Abstract
The Arabidopsis thaliana floral homeotic gene AGAMOUS (AG) plays a central role in reproductive organ (stamen and carpel) development. AG RNA is expressed in the center of floral primordia from a time prior to the initiation of stamen and carpel primordia until late in flower development. While early AG expression acts in specification of stamens and carpels, the role, if any, of continued AG expression in later flower development is unknown. To examine the timing of AG action and its possible late-stage functions, we performed a series of time-course experiments using a transgenic line with inducible AG activity in an ag homozygous mutant background. We show that AG controls late-stage stamen development, including anther morphogenesis and dehiscence, as well as filament formation and elongation. We further show that AG coordinates late stamen maturation by controlling a biosynthetic gene of the lipid-derived phytohormone jasmonic acid (JA). Expression analysis and in vivo binding of AG indicate that AG directly regulates the transcription of a catalytic enzyme of JA, DEFECTIVE IN ANTHER DEHISCENCE1. Our results indicate that stamen identity and differentiation control by AG is achieved by the regulation of different transcriptional cascades in different floral stages, with organ specification induced early, followed by phytohormone biosynthesis to coordinate stamen maturation.
INTRODUCTION
The term homeotic selector gene has been used to describe genes that are able to trigger an entire developmental pathway (Garcia-Bellido, 1975). Plants and animals are considered to have evolved multicellular developmental processes independently, but homeotic genes in both kingdoms code for transcriptional factors, suggesting there is parallel logic in the developmental process (Meyerowitz, 2002). In animal development, Drosophila melanogaster homeotic HOX genes are suggested to regulate target genes at several levels of a regulatory hierarchy (Weatherbee et al., 1998). The HOX gene Ultrabithorax negatively regulates the expression of the signaling molecules Wingless, selected targets of Decapentaplegic, and its receptor Thickveins for organ identity control as well as for control of organ size (Capovilla and Botas, 1998; Weatherbee et al., 1998; Crickmore and Mann, 2006).
In plants, floral organ identity is controlled by different families of homeotic transcription factors. The ABC model predicts that floral organ identity is controlled by the combinatorial action of three classes of transcription factors (Coen and Meyerowitz, 1991). The Arabidopsis thaliana floral homeotic gene AGAMOUS (AG) is a C-class gene, and it encodes a MADS box transcription factor (Yanofsky et al., 1990). AG together with other MADS box proteins is necessary and sufficient for induction of reproductive organ development in Arabidopsis. For example, stamen identity is dependent on the expression of AG, the B-class genes APETALA3 (AP3) and PISTILLATA (PI), and SEPALLATA1/2/3 genes (Coen and Meyerowitz, 1991; Pelaz et al., 2000; Honma and Goto, 2001).
AG is thought to control developmental pathways by regulating downstream target genes responsible for stamen and carpel identity. Genome-wide analysis comparing the expression profiles of wild-type and ag mutant inflorescences showed that more than one thousand genes are regulated downstream of AG, especially in stamens (Wellmer et al., 2004). Microarray screening using inducible lines of AG activity enabled examination of immediate targets of AG during early organogenesis (Ito et al., 2004; Gomez-Mena et al., 2005). For example, there exists a positive-feedback regulation of AG itself and other MADS genes by AG (Gomez-Mena et al., 2005). It was also shown that AG controls microsporogenesis (pollen induction) by its regulation of the SPOROCYTELESS (SPL; NOZZLE) gene encoding a putative transcription factor (Ito et al., 2004). The induction of SPL by AG occurs immediately after ectopic AG activation and is causal in microsporogenesis, which happens several days later (Ito et al., 2004). However, the mechanisms by which an undifferentiated floral primordium responds to the genetic activities initiated by the homeotic protein AG and thereby develops mature stamens are still largely unknown, as is the type of regulatory network by which AG can orchestrate the expression of a large number of downstream genes necessary to produce functional mature stamens.
The plant phytohormone jasmonate or jasmonic acid (JA) plays an important function in stamen maturation (Feys et al., 1994; Xie et al., 1998; Sanders et al., 2000; Stintzi and Browse, 2000; Ishiguro et al., 2001; Park et al., 2002; von Malek et al., 2002). JA is a lipid-derived signaling molecule and functions in stress response and development in plants. Mutants that are deficient in the JA biosynthetic pathway, including defective in anther dehiscence1 (dad1), delayed dehiscence1 (dde1, opr3), and the JA response mutant coronatine insensitive1 (coi1), are male-sterile, with immature pollen in indehiscent (unopened) anthers. In these mutants, filament elongation is also inhibited or delayed. Therefore, JA is considered to coordinate such processes as anther dehiscence, filament elongation, and pollen maturation (Stintzi and Browse, 2000). The DAD1 gene encodes a chloroplastic lipase A1 that catalyzes the first step of JA biosynthesis (Ishiguro et al., 2001). DDE1/OPR3 encodes an isozyme of 12-oxo-phytodienoate (OPDA) reductase that reduces OPDA in JA biosynthesis (Sanders et al., 2000; Stintzi and Browse, 2000). Mutants of these catalytic genes can be rescued by exogenous JA application to developing floral buds. By contrast, coi1 mutants are insensitive to JA treatment, and the gene encodes an F-box protein that is thought to function in JA signal transduction (Xie et al., 1998). Thus, the synthesis and signaling of JA plays important functions in the temporal coordination of late stamen development.
