the MS1 protein suggests that an active developmentally regulated process of MS1 protein breakdown may be occurring within the tapetum. Ubiquitin-mediated proteolysis is emerging as the main nonlysosomal proteolytic pathway for the rapid degradation of intracellular proteins in eukaryotic cells (Kepinski and Leyser, 2002); therefore, MS1 protein degradation may be occurring in this manner. It is possible that MS1 breakdown may be critical for the progression of pollen development and that the persistence of the MS1 protein may serve to downregulate genes that are required for continued development or to maintain expression of repressors of microspore maturation.
The ms1 Mutant Shows Abnormal Exine Development
The pollen wall can be divided into two principle parts: the exine, composed mainly of polymerized sporopollenin; and the intine. Initially, lipidic precursors of sporopollenin are secreted by the tapetum and in association with the microspore are deposited between the microspore plasma membrane and the callose wall to form the primexine matrix. This then serves as a scaffold for the deposition and polymerization of sporopollenin, resulting in the sculptured exine wall (Ariizumi et al., 2004). Primexine development and limited exine formation is seen in the ms1 mutant (Figure 6); however, exine deposition is reduced and the agglutination of the deposits and stickiness of the mutant microspores suggest an unusual chemical composition of the sporopollenin, which is supported by the downregulation in ms1 of genes associated with acyl lipids and phenyl propanoid precursors.
Aberrations in exine structure have been linked to defects in the biosynthesis of sporopollenin (e.g., ms2 and flp1; Aarts et al., 1997; Ariizumi et al., 2003) or due to defective exine patterning (e.g., the dex1 mutant; Paxson-Sowders et al., 2001). Random deposition and apparent lack of anchoring of sporopollenin is seen in the dex1 mutant, and it is speculated that DEX1 may play a role as a nucleation point for sporopollenin deposition (Paxson-Sowders et al., 2001). The patterning of primexine deposition is unchanged in the ms1 mutant, and no expression changes are observed in the DEX1 and NEF1 genes, indicating that MS1 acts independently of these pathways.
During normal wall development Pro-orbicules (Ubisch) osmiophilic lipid bodies form in the tapetum and are exported into the locule by exocytosis, and the acyl precursors polymerize to form the sculptured exine. The tapetum in Arabidopsis is of the secretory type (Pacini et al., 1985), and as in most species exhibiting secretory tapetum, the cells retain their shape and position as they lose their cell walls. In Ler, as seen in Columbia (Regan and Moffatt, 1990), only the inner tangential and radial surfaces seem to have secretory properties. Changes in the pattern of tapetal secretion are observed in the ms1 mutant, and transmission electron microscopy observations in ms1 tapetal cells suggest that the inner tangential membrane persists, somehow preventing the secretion (Figure 6). In the wild type, vesicles fuse to the radial and tangential membranes, releasing their contents into the anther locule; however, this fails to occur in the mutant. Instead, they fuse to others situated in the inside of the cell, thus failing to provide crucial components of the pollen wall to the maturing microspores. This change in the polarity of the secretion might explain the decreased amount of material being deposited onto the ms1 pollen.
Lipid transfer proteins are thought to be involved in the trafficking of fatty acids and other sporopollenin precursors from the tapetum to the microspore. The LTp-1 promoter has been shown to have similar regulatory sequences to genes in the phenyl propanoid pathway, suggesting a common mechanism for expression of both genes (Piffanelli et al., 1998). Similar coordinated downregulation of LTP and genes linked to the PAL pathway are seen in the ms1 mutant (see Supplemental Data Set 1 online), indicating that these genes may be controlled directly or indirectly by MS1.
The At GPAT1 male-sterile mutant shows many similarities in tapetal phenotype to the ms1 mutant, in that the tapetal cell membrane persists and there is a decrease in secretion in the tapetum due to a reduction in the fusion of dilated endoplasmic reticulum (ER) with the tapetal plasma membrane possibly as a consequence of impaired ER membrane biogenesis (Zheng et al., 2003). They speculate that the impaired membrane biosynthesis in the mitochondria may be responsible for the lack of initiation of tapetal PCD and the resultant necrotic degeneration of the tapetum. A similar alteration in the secretion of pollen wall materials and lack of PCD is observed in the ms1 mutant (Vizcay-Barrena and Wilson, 2006). Although the At GPAT gene family members do not show altered expression in the ms1 mutant, it is interesting to speculated whether the changes in the secretion of pollen wall materials, possibly as a consequence of impaired biosynthesis of the various components of the pollen wall and exine, result in the failure of triggering of PCD.
The ms1 Mutant Has Impaired Pollen Coat Development
After deposition of the prime-exine, the biosynthesis of pollen coat material occurs in the tapetum; this comprises >90% lipase and oleosins and is involved in the pollen–stigma interaction for efficient pollination. All of the gene families associated with pollen coat formation (Mayfield et al., 2001) are downregulated in ms1 (Figure 3), indicating that they are controlled by MS1 expression. In the wild type prior to tapetal lysis, the tapetum contains abundant elaioplasts and tapetosomes, which are enriched for steryl esters, triacylglycerols (TAGs), and oleosin-coated droplets, respectively. These are released as the tapetum disintegrates and form the pollen coat (Piffanelli et al., 1998). In the ms1 tapetum, very few elaioplasts can be seen, indicating that the production of steryl esters is also compromised in ms1. The tapetosomes are formed from the rough endoplasmic reticulum in the tapetum in a process similar to that involved in maturing seed (Hsieh and Huang, 2007). TAGs become coated with phospholipids and oleosins/oleosin-like proteins and converge together, combined with vesicles derived from the ER. Oleosins are involved in the stabilization of lipid granules by the interaction of a conserved hydrophilic hairpin and amphipathic amino and carboxylic regions that allow the oleosin molecule to reside on the surface of the lipid granule. They are then transferred to the mature pollen where they play a role in pollen surface attachment to the female stigma and hydration; for example, the Arabidopsis At GRP17 mutant exhibits delayed pollen hydration (Mayfield and Preuss, 2000).