In the developing stamen, the homeotic gene AG continues to be expressed after it specifies stamen identity (Bowman et al., 1991; Sieburth and Meyerowitz, 1997). AG RNA starts to be expressed in the floral primordia in whorls 3 and 4, where stamens and carpels will later form, at stage 3, when no organ primordia of stamens and carpels are yet initiated (Smyth et al., 1990; Yanofsky et al., 1990; Drews et al., 1991). The AG RNA remains present in filaments, connectives, and anther walls of developing stamens (Bowman et al., 1991). The later functions of AG in stamen development are not clear because eliminating AG activity prevents the specification of stamens and carpels and therefore eliminates the cell types in which later expression is found. Here, we examine the late-stage functions of AG using transgenic plants with inducible AG activity in an ag homozygous background and demonstrate that prolonged AG activity is necessary for the production of normal mature stamens. We propose that organ identity and differentiation control by plant homeotic proteins is achieved by the regulation of different transcriptional cascades in different floral stages, with organ specification induced early, followed by phytohormone biosynthesis to coordinate organ maturation.
RESULTS
Timing of AG-Induced Stamen Development
To examine the action of AG during different stages of reproductive organ development, we performed a series of timed activation experiments using a line transgenic for 35S:AG-GR in the ag-1 mutant background (Ito et al., 2004). This line contains a gene coding for a fusion of the AG protein and the steroid binding domain of the rat glucocorticoid receptor (Lloyd et al., 1994). The encoded fusion protein is held inactive in the cytoplasm of plant cells when glucocorticoids are absent. Upon application of the synthetic glucocorticoid dexamethasone (DEX), the fusion protein enters the nucleus, where it provides AG activity. The fusion gene is expressed ubiquitously under the cauliflower mosaic virus 35S protein promoter independently of floral stage and shows rescue of the ag-1 mutant phenotypes of homeotic conversion of stamens to petals and conversion of carpels to a new flower when DEX is provided continuously (Ito et al., 2004) (Figure 1A). As the transgene is expressed in all floral tissues, activation of its encoded protein by DEX also leads to the gain-of-function phenotypes of formation of carpelloid sepals in the first whorl of the developing flowers and of stamens in the second whorl (Mandel et al., 1992; Mizukami and Ma, 1992).
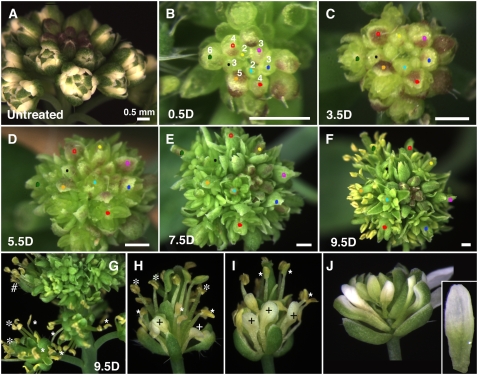
Figure 1.
Timing of AG-Induced Stamen Development.
(A) Intact inflorescence of ag-1 35S:AG-GR before DEX treatments.
(B) to (F) A dissected inflorescence of ag-1 35S:AG-GR to identify floral buds at stages 2 to 6 were treated with 10 μM DEX solution at 12-h intervals up to four times and observed at 0.5 d (B), 3.5 d (C), 5.5 d (D), 7.5 d (E), and 9.5 d (F) after the initial DEX treatment. The same colored dot represents the same floral bud. Floral stages at the time of initial DEX treatment (day 0) are indicated as follows: green, 6; orange, 5; red and open red, 4; black, blue, and purple, 3; yellow and light blue, 2.
(G) to (J) Intact inflorescences were treated with 10 μM DEX solution at 12-h intervals four times and observed 9.5 d after the initial DEX treatment.
(G) Flower (#) that had been at stage 6 in day 0 shows normal mature stamens. Flowers at approximately stage 7 in day 0 show incompletely developed stamens with sterile anthers (smaller asterisks) in whorls 2 and 3 even though inner stamens (larger asterisks) were normal.
(H) and (I) Flowers at later than stage 7 in day 0, which were produced at lower positions on the stem shown in (G), show organs varied from fertile stamens (larger asterisks), sterile stamens (smaller asterisks), to stalked petals (+).
(J) Mature petals did not show morphological change 9.5 d after DEX treatments. Inset shows a close-up of the petal.
Bars = 0.5 mm.
The inflorescences of ag-1 35S:AG-GR plants were dissected under a stereomicroscope to observe floral buds at early stages. Floral buds at stage 3 or 4 were identified based on morphology of sepal primordia (Smyth et al., 1990). Based on the size increment and growth duration relative to wild-type floral buds, sequential stages up to stage 6 were defined. The inflorescences were then treated with 10 μM DEX every day and observed daily after the treatments (Figures 1B to 1J). For the first 2 d after the initial DEX treatment, no visible morphological changes were observed except for the stage-dependent increased size of floral buds (Figure 1B; data not shown). Young floral buds that had been at stage 3∼4 at the time of initial DEX treatment started to show anther structures by 5.5 d, and these anther structures with locules became distinct in 7.5 d (Figures 1D and 1E; data not shown). These developing stamens were composed of immature anthers and short filaments by 9.5 d (Figure 1F). These floral buds reached anthesis with mature, normal stamens in 11 to 12 d (see below).
By contrast, floral buds that had been at stage 5∼6 at the time of initial DEX treatment showed stamen-like characters by 3.5 d (Figure 1C). Locule-like compartments started to be observed in the developing organs of these floral buds (Figure 1C). At 5.5 d, these floral buds showed clear anther structures (Figure 1D). By 7.5 d, developing stamens and carpels became very distinct (Figure 1E). The stamens showed markedly elongating filaments. At 9.5 d, the first fully developed stamens with dehisced anthers and fully elongated filaments were observed in open flowers (Figure 1F). These results suggest that petal identity in floral buds at stage 5∼6 is not determined irreversibly and that stage 5∼6 second and third whorl organs, originally fated to be petals, can be respecified to become normal stamens.
AG Has Stage-Specific Functions in Stamen Organogenesis
Next, we examined the effect of induced AG activity in floral buds at stage 7 or older in undissected inflorescences (Figure 1G). Flowers that were at stage 7 or older at the time of initial DEX treatment reached anthesis earlier than 9.5 d, but no mature stamens were observed in these inflorescences until 9.5 d after treatment. They started to show normal-looking dehisced stamens in whorls more central than the second and third in 9.5 d (larger asterisks in Figures 1G and 1H). However, incomplete stamens were observed in whorls 2 and 3 of these flowers, which were produced earlier than the inner organs. These organs varied from stamens with sterile anthers to stalked petals with different degrees of filament development (smaller asterisks and + in Figures 1G to 1I). None of these showed microsporogenesis. Thus, in the second- and third-whorl organs at stage 7, the regulatory cascades for microsporogenesis can no longer be induced by ectopic AG, although anther morphogenesis and filament elongation are still induced (Figure 1G). At later than stage 7, the cascade for filament elongation still can be induced even after anther morphogenesis is not any longer induced (Figures 1H and 1I). AG activation in nearly or fully developed petals in the floral buds at much later stages had no effect on petal morphology (Figure 1J, inset). These results show that the microsporogenesis function of AG needs to be activated by stage 6 but that anther and filament morphology can be induced in developing petals even at later stages. In summary, AG has stage-specific functions in stamen organogenesis.
Timed Activation Experiments of AG
To examine the phenotypic effects of timed AG activity in detail, we performed a series of timed activation experiments instead of continuous induction of AG. Inflorescences of ag-1 35S:AG-GR were treated with DEX once, twice, three times, or four times at 12-h intervals. Floral buds that had been at stage 3 at the time of initial DEX treatment were observed 12 d later when they reached anthesis (Figures 2A to 2E and 3A to 3E; see Supplemental Figures 1A to 1F online). All of these flowers produced carpels in whorl 4, suggesting that a single induction of AG activity is sufficient to induce carpels (Figures 2B to 2E and 3F). By contrast, in the organs produced in whorls 2 and 3 of these flowers, stamen morphology showed clear correlation with the number of DEX treatments. For example, in inflorescences treated once with 10 μM DEX, we observed petals with two-loculed structures (loculed petals) on the adaxial surface, in addition to normal petals (Ito et al., 2004; Figures 2B and 3B; see Supplemental Figure 1C online). Inside the locules of such a laminar stamen, normal-looking pollen and tapetum structure were observed (Figure 3B, inset). A single treatment with 10-fold less concentrated DEX solution led to petals with a smaller proportion bearing locules (see Supplemental Figure 1B online). If two 10 μM DEX treatments were given, flowers showed structures with more stamenoid features (Figures 2C and 3C; see Supplemental Figure 1D online). In addition to the loculed petals, some organs showed basal narrowing, thereby forming a filament (Figure 3C). Three DEX treatments resulted in the complete reduction of the basal part in all second- and third-whorl organs (Figures 2D and 3D; see Supplemental Figure 1E online). The anther appears to contain normal cell types, including stomium (Figure 3D, inset). Further induction of AG activity resulted in filament elongation and the eventual dehiscence of anthers to release pollen (Figures 2E and 3E; see Supplemental Figure 1F online). These results suggest that normal Arabidopsis stamen development in the ag-1 35S:AG-GR strain requires repeated AG activation. Furthermore, we conclude that AG controls various aspects of stamen morphogenesis.
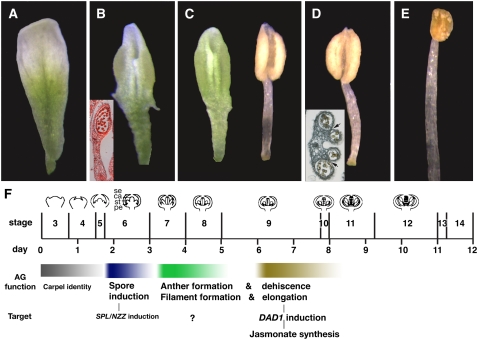
Figure 2.
Timed Activation Experiments.
ag-1 35S:AG-GR floral buds that had been at stage 3 at the time of initial DEX treatment were observed 12 d after a series of timed treatments with 10 μM DEX as shown below: (A) untreated, (B) one treatment, (C) two treatments at 12-h intervals, (D) three treatments at 12-h intervals, (E) four treatments at 12-h intervals, (F) two treatments at 1-d intervals, (G) three treatments at 1-d intervals, (H) four treatments at 1-d intervals, (I) two treatments at 2-d intervals, and (J) three treatments at 2-d intervals. Normal-looking stamens with dehiscent anthers and elongated filaments were observed in (E), (H), and (J).
Figure 3.