Sixteen oleosin genes have been identified in Arabidopsis, and eight have been shown to have tapetal-specific expression, with six of these arranged in tandem on the top arm of chromosome 5 (Kim et al., 2002), and all of these show coordinate downregulation in ms1. In the tapetosomes it is speculated that oleosins play an important role in stabilizing the lipid globules, ensuring that they remain distinct and that polar secretion toward the inside of the locule and microspore occurs (Hsieh and Huang, 2004). It has been shown that the size and production of oil bodies are affected by the oleosin abundance (Siloto et al., 2006). We propose that the loss of these proteins in the ms1 mutant may effect the production and release of the TAGs from the rough endoplasmic reticulum, resulting in the accumulation of lipids and impaired secretion, rather than the normal process of tapetosome formation and deposition onto the developing pollen.
MS1 Transcriptionally Controls Anther Gene Expression
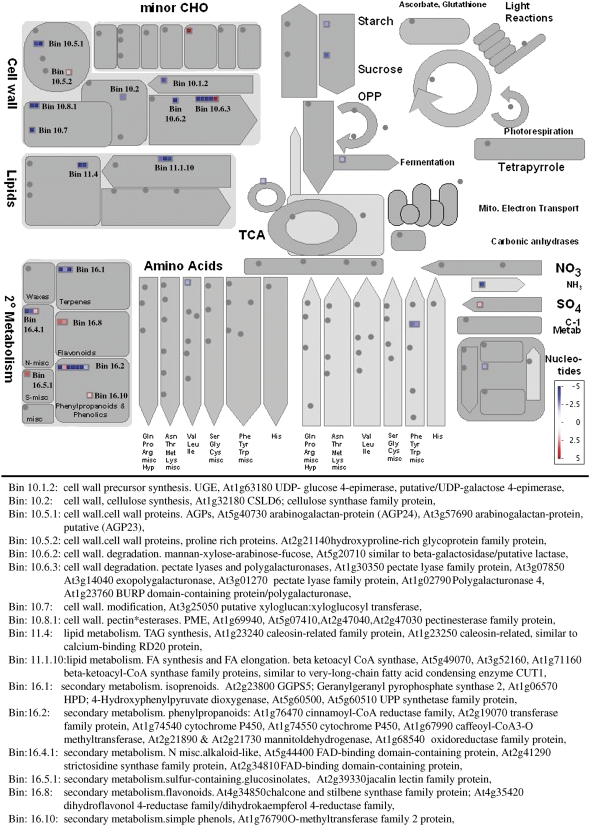
The MS1 protein has homology to a group of PHD-finger transcription factors (Asaland et al., 1995), which are hypothesized to regulate transcription by the modification of chromatin. While this putative role is supported by the nuclear localization of the MS1 protein (Figure 2), the most compelling evidence arises from the downregulation of floral-specific transcripts in the ms1 mutant buds and ectopic induction of anther-specific genes in MS1-overexpressing lines. In young buds (meiosis–mitosis 1 stage), an extensive change in the expression of 966 genes was observed, and of these, 228 are present in anthers and downregulated at >log2 fold level, and 94 of these (see Supplemental Data Set 2 online) show changes in expression in spl/exs mutants (Wijeratne et al., 2007), indicating that they are likely to be downstream factors in the same pathway. A high proportion of the genes changing in ms1 are associated with wall biosynthesis, lipid metabolism, and secondary metabolism as indicated by overlays against general metabolism pathways (Figure 9; MAPMAN). The composition of sporopollenin has still not been fully defined but is thought to be derived from long-chain fatty acids combined with oxygenated aromatic rings and phenyl propanoids (Piffanelli et al., 1998). Therefore, the downregulation of components of the phenyl propanoid pathway, lipid metabolism, and cell wall biosynthesis pathways, lipid transfer proteins and Hyp-rich glycoproteins (oleosins/APGs) (Figure 9), which are thought to be involved in the biosynthesis of sporopollenin and pollen coat materials, confirms the major role that MS1 plays in the control of late pollen development and specifically pollen wall and coat formation.
Figure 9.
Cell Function Classification Analysis of the 260 Genes That Show Up-/Downregulation in Young ms1 Buds Compared with the Wild Type (MAPMAN; Thimm et al., 2004).
Genes associated with the cell wall, lipids, and secondary metabolism are listed in the MAPMAN allocated bins.
The CaMV35S:MS1-overexpressing lines show phenotypes of stunted growth and late flowering, frequently forming flowering stems with a minimal rosette, increased branching, distorted organs, and reduced fertility (Figures 7A to 7E). The genes associated with pollen wall formation are generally specific to reproductive tissues, for example, At5g07550 and At5g07560, which are both oleosins, and At5g07410, which is a pectin methylesterases. Pectin methylesterases have been indicated as important both in pollen tube formation and at the earlier step of wall development. Antisense downregulation of a flax pectin methylesterase in tobacco (Nicotiana tabacum) resulted in modification of pollen cell wall sculpturing, suggesting the involvement of the demethylation of pectin in the pollen cell wall–specific structure (Lacoux et al., 2003). These genes show reproducible downregulation in the ms1 mutant but are ectopically induced in leaves in the MS1-overexpressing lines (Figures 8A to 8C), indicating that expression of MS1 results in transcriptional activation of genes that are normally not actively expressed in the vegetative tissues.
Pollen from the overexpression lines is generally fertile but appears to have increased deposition of pollen coat material and exhibits a similar, although less severe, phenotype to the faceless pollen1 (flp1) mutant (Ariizumi et al., 2003). In the flp1 mutant, they proposed that the smooth appearance of the pollen is due to altered sporopollenin composition and increased deposition of tryphine, which had a decrease in lipid droplet size. In the ms1 mutant, there is a decline in pollen wall material, including components of sporopollenin and the pollen coat. A corresponding increase in expression of wall components, specifically oleosins (as exemplified by At5g07550 and At5g07560), is seen in the 35S:MS1 overexpression lines. This correlates with the alterations in the observed phenotypes and suggests that ectopic expression of MS1 significantly alters gene developmental programs in the vegetative tissues of the plant and implies that MS1 may play a major role in the control of wall components in the tapetum. This is supported by observations of wall development in the ms1-1 mutant in which aberrant exine and intine formation occurs.
Fifteen transcription factors are downregulated in ms1 buds (SAM analysis FDR 5%, >log2 fold; see Supplemental Table 2 online); however, five of these show expression early in the wild-type buds and coordinated expression profiles, indicating they are likely primary targets for MS1 regulation. Two of these are NAM proteins (At1g61110 and At5g61430), of which At1g61110 exhibits the highest level of repression in ms1. The NAC domain consists of a twisted antiparallel β-sheet, next to an N-terminal α-helix on one side and a shorter helix on the other side surrounded by helical elements, which is likely to mediate dimerization through conserved interactions and DNA binding. NAM proteins have been linked to many aspects of plant development, including pollen production (Mitsuda et al., 2005, 2007). The other three are At4g29190, a putative zinc-finger transcription factor; At4g34000, an abscisic acid–responsive element binding factor (ABF3); and At5g62320, which encodes for MYB99. MYB factors have been proposed as key regulators of many aspects of anther development (Wijeratne et al., 2007). The regulation of MYB99 by MS1 has been further confirmed by Ito et al. (2007), who have shown using a dexamethasone-inducible MS1 construct that MYB99 is induced by MS1 expression without the requirement for de novo protein synthesis, suggesting that MYB99 may be a direct target for MS1 and that MYB99 may act via At1g61110 to regulate components of sporopollenin biosynethsis.
A class of Cys proteases (caspases) has been linked to PCD processes in animals, and caspase-like activity has also been associated with PCD in plants (Solomon et al., 1999; Lam and del Pozo, 2000; Hoeberichts and Woltering, 2003); however, the exact nature of these plant Cys proteases have yet to be established. A number of Cys proteases show coordinated downregulation of expression in ms1 buds (Figure 5). We have previously shown that tapetal PCD does not occur in the ms1 mutant (Vizcay-Barrena and Wilson, 2006). It is likely that these Cys proteases may play a role in the induction of PCD in the wild-type tapetum.
Potential Role of MS1 in Anthers
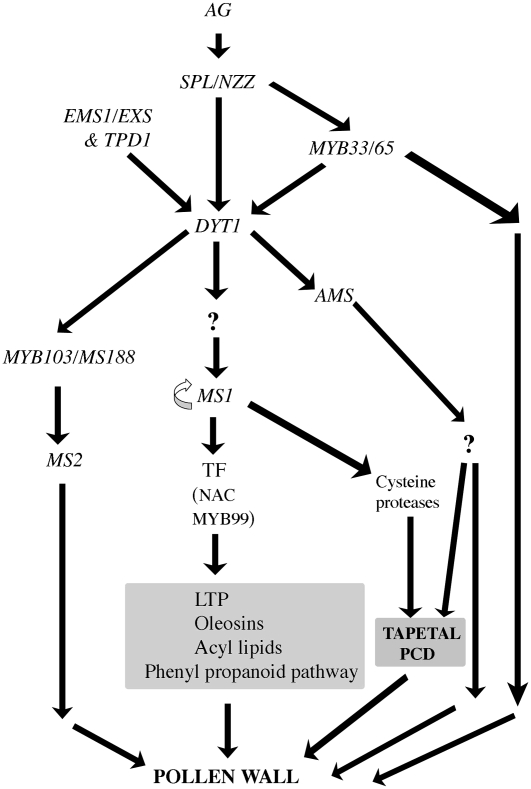
Early events in pollen development and tapetal initiation proceed normally in the ms1 mutant, and expression data indicate that MS1 acts late in pollen development after tapetal initiation and is downstream of DYT1 (Zhang et al., 2006). Our microarray data indicate that MS1 coordinates the expression of late genes associated with pollen wall formation, which are involved in the biosynthesis of components of the phenyl propanoid pathway, long-chain fatty acids, and phenolics, which are required for sporopollenin biosynthesis (Figures 9 and 10). We have previously shown that in the ms1 mutant, tapetal PCD does not occur and that tapetal degeneration occurs by necrotic breakdown (Vizcay-Barrena and Wilson, 2006) and have observed downregulation in the expression of a number of Cys proteases in the ms1 mutant buds. We propose that the induction of these proteases may be critical to the progression of PCD and that in their absence, possibly in association with a lack of secretion from the tapetal cells, PCD does not occur (Figure 10). MS1 also controls the synthesis of the pollen coat, including the tapetally expressed oleosin gene family, which mediate pollen coat behavior by altering the size of lipid bodies, and the production of lipid transfer proteins. The altered expression of pollen wall material either directly or indirectly affects the secretion and deposition of such materials during the early stages of microspore maturation. However, it is envisaged that MS1 does not directly regulate genes associated with pollen wall biosynthesis, due to the timing of MS1 expression and the subsequent phenotypic changes seen in pollen wall development, but acts via one or a number of additional transcription factors, including MYB99 and two NAM genes (At1g61110 and At5g61430) that contain a conserved NAC domain (Figure 10). This is further supported by Ito et al. (2007) using a dexamethasone-inducible MS1 construct.