Gradual Transformation from Petals to Stamens by Continued AG Activity.
(A) Petal produced in whorl 3 in the ag-1 mutant.
(B) Locule-bearing petal in flowers at anthesis 12 d after a single 10 μM DEX treatment. Inset, an aceto-orcein–stained paraffin section of a laminar stamen.
(C) to (E) Floral organs observed at second or third whorl positions in flowers at anthesis 12 d after two (C), three (D), or four (E) 10 μM DEX treatments at 12-h intervals. Inset in (D) shows a transverse section of an indehiscent anther with normal-looking stomium (arrows).
(F) Timeline of functional AG activity. Duration of floral stages was referred from Smyth et al. (1990). Diagrams of developing flower are shown at the top. We confirmed that similar timing of morphological stages was observed under our growth conditions by observing the growth of ag-1 35S:AG-GR and wild-type inflorescences (as shown in Figure 1). Duration of AG expression necessary for each function was estimated from the experiments described here. AG controls SPL (NZZ) and DAD1 for spore formation and stamen maturation, respectively (Ito et al., 2004; this work). se, sepal primordia; ca, carpel primordia; st, stamen primordia; pe, petal primordia.
Prolonged AG Activity Is Necessary for Normal Stamen Development
To examine whether the effect of repeated DEX treatments at 12-h intervals is due to an increase in the duration, or the level, of AG activity, inflorescences were treated at 1- or 2-d intervals (Figures 2F to 2J). If prolonged AG activity is necessary, treatments at longer intervals should rescue the mutant phenotype. If quantitatively higher levels of nuclear AG protein is produced by the repeated treatments, and this affects different AG downstream cascades, treatments at longer intervals should not efficiently rescue the mutant phenotype.
DEX treatments at 2-d intervals clearly showed stronger effects than the same number of treatments at 12-h or 1-d intervals. In inflorescences treated twice with DEX at 2-d intervals, the flowers showed stamens with locules and with partly elongated filaments, which was similar to three treatments at 0.5- or 1-d intervals (cf. Figures 2I with 2D and 2G). Three DEX treatments at 2-d intervals were sufficient to induce normal stamen structures (Figure 2J); three treatments at 2-d intervals had a similar effect to four treatments at 12-h or 1-d intervals (Figures 2E and 2H). These results suggest that prolonged duration, and not a higher level, of AG activity was responsible for the differences in stamen development seen after different numbers of DEX applications. Furthermore, we can conclude that normal stamen development requires prolonged AG activity.
Late Expression of AG Is Functional for Stamen Organogenesis and Maturation
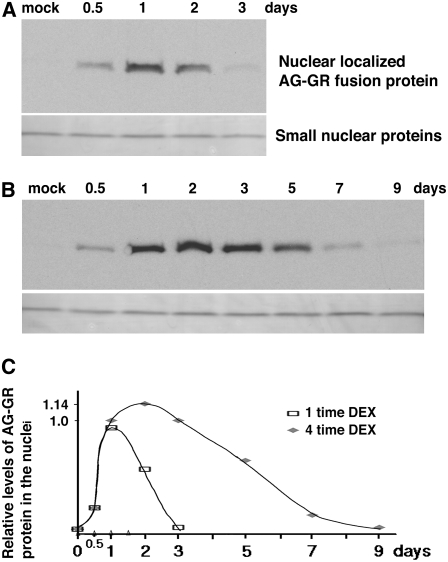
To check the time course of AG function after DEX treatment, we examined dynamic accumulation of AG-GR protein in nuclei caused by repeated DEX treatment at 12-h intervals. ag-1 35S:AG-GR transgenic plants were treated once or four times with DEX, and the inflorescences were harvested after 0.5, 1, 2, 3, 5, 7, and 9 d (Figure 4). A single DEX treatment starting from stage 3 floral buds leads to loculed petals, and four treatments lead to normal stamens (Figures 3B and 3E). The AG-GR fusion protein localized in nuclei was examined by protein gel blotting using an AG-specific antiserum. Barely detectable signal was observed in the nuclear extract of the mock-treated inflorescences (Figures 4A and 4B). In the inflorescences treated once with DEX, the signal for AG-GR fusion protein was detected at the 0.5-d time point, increased at the 1-d time point, and then decreased to lower levels at the 2-d time points, being barely detectable afterward (Figures 4A and 4C). In inflorescences treated four times with DEX, levels of AG fusion protein increased at the 1-d time point (similarly to what was observed with the single DEX treatment) and further increased ∼14% when reaching the maximum at the 2-d time points (Figures 4B and 4C). After the 2-d time point, AG levels gradually declined, reaching similar levels by 7 d to those observed at the 0.5-d time point (Figures 4B and 4C). These data showed that four DEX treatments at 12-h intervals provide for presence of nuclear AG protein for longer than 7 d. The duration of AG activity is sufficient to fully rescue the ag-1 mutant phenotype if it is present from stage 3 (Figure 2E). However, floral buds that are treated with DEX at earlier stages (that reached anthesis in 13 or 14 d) showed stamens with short filaments and indehiscent anthers, evidently as effects of a deficit in AG activity late in development (see Supplemental Figures 2A and 2B online). This suggests that this duration of AG activity is not quite sufficient to fully perform AG functions when started earlier than stage 3. In our growth conditions, a floral bud at stage 3 reached stage 11 in 8 d (Figure 3F). This duration matches that of endogenous AG expression; AG starts to be expressed at stage 3 (Yanofsky et al., 1990; Drews et al., 1991). AG continues to be expressed in filaments, connectives, and anther walls of developing stamens at stages 10 to 11 (Bowman et al., 1991). The observation that fewer than four DEX treatments or treatment earlier than stage 3 are ineffective in completely complementing the ag mutant phenotypes in stamens implies that late expression of AG is important in filament elongation and anther dehiscence (Figure 3F). In summary, prolonged AG activity until late stamen development is necessary to produce normal mature stamens. Stamen identity and differentiation control by plant homeotic protein AG is achieved by the regulation of different transcriptional cascades in different floral stages, with organ specification induced early, followed by the activities for organogenesis and maturation.