Figure 10.
Model for the Role of MS1 during Anther and Pollen Development.
TF, transcription factor; LTP, lipid transfer protein; AG, AGAMOUS.
METHODS
Plant Material and Growth Conditions
Ler seeds carrying the ms1 ttg mutations (N261) were provided by the Nottingham Arabidopsis Stock Centre (NASC). Plant material was sown on Levington M3:John Innes No. 3:vermiculite:perlite (6:6:1:1) compost mix supplemented with 0.2 g L−1 of Intercept and grown in controlled environmental chambers at 21 to 22°C under illumination of 167 μmol·m−2·s−1 with a 22-h photoperiod. Seedlings were selected by expression of the ttg mutation and the male-sterile phenotype confirmed after flowering. Lines were maintained and heterozygotes generated by crossing ms1 ttg plants with wild-type Ler pollen.
Constructs
Three constructs were produced that contained the GFP-coding region in three separate regions of the MS1 gene: C- and T-terminal fusions and centrally within the third exon of the MS1 gene. The 2.9-kb upstream region of MS1, combined with the complete MS1 gene and 333-bp untranslated 3′ region, was cloned into the pBluescript KS+ vector (Stratagene) to form pBKSMS1.
C-Terminal Construct
PCR was used to amplify a 1.1-kb BamHI fragment from the 3′ end of the MS1 gene, delete the MS1 stop codon, and add three extra amino acids (SSG) and a BglII site to the 3′ end. The GFP coding region (kindly provided by M. Bennett's laboratory, University of Nottingham) was amplified using primers with a 5′ BglII site and 3′ XbaI and SacII restriction sites. The MS1 terminal region and the GFP fragment were cut with BglII and then ligated together. This fragment was then digested with BamHI and SacII and ligated into the pBKSMS1 clone that had been previously digested with BamHI and SacII to produce pBKSMS1d. The 3′ untranslated region of the wild-type MS1 gene was PCR amplified from the stop codon to 329 bp downstream and an AvrII site added to the 5′ end and XbaI and SacII restriction sites to the 3′ end. This fragment was then digested with AvrII and SacII and ligated to the pBKSMS1d clone that had been previously digested with XbaI and SacII. This resulted in an MS1 genomic insert containing 2.9 kb upstream of the translational start, the MS1 genomic coding sequence, and the GFP sequence, followed by the MS1 endogenous stop and 329 bp of the MS1 untranslated 3′ region (see Supplemental Figure 2 online) to form pBKSMS1-C-GFP.
N-Terminal Construct
Inverse PCR was performed to insert BglII, SalI, and XbaI restriction sites at the MS1 translational start site. The pBKSMS1 plasmid was amplified using primers flanking the translation start site carrying BglII and SalI sites, and SalI and XbaI sites, respectively. The amplified fragment was digested with SalI and ligated and cloned into Escherichia coli D5Hα cells. The GFP coding sequence was then amplified with flanking BglII and XbaI restriction sites and then ligated into the IPCR-derived clone that had been previously digested with BglII and XbaI. This plasmid was then digested with SphI and HindIII to generate a 1.7-kb MS1 fragment containing the inserted GFP coding region. This MS1-GFP region was then ligated into the pBKSMS1 plasmid (previously digested with SphI and HindIII) to form pBKSMS1-N-GFP.
Internal Construct
Inverse PCR was repeated to insert BglII, SalI, and XbaI restriction sites in the MS1 third exon at base 1538. The pBKSMS1 plasmid was amplified using primers flanking the base 1538 carrying BglII and SalI sites, and SalI and XbaI sites, respectively. The amplified fragment was digested with SalI and ligated and cloned into E. coli D5Hα cells. This was then digested with BglII and XbaI and ligated with the GFP coding sequence, which had been previously amplified with flanking BglII and XbaI restriction sites. This plasmid was then digested with BamHI and SacII and the MS1-GFP region (1.9 kb) ligated into pBKSMS1 plasmid that had also previously been digested with BamHI and SacII to form pBKSMS1-int-GFP.
Overexpressing Lines
The MS1 cDNA was PCR amplified using primers FGWMS1 and RGWMS1 (see Supplemental Table 3 online) and cloned by recombination into Gateway pDONR201 entry vector (Invitrogen) and then transferred into pGWB5 (Karimi et al., 2002) under the control of the CaMV35S promoter. The construct was then transferred into Agrobacterium tumefaciens C58 and wild-type Arabidopsis thaliana Ler plants. Plants were screened for presence of the transgene on kanamycin and by PCR. RNA was isolated from buds and leaves of the T1 lines carrying the CaMV35S:MS1 gene (RNeasy; Qiagen) and cDNA prepared using 2.5 μg of total RNA in a 20-μL reaction (Superscript II reverse transcriptase; Invitrogen). This was used for RT-PCR analysis of MS1 expression and three genes that had shown downregulation in ms1 buds (At5g07550, At5g07410, and At5g07560; see Supplemental Table 3 online). Amplification was performed using 0.5 μL of cDNA template and the appropriate primer pairs (see Supplemental Table 3 online) for 30 cycles at 94°C for 30s, 58°C for 30s, and 72°C for 1 min. Control RT-PCR analyses were conducted using Arabidopsis actin primers (Wilson et al., 2001).
Transformation
All constructs were checked by sequencing. The GFP fusions were ligated into pBIN19 after digestion with KpnI and SacII. The binary vectors were then transferred into Agrobacterium C58 by electroporation (Sambrook et al., 1989). Heterozygous ms1 ttg plants were separately transformed with the three GFP constructs using the floral dip method (Clough and Bent, 1998). Seeds were collected and screened on Murashige and Skoog medium (Murashige and Skoog, 1962) containing 50 μg/mL of kanamycin. T1 seedlings were selected for the presence of the transgene, transferred to soil, and allowed to self. T1 plants were screened for GFP expression by PCR and microscopy and for the ms1-linked ttg marker and male fertility.
Seeds from the T1 plants were harvested individually. Complementation of the ms1 mutation by a functional MS1:GFP fusion protein was indicated by kanamycin resistance, the ttg phenotype, RT-PCR, and restoration to full fertility. To ensure that we were observing complementation of the ms1 mutation and not recombination between the ttg and ms1 mutations, an excess of 40 plants from each line were analyzed per generation, over a minimum of three generations; in all cases, the transgene cosegregated with the ttg marker and restoration of fertility. Lines that were complemented by the MS1P:MS1:GFP constructs were also analyzed in the T2 generation without kanamycin selection. These lines showed distorted segregation of fertility, with presence of functional transgenes cosegregating with fertility; in all cases, the lines exhibiting the ttg phenotype were fully fertile and carried the transgene.
Microscopy
Inflorescences selected from homozygous lines carrying the transgene were analyzed for GFP expression by fluorescence (Nikon) and confocal microscopy (TCS SP2; Leica). A minimum of 10 independent transformants were analyzed (5 to 10 inflorescences per line). Pollen viability was determined by Alexander staining (Alexander, 1969). For ultrastructure analysis, whole inflorescences were removed from Ler and ms1 mutant plants, fixed overnight in 4% paraformaldehyde in phosphate buffer, pH 7.2, rinsed twice in phosphate buffer, and fixed in 1% (w/v) OsO4 (in phosphate buffer) for 3 h. Tissue was dehydrated in an ethanol series and infiltrated with Spurr's resin. Individual buds were then dissected and embedded in Spurr's resin. Ultrathin sections (50 to 70 nm) were cut using a diamond knife on a Reichert-Jung Ultracut ultramicrotome and picked up on 200 mesh copper grids. Sections were double stained with saturated uranyl acetate in 50% ethanol and lead citrate and examined with a Jeol JEM 1010 electron microscope. For scanning electron microscopy, material was fixed in FAA (5% [v/v] formalin, 5% [v/v] acetic acid, and 50% [v/v] ethanol), dried in an ethanol series, coated with gold particles, and observed using a Jeol JSM 840 scanning electron microscope.
Detection of GFP Fusion Protein by Protein Gel Blot Hybridization
Total protein was isolated by trichloroacetic acid precipitation (http://www.biotech.unl.edu/proteomics/mercaptoethanol.html), separated on NuPAGE 4 to 12% Bis-Tris gels (Invitrogen), and transferred to nitrocellulose membrane in 1× NuPAGE transfer buffer at a constant voltage (30 V; room temperature). The membranes were blocked in 5% milk powder (Marvell) in TBS-T (0.01% Tween 20 in TBS) buffer for 2 h (room temperature) or overnight at 4°C. They were then hybridized with rabbit anti-GFP antibody (1:2000 dilution; Molecular Probes) in blocking buffer for 1 h at room temperature, washed four times with 1% milk powder in TBS-T, and reacted with horseradish peroxidase–conjugated anti-rabbit IgG (1:2500 dilution; Sigma-Aldrich). The cross-reacting proteins were visualized by chemiluminescence using ECL protein gel blotting detection reagents according to the manufacturer's instructions (Amersham Pharmacia Biotech).