Figure 4.
Repeated DEX Treatments at 12-h Intervals Result in Prolonged Nuclear Accumulation of AG-GR Protein.
(A) and (B) Protein gel blot detection of AG-GR fusion protein using crude nuclear extract isolated from ag-1 35S:AG-GR plants treated once (A) or four times (B) with 10 μM DEX at 12-h intervals (as shown by Δ in [C]). Samples were harvested 0.5, 1, 2, and 3 d after initial treatment for (A) or 0.5, 1, 2, 3, 5, 7, and 9 d after the treatment for (B). Nuclear protein was loaded on polyacrylamide gels, and AG-GR fusion protein was detected by protein gel blot analysis. Control panels show a band of small nuclear proteins, which are <25 kD.
(C) Graph of relative amount of AG-GR fusion protein localized in the nuclei after a single DEX treatment (open squares) or four DEX treatments (gray diamonds). Band intensities of AG-GR signal were measured by NIH ImageJ and normalized using the band intensities of the small nuclear proteins. The 12-h point was used to normalize the two treatment values.
A Catalytic Enzyme for JA Biosynthesis Shows AG-Dependent mRNA Expression in Late Stamen Development
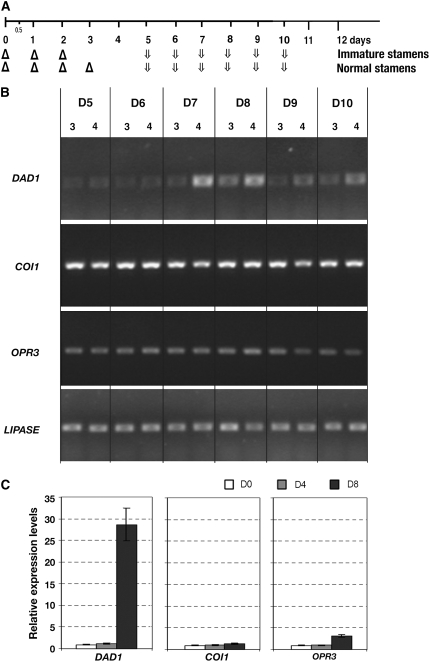
Loss-of-function mutants of genes involved in JA biosynthesis or its signal transduction pathway show similar defects in stamens to those caused by a short duration of AG activity (as observed in Figure 3D). To examine the link between the JA-related genes and AG, we examined the AG-dependent mRNA expression of JA-related genes, including two JA catalytic enzymes, DAD1, a phospholipase A1 that catalyzes the initial step of JA biosynthesis, and OPR3, which reduces OPDA for JA synthesis, and one F-box JA signal transducer, COI1 (Xie et al., 1998; Stintzi and Browse, 2000; Ishiguro et al., 2001). ag-1 35S:AG-GR transgenic plants were treated three or four times with DEX at 1-d intervals, which would give immature or mature stamens in day 12, respectively (Figures 2G and 2H), and the developing inflorescences were harvested (Figure 5A). The RNA samples were used for semiquantitative RT-PCR and real-time PCR to test the mRNA expression of DAD1, COI1, and OPR3 (Figures 5B and 5C). AG-dependent upregulation of DAD1 mRNA was detected at the day 7 time point in inflorescences treated four times with DEX, and the signals remain relatively high in day 8 and then slightly declined in days 9 and 10 (Figure 5B). In samples treated three times with DEX, slight induction of DAD1 mRNA level was detected in day 8, but such expression reduced to the background level in days 9 and 10 (Figure 5B). According to real-time PCR analysis using independently prepared samples treated four times with DEX, the DAD1 transcripts stayed low in day 0 and day 4 samples but was induced ∼28-fold in day 8 compared with day 0 (Figure 5C). By contrast, COI1 and OPR3 showed relatively stable expression or a low level of induction after AG activation, respectively (Figures 5B and 5C). These results show that AG strongly upregulates DAD1 expression in stamen maturation.
Figure 5.
DAD1 Is Induced in an AG-Dependent Manner.
(A) Inflorescences of ag-1 35S:AG-GR plants were treated (Δ) three or four times with 10 μM DEX at 1-d intervals and were harvested (arrows) 5, 6, 7, 8, 9, and 10 d after the initial treatments.
(B) Semiquantitative RT-PCR using specific primers for DAD1, COI1, OPR3, and a control lipase gene to detect the RNA expression levels. Primers used in this assay are shown in Supplemental Table 1 online.
(C) Real-time PCR for DAD1, COI1, and OPR3 using inflorescences of ag-1 35S:AG-GR in days 0, 4, and 8 treated four times with DEX. Primers used for analyses are shown in Supplemental Table 1 online. The y axis shows expression levels relative to day 0. The PCR reaction was done in triplicate, and the results are shown as means ± se.