Microarrays
Closed buds were collection as soon as the plants showed three fertile (wild type) or sterile siliques (ms1) (∼27 to 29 d old). Buds were dissected and separated into different groups according to their relative position within the inflorescence, taking bud 1 as the oldest unopened bud. All opened flowers and buds that were about to open (petals clearly visible) were discarded. The buds were staged by light microscopy and by RT-PCR analysis of MS1 expression. Two pools of unopened buds, representing young buds (PMC–mitosis I) and old buds (mitosis II), were collected from Ler, Ler ttg, and ms1 ttg plants, immediately frozen in liquid nitrogen, and stored at −80°C until required. The number of buds in the inflorescence varies between the wild type and the ms1 mutant; therefore, the number of buds included in each wild type/mutant group was selected to reflect developmental stage rather than numerical bud classes. For the wild type, the old bud group comprised buds 1 to 4, and young buds consisted of buds 5 and younger. For the mutant, buds 1 to 6 were included in the old buds group; buds 7 and onwards formed the young bud group. The ttg mutation was used as an early marker to help identify homozygous ms1 lines for array analysis; therefore, control arrays were performed using both Ler and ttg bud material. Total RNA was extracted from 20 to 40 mg of buds (RNeasy plant kit; Qiagen). RNA samples were DNase treated and then purified using RNeasy spin columns (Qiagen). Affymetrix microarray analysis was performed by NASC using 10 μg of total RNA per sample as described in the Affymetrix GeneChip technical analysis manual (Affymetrix). Double-stranded cRNA was synthesized using the SuperScript Choice system (Invitrogen) with oligo(dT)24 primer fused to a T7 RNA polymerase promoter. Biotin-labeled cRNA was prepared by cDNA in vitro transcription using the BioArray High-Yield RNA transcript labeling kit (Enzo Biochem) with biotinylated UTP or CTP. The Arabidopsis ATH1 Genome array (Affymetrix) was screened using 15 μg of labeled target cRNA according to the Affymetrix GeneChip Technical Analysis Manual. GeneChips were then stained using streptavadin-phycoerythrin solution and scanned using an Agilent 2500A GeneArray scanner (Agilent Technologies). Two to three replicates of independently grown material were used.
Data Analysis and Gene Annotation
The raw hybridization data were provided by NASC and were normalized using robust multichip average analysis (Bolstad et al., 2003). Reproducibility was tested using the standard error /average *100% calculation to give a measurement of the variability that is independent of the size of the average. A small value (10 to 20%) indicated that the replicate data were consistent, with a fold change between replicates no bigger than 2.
SAM (Tusher et al., 2001) was used to analyze the microarray data. Triplicates were used, and any genes with absent or marginal signal in all of the samples were removed from the analysis. Samples were compared across the two developmental stages. A false discovery cutoff of 5% was used for the initial selection of candidate genes, and a secondary filtering of log2 fold difference was then applied. Genes were also sorted according to their biological and molecular function using information from Gene Ontology (Netaffix), KEGG, and MAPMAN (http://gabi.rzpd.de/projects/MapMan/).
RT-PCR
First-strand cDNAs were synthesized from 5 μg of total RNA isolated from different Ler and ms1 staged buds (old and young stages) and vegetative and reproductive tissues. RT was performed using Superscript II reverse transcriptase (Invitrogen) and an oligo(dT) primer (Invitrogen) according to the manufacturer's instructions. Amplification was conducted in a 25-μL volume reaction using 0.5 μL of the cDNA template, or equivalent RT control, which lacked Superscript II reverse transcriptase, gene-specific primers, and Taq DNA polymerase (Promega) for one cycle of 2 min at 94°C, followed by 30 cycles of 94°C for 30 s, 56 to 58°C for 30 s, 72°C for 1 min, and a final elongation cycle at 72°C for 4 min. The sequences of all primers used are presented in Supplemental Table 3 online.
Quantitative RT-PCR analyses were performed using the Mx4000 multiplex quantitative PCR system (Stratagene). To quantify copy number, a standard curve was produced using serial dilutions of cloned MS1 cDNA. Reactions were set up using Brilliant SYBR Green QPCR master mix (Stratagene) in a final volume of 25 μL containing 0.5 μL of cDNA and 0.5 μL of the appropriate primers (see Supplemental Table 3 online). PCR cycling conditions for amplification were 95°C for 10 min and then 40 cycles of 95°C for 30 s, 58°C for 1 min, and 72°C for 1 min. Data acquisition and analyses were performed using Mx4000 real-time multiplex quantitative PCR system software. All samples were tested at least twice.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RT-PCR of the MS1 Gene in the Different ms1 Alleles.
Supplemental Figure 2. Schematic Representations of the MS1:GFP Fusion Constructs.
Supplemental Figure 3. Protein Gel Blot Hybridization of MS1:GFP Fusion Protein Using Anti-GFP Antibody.
Supplemental Figure 4. Functional Grouping and Comparison of Genes Changing in ms1 with the Pollen Transcriptome.
Supplemental Figure 5. RT-PCR Analysis of Genes Showing High and Low-Level Expression Changes in ms1 Buds.
Supplemental Figure 6. RT-PCR Validation of a Number of Genes Involved in Pollen Development in Wild-Type and ms1 Mutant Buds.
Supplemental Table 1. Oleosin Genes in Arabidopsis.
Supplemental Table 2. All Putative Transcription Factors Changing at <5% FDR by SAM Analysis and >Log2 Fold in ms1 Young Buds.
Supplemental Table 3. Primer Sequences Used in the Analyses.
Supplemental Data Set 1. Genes That Are Changing in ms1 Young Buds Compared with the Wild Type as Identified by SAM Analysis (5% FDR) Show a More Than Twofold Change and Are Present in Anther Array Samples.
Supplemental Data Set 2. Genes That Are Changing in ms1 Young Buds Compared with the Wild Type as Identified by SAM Analysis (5% FDR) and Show a More Than Twofold Change That Are Also Changing in the spl/nzz and ems1 Mutants (Wijeratne et al., 2007).
Supplementary Material
Acknowledgments
We thank Hong Ma and Takaya Ito for sharing results prior to publication. We also thank Ranjan Swarup for advice with construct preparation and confocal microscopy, Malcolm Bennett and Jerry Roberts for critical reading of the manuscript, and to Susan Anderson, Ian Ward, Trevor Gray, and Ilda Casimiro for support with the transmission electron microscopy. Seed stocks, bioinformatics information, and Affymetrix analysis were provided by NASC. This work was funded by the Biotechnology and Biological Science Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zoe A. Wilson (zoe.wilson@nottingham.ac.uk).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aarts, M.G., Hodge, R., Kalantidis, K., Florack, D., Wilson, Z.A., Mulligan, B.J., Stiekema, W.J., Scott, R., and Pereira, A. (1997). The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12 615–623. [DOI] [PubMed] [Google Scholar]
- Aarts, M.G.M., Dirkse, W.G., Stiekema, W.J., and Pereira, A. (1993). Transposon tagging of a male sterility gene in Arabidopsis. Nature 363 715–717. [DOI] [PubMed] [Google Scholar]
- Albrecht, C., Russinova, E., Hecht, V., Baaijens, E., and de Vries, S. (2005). The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17 3337–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44 117–122. [DOI] [PubMed] [Google Scholar]
- Ariizumi, T., Hatakeyama, K., Hinata, K., Inatsugi, R., Nishida, I., Sato, S., Kato, T., Tabata, S., and Toriyama, K. (2004). Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39 170–181. [DOI] [PubMed] [Google Scholar]
- Ariizumi, T., Hatakeyama, K., Hinata, K., Sato, S., Kato, T., Tabata, S., and Toriyama, K. (2003). A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol. Biol. 53 107–116. [DOI] [PubMed] [Google Scholar]
- Ariizumi, T., Hatakeyama, K., Hinata, K., Sato, S., Kato, T., Tabata, S., and Toriyama, K. (2005). The HKM gene, which is identical to the MS1 gene of Arabidopsis thaliana, is essential for primexine formation and exine pattern formation. Sex. Plant Reprod. 18 1–7. [Google Scholar]
- Asaland, R., Gibson, T.J., and Stewart, A.F. (1995). The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biotechnol. 20 56–59. [DOI] [PubMed] [Google Scholar]
- Bhatt, A.M., Canales, C., and Dickinson, H.G. (2001). Plant meiosis: The means to 1N. Trends Plant Sci. 6 114–121. [DOI] [PubMed] [Google Scholar]
- Bolstad, B.M., Irizarry, R.A., Astrand, M., and Speed, T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 185–193. [DOI] [PubMed] [Google Scholar]
- Canales, C., Bhatt, A.M., Scott, R., and Dickinson, H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 12 1718–1727. [DOI] [PubMed] [Google Scholar]
- Chasan, R. (1992). Breaching the callose wall. Plant Cell 4 745–746. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Colcombet, J., Boisson-Dernier, A., Ros-Palau, R., Vera, C.E., and Schroeder, J.I. (2005). Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17 3350–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, J., Wilson, Z.A., Aarts, M.G.M., Braithwaite, A.F., Briarty, L.G., and Mulligan, B.J. (1993). Microspore and pollen development in six male-sterile mutants of Arabidopsis thaliana. Can. J. Bot. 71 629–638. [Google Scholar]
- Dong, X., Hong, Z., Sivaramakrishnan, M., Mahfouz, M., and Verma, D.P. (2005). Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 42 315–328. [DOI] [PubMed] [Google Scholar]
- Goldberg, R., Beals, T., and Sanders, P. (1993). Anther development: Basic principles and practical applications. Plant Cell 5 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeberichts, F.A., and Woltering, E.J. (2003). Multiple mediators of plant programmed cell death: Interplay of conserved cell death mechanisms and plant-specific regulators. Bioessays 25 47–57. [DOI] [PubMed] [Google Scholar]
- Honys, D., and Twell, D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5 R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker, T.S., Millar, A.A., and Kunst, L. (2002). Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 129 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, K., and Huang, A.H. (2004). Endoplasmic reticulum, oleosins, and oils in seeds and tapetum cells. Plant Physiol. 136 3427–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, K., and Huang, A.H. (2007). Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 19 582–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Nagata, N., Yoshiba, Y., Ohme-Takagi, M., Hong, M., and Shinozaki, K. (2007). Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19 3549–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., and Shinozaki, K. (2002). The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol. 43 1285–1292. [DOI] [PubMed] [Google Scholar]
- Jung, K.H., Han, M.J., Lee, Y.S., Kim, Y.W., Hwang, I., Kim, M.J., Kim, Y.K., Nahm, B.H., and An, G. (2005). Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17 2705–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kepinski, S., and Leyser, O. (2002). Ubiquitination and auxin signaling: A degrading story. Plant Cell 14(Suppl): S81–S95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.U., Hsieh, K., Ratnayake, C., and Huang, A.H. (2002). A novel group of oleosins is present inside the pollen of Arabidopsis. J. Biol. Chem. 277 22677–22684. [DOI] [PubMed] [Google Scholar]
- Lacoux, J., Gutierrez, L., Dantin, F., Beaudoin, B., Roger, D., and Laine, E. (2003). Antisense transgenesis of tobacco with a flax pectin methylesterase affects pollen ornamentation. Protoplasma 222 205–209. [DOI] [PubMed] [Google Scholar]