Catalytic Activity of DAD1 Is Responsible for AG-Dependent Stamen Maturation
To determine whether DAD1 expression is responsible for AG-dependent stamen maturation, we first examined the effect of exogenously applied JA to immature stamens caused by short duration of AG activity. If the late function of AG in stamens is to induce JA by transcriptionally upregulating DAD1, JA treatment should be sufficient to rescue the immature stamens. By contrast, if the DAD1 transcript is induced as one of the consequences of late stamen development, JA treatment may not be sufficient to complement a stamen defect in maturation. ag-1 35S:AG-GR transgenic plants were treated three times with DEX at 1-d intervals, further treated with methyl jasmonate in days 9 and 10, and flower phenotypes were observed in day 12. The concentration of exogenously applied methyl jasmonate in different experiments varies in an ∼10-fold range in previous publications (Feys et al., 1994; Xie et al., 1998; Sanders et al., 2000; Stintzi and Browse, 2000; Ishiguro et al., 2001; Park et al., 2002; von Malek et al., 2002; Nagpal et al., 2005; Sasaki-Sekimoto et al., 2005). Thus, we tested two different concentrations, 50 and 500 μM, of JA. The JA treatments at both concentrations to inflorescences treated three times with DEX efficiently rescued immature stamen phenotypes of short filaments and indehiscent anthers, leading to dehiscent anthers with elongated filaments resembling those treated four times with DEX (Figures 6A to 6E). This suggests that one late function of AG in stamens is to produce JA by activating its biosynthetic pathway.
Figure 6.
Methyl-Jasmonate and Its Precursor LA Can Rescue the Immature Stamens Caused by an Insufficient Duration of AG Activity.
(A) Flower of untreated inflorescence of ag-1 35S:AG-GR plants.
(B) Flower produced 12 d (stage 3 in day 0) after four treatments with 10 μM DEX at 1-d intervals.
(C) to (J) Flowers from the inflorescences treated three times with 10 μM DEX at 1-d intervals and either untreated (C) or further treated with methyl jasmonate, GA (GA3), LA, and/or linoleic acid once in day 9 and once in day 10 as shown below: (C) mock control, (D) 500 μM JA, (E) 50 μM JA, (F) 100 μM GA, (G) 500 μM JA and 100 μM GA, (H) 50 μM JA and 100 μM GA, (I) 0.1% (v/v) LA, (J) 0.1% (v/v) linoleic acid. Insets in (C) and (E) show close-up images of stamens. Treatments by LA and linoleic acid caused partial drying and withering effects on plants at the same level, but the stamen maturation was induced only by LA treatments.
AG was previously shown to regulate the expression of GA4, which encodes GA3-β-hydroxylase, an enzyme that catalyzes the last step of another phytohormone gibberellic acid (GA) biosynthesis (Gomez-Mena et al., 2005), suggesting that GA might also be involved in late stamen development. Thus, we tested if the exogenous application of GA could also complement the immature stamen phenotype. However, treatment with 100 μM GA in the same way showed no visible effects in stamen maturation (Figure 6F). A combined application of GA and JA showed similar effects to JA treatments alone (Figures 6G and 6H). This result suggests that AG controls late stamen maturation solely through JA and not with GA.
DAD1 protein catalyzes the initial step of JA biosynthesis, and application of the catalytic product of DAD1, α-linolenic acid (LA; 18:3) rescues the dad1 floral buds but cannot rescue the mutant flowers of other downstream catalytic enzymes of JA (e.g., OPR3 or allene oxide synthase) (Ishiguro et al., 2001; Park et al., 2002). To examine whether AG also controls the transcription of multiple genes involved in later steps of the JA biosynthesis, we tested if LA can rescue the immature stamens caused by short duration of AG activity. Two days after LA treatments to inflorescences treated three times with DEX, the newly opened flower showed normal-looking stamens with elongated filaments and dehiscent anthers (Figure 6I). By contrast, the inflorescences treated with a lipid of similar structure, linoleic acid (18:2), which cannot be used as a JA precursor, did not rescue the immature stamens (Figure 6J). Therefore, the other JA-synthetic enzymatic activities that function downstream of DAD1 seem to be AG independent. Our results show that the DAD1 activity leading to JA production is responsible for AG-dependent stamen maturation (Figure 3F).
The DAD1 Promoter Is Ectopically Induced by Ectopic AG Expression
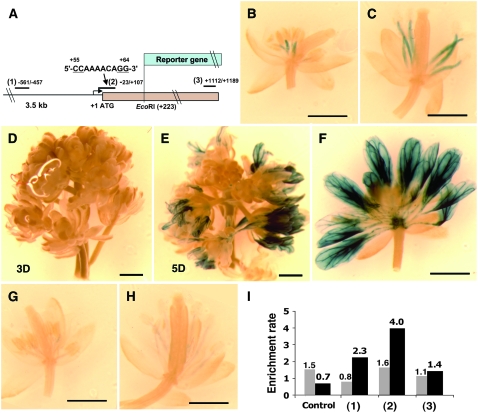
To examine how AG regulates the transcription of DAD1, the DAD1 promoter was fused with a bacterial β-glucuronidase (GUS) reporter gene, and AG-dependent reporter gene expression was examined. The 3.5-kb 5′ upstream region and 0.2-kb coding region of DAD1 was fused in frame to the β-glucuronidase coding region (Figure 7A). GUS activity was detected in stamen filaments in the middle and late stages of stamen development (Ishiguro et al., 2001) (Figures 7B and 7C). AG is continuously expressed in the filaments of developing stamens till the late stamen development (Bowman et al., 1991; Sieburth and Meyerowitz, 1997). These overlapping expression patterns are in agreement with the idea that AG is required to activate DAD1 expression.
Figure 7.
AG Directly Regulates DAD1.
(A) Promoter and gene structure of DAD1. A CArG-box sequence is located in the coding region near the initiation codon (+55/+64). The promoter-GUS construct (pDAD-GUS) was produced as a translational fusion to the N-terminal part of DAD1 protein (amino acids 1 to 72). Black bars numbered 1 to 3 indicate the regions used for real-time PCR analysis to detect enrichments in the ChIP assay shown in (I).
(B) and (C) Flowers transgenic for pDAD-GUS at stage 10 (B) and at anthesis (C).
(D) to (F) Inflorescences of ag-1 35S:AG-GR and pDAD1:GUS 3 d (D) or 5 d ([E] and [F]) after four daily treatments with 10 μM DEX.
(G) and (H) Flowers at stage 10 (G) and at anthesis (H) transgenic for mutated pDAD:GUS. Bars in (B) to (H) = 1 mm.
(I) ChIP using ag-1 35S:AG-GR inflorescences in day 7 after four DEX treatments. Nuclear extract was immunopurified by AG-specific antibody (shown as black bars) or IgG control (gray bars). Primers used for analysis are shown in (A) and in Supplemental Table 1 online. The y axis shows enrichment rates compared with an unrelated control gene.
To test the inducibility of the DAD1 promoter by AG, we transformed the DAD1 reporter construct into the AG-inducible line ag-1 35S:AG-GR. No reporter gene expression was detected in the uninduced condition, suggesting that DAD1 expression is AG dependent. The plants were treated with DEX daily, and GUS activities were examined daily. We did not see any GUS staining in inflorescences of day 0 and 3 (Figure 7D). In day 5 samples, strong GUS activity starts to be observed in developing petals of maturing flowers (Figure 7E). The strong GUS staining was also observed in vasculature of mature petals (Figure 7F). In normal flower development, DAD1 is expressed in filaments of stamens but not in petals. Our results showed that ectopic expression of AG is sufficient to ectopically induce DAD1 in mature petals. This suggests that AG closely regulates DAD1 expression in a tissue-specific manner. In day 5, reporter gene activity was observed only in mature petals but not in early organs produced in the central region of the flower (Figure 7F). These results suggest that induction of DAD1 by AG is stage specific and that AG-dependent and -independent cues are necessary for the stage- and tissue-specific induction of DAD1.
DAD1 Is a Late Target of AG
To examine whether AG directly regulates the expression of DAD1, we first examined if there are putative binding sites of AG in the DAD1 genomic region used in the promoter constructs. The consensus binding sequence of MADS proteins, which consists of the 10-bp CArG-box [5′-CC(A/T)4NNGG-3′] (Riechmann et al., 1996) was not found in the upstream region but was found in the protein coding region at ∼60 bp from the initiation codon (Figure 7A). Another floral regulator, SUPERMAN, contains cis-regulatory elements in the protein coding region near its N terminus (Ito et al., 2003), so it is possible that this CArG-box is functional. To test if this CArG-box sequence is necessary for the proper expression of DAD1, we mutagenized the site in the promoter construct to prevent the binding of MADS proteins [5′-CC(A/T)4NNGG-3′ was changed to 5′-GG(A/T)4NNCC-3′] to abolish the binding of AG protein (Ito et al., 2004) and transformed into plants. The mutation in the CArG-box causes amino acid change from LPKQDV to LGKHHV, but both of the codon usages of the new sites are common in Arabidopsis (GGA for G 36.7%; CAC for H 38.6%; from www.kazusa.or.jp/codon). The majority of T1 plants with mutated construct showed reduced or barely detectable GUS expression (Figures 7G and 7H). Nine out of 14 (64%) of T1 transgenic plants for the wild-type construct conferred the GUS expression in filaments of developing stamens (Figures 7B and 7C). The rest of the T1 plants showed no, or abnormal, expression, most possibly due to the positional effect of insertion sites. By contrast, only three out of 14 (21%) of T1 lines with the mutant construct showed normal levels of expression in filaments, and five out of 14 (35%) of lines showed reduced or barely detectable expression in filaments (Figures 7G and 7H). This suggests that the CArG-box located in the DAD1 coding region is necessary for the proper expression of the DAD1 gene.