- Lam, E., and del Pozo, O. (2000). Caspase-like protease involvement in the control of plant cell death. Plant Mol. Biol. 44 417–428. [DOI] [PubMed] [Google Scholar]
- Li, N., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18 2999–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H. (2005). Molecular genetic analysis of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56 393–434. [DOI] [PubMed] [Google Scholar]
- Mayfield, J.A., Fiebig, A., Johnstone, S.E., and Preuss, D. (2001). Gene families from the Arabidopsis thaliana pollen coat proteome. Science 292 2482–2485. [DOI] [PubMed] [Google Scholar]
- Mayfield, J.A., and Preuss, D. (2000). Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nat. Cell Biol. 2 128–130. [DOI] [PubMed] [Google Scholar]
- McCormick, S. (2004). Control of male gametophyte development. Plant Cell 16 (suppl.): S142–S153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.A., and Gubler, F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda, N., Iwase, A., Yamamoto, H., Yoshida, M., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17 2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassay with tobacco pith cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Pacini, E., Franchi, G., and Hesse, M. (1985). The tapetum: Its form, function, and possible phylogeny in embryophyta. Plant Syst. Evol. 149 155–185. [Google Scholar]
- Paxson-Sowders, D.M., Dodrill, C.H., Owen, H.A., and Makaroff, C.A. (2001). DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol. 127 1739–1749. [PMC free article] [PubMed] [Google Scholar]
- Piffanelli, P., Ross, J.H.E., and Murphy, D.J. (1998). Biogenesis and function of the lipidic structures of pollen grains. Sex. Plant Reprod. 11 65–80. [Google Scholar]
- Quesada, V., Macknight, R., Dean, C., and Simpson, G.G. (2003). Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J. 22 3142–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, T.V., Kaur, J., Agashe, B., Sundaresan, V., and Siddiqi, I. (2003). The DUET gene is necessary for chromosome organization and progression during male meiosis in Arabidopsis and encodes a PHD finger protein. Development 130 5975–5987. [DOI] [PubMed] [Google Scholar]
- Regan, S.M., and Moffatt, B.A. (1990). Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell 2 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinelli, P., Hu, Y., and Ma, H. (1998). Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Mol. Biol. 37 607–619. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schiefthaler, U., Balasubramanian, S., Sieber, P., Chevalier, D., Wisman, E., and Schneitz, K. (1999). Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, R.J., Spielman, M., and Dickinson, H.G. (2004). Stamen structure and function. Plant Cell 16(Suppl 1): S46–S60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X., et al. (2006). ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siloto, R.M., Findlay, K., Lopez-Villalobos, A., Yeung, E.C., Nykiforuk, C.L., and Moloney, M.M. (2006). The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18 1961–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, M., Belenghi, B., Delledonne, M., Menachem, E., and Levine, A. (1999). The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 11 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, A.M., Krober, S., Unte, U.S., Huijser, P., Dekker, K., and Saedler, H. (2003). The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33 413–423. [DOI] [PubMed] [Google Scholar]
- Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., Selbig, J., Muller, L.A., Rhee, S.Y., and Stitt, M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 914–939. [DOI] [PubMed] [Google Scholar]
- Tusher, V.G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcay-Barrena, G., and Wilson, Z.A. (2006). Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J. Exp. Bot. 57 2709–2717. [DOI] [PubMed] [Google Scholar]
- Wijeratne, A.J., Zhang, W., Sun, Y., Liu, W., Albert, R., Zheng, Z., Oppenheimer, D.G., Zhao, D., and Ma, H. (2007). Differential gene expression in Arabidopsis wild-type and mutant anthers: Insights into anther cell differentiation and regulatory networks. Plant J. 52 14–29. [DOI] [PubMed] [Google Scholar]
- Wilson, Z.A., Morroll, S.M., Dawson, J., Swarup, R., and Tighe, P.J. (2001). The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28 27–39. [DOI] [PubMed] [Google Scholar]
- Wysocka, J., Swigut, T., Xiao, H., Milne, T.A., Kwon, S.Y., Landry, J., Kaue, M., Tackett, A.J., Chait, B.T., Badenhorst, P., Wu, C., and Allis, C.D. (2006). A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442 86–90. [DOI] [PubMed] [Google Scholar]
- Yang, S.L., Xie, L.F., Mao, H.Z., Puah, C.S., Yang, W.C., Jiang, L., Sundaresan, V., and Ye, D. (2003. a). TAPETUM DETERMINANT1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15 2792–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W.C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Makaroff, C.A., and Ma, H. (2003. b). The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15 1281–1295. [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Sun, Y.L., Timofejeva, L., Chen, C., Grossniklaus, U., and Ma, H. (2006). Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM (DYT1) encoding a putative bHLH transcription factor. Development 133 3085–3095. [DOI] [PubMed] [Google Scholar]
- Zhao, D.Z., Wang, G.F., Speal, B., and Ma, H. (2002). The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z., Xia, Q., Dauk, M., Shen, W., Selvaraj, G., and Zou, J. (2003). Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 15 1872–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.