To test if AG directly binds this site, we performed a chromatin immunoprecipitation (ChIP) assay. The ag-1 35S:AG-GR plants were treated four times with DEX daily, and the inflorescences were harvested 3 and 7 d after the initial DEX treatments. Nuclear extract was immunopurified using an AG-specific antibody (Ito et al., 1997). Thereafter, the purified DNA was used as a template for real-time PCR with primer sets located in the DAD1 genome or in control genes (three primer sets in the DAD1 genomic region and two negative controls, phosphofructokinase [PFK] and Mu-like transposon; Mu was used as a standard). In day 3, no significant enrichment was observed in the assay using AG-specific antibody (see Supplemental Figure 3 online). By contrast, in day 7 samples precipitated with AG-antibody, primer sets for a control gene and 3′ coding region of the DAD1 gene did not show any significant enrichment, but the primer sets around the CArG-box gave fourfold enrichment (Figure 7I). The same assay in day 7 samples using preimmune serum showed substantially lower levels of enrichment rate. The primer set at −500 bp upstream showed a weak level of enrichment. This may be due to the AG binding to the CArG-box sequences or due to other putative in vivo binding sites located in the upstream promoter. These results of AG binding to the DAD1 genome were confirmed using the ap1 cal 35S:AP1-GR transgenic plants, in which flower development can be synchronized (Wellmer et al., 2006). The plants were treated once with DEX, and the inflorescences were harvested 2 and 6 d later, which corresponds to stage 3 and stages 8 to 9, respectively. We performed the ChIP assay using the same AG-specific antibody. In day 2, no enrichment was observed (see Supplemental Figure 4 online). By contrast, in day 6, significant enrichment was observed around the CArG-box sequences (see Supplemental Figure 4 online). These results indicate that AG directly binds the 5′ coding region of DAD1 and upregulates expression in late stamen development.
DISCUSSION
We examined the stage-specific effects of induced AG activity in flowers using a constitutively expressed AG-GR fusion protein, whose activation is regulated by exogenous application of DEX. It was not clearly known if AG acts transiently at the top of a cascade of other regulators, or if it acts persistently to regulate different downstream genes at different developmental times. We showed that the mode of AG function is a persistent action throughout much of stamen development by controlling different steps at different developmental stages. Previous morphological assays using temperature-sensitive B class mutants, ap3-1 in Arabidopsis and deficiens-101 in Antirrhinum majus, suggested that B-class gene activities are necessary not only for early stages of petal and stamen development but also for late organogenesis (Bowman et al., 1989; Zachgo et al., 1995). Genome-wide screening for downstream genes of such B-class MADS proteins identified genes required for the basic cellular processes responsible for petal and stamen morphogenesis, but a very low percentage of the downstream genes code for transcription factors, suggesting that that B-class genes exert their regulatory control rather directly at late stages (Zik and Irish, 2003; Bey et al., 2004). Our study, together with others, provides direct evidence to support the idea that plant homeotic selector genes, like Drosophila HOX genes (Weatherbee et al., 1998), regulate target genes to control morphogenesis at several levels of a regulatory hierarchy.
Early and Late Targets of AG in Stamen Development
We show that petal primordia in floral buds as late as stage 6 can be transformed into normal stamens by ectopic AG activity. It was recently shown that AG directly regulates SPL (NOZZLE), a gene necessary for microsporocyte formation (Ito et al., 2004). The SPL RNA begins to be expressed in stage 5 in the stamen primordia of wild-type flowers (Yang et al., 1999; Ito et al., 2004). In the inducible line ag-1 35S:AG-GR, induction of SPL expression was observed rapidly after DEX treatments in limited regions of organ primordia at certain stages (Ito et al., 2004). In the experiments described here, petals with locules induced 12 d after a single DEX treatment showed pollen grains inside their locules. This indicates that a short burst of AG activity at stage 5∼6 is sufficient to functionally activate SPL. On the other hand, the ectopic AG activity initiated in developing petals at later stages leads to sterile stalked petals with filamentous structures. This suggests that some late targets of AG can be induced independently of early targets. In addition, prolonged AG activity is necessary for anther morphogenesis and dehiscence as well as for filament formation and elongation, indicating that some steps of stamen development, unlike formation of microspores, require long-term AG activation.
Regulation of DAD1
We further showed that AG regulates the DAD1 gene encoding the catalytic enzyme for the initial step of JA biosynthesis in late stamen development. The induction of DAD1 needs several days of prolonged AG activity. For mRNA expression analysis using floral buds smaller than 0.5 mm in diameter (younger than at ∼ stage 10) of ag-1 35S:AG-GR plants, the AG-dependent induction of DAD1 was observed in day 7 after the activation of AG. This induction of DAD1 leads to mature stamens in day 11∼12. By contrast, in reporter gene assays using whole inflorescences of the same AG-inducible lines, ectopic induction of DAD1 was observed even in mature petals in 5 d. Ectopic induction of DAD1 in mature petals, in vivo binding by ChIP, and promoter mutagenesis all indicate that DAD1 is a late target of AG in stamen development.
Why does AG induce DAD1 only in late stamen development? Our ChIP data showed that AG cannot bind to the DAD1 genomic region in early flower development, suggesting that accessibility of AG protein to DAD1 genomic sequences is developmentally regulated. The binding site of AG in the DAD1 gene contains 10- bp CArG-box core sequences but has three mismatches from the 16-bp consensus binding sequence bound by an AG homodimer in vitro (Huang et al., 1993; Shiraishi et al., 1993). Thus, DAD1 expression might need a late-stage-specific coregulator, which enhances the AG binding to the target site. DAD1 genomic DNA also might be regulated by chromatin modification during stamen development.
The B-class genes AP3 and PI are also necessary for stamen identity (Meyerowitz et al., 1991). Phenotypic analysis of the ap3-1 temperature-sensitive mutant showed that AP3 activity is necessary from stages 5 to 6 to stages 7 to 8 in specifying stamen identity (Bowman et al., 1989). Our results on the initial timing of AG activity match with this; AG activity starting from stage 6 is sufficient to induce stamens. However, our results also showed that prolonged AG expression up to stage 11 is necessary to produce fully mature stamens with dehiscent anthers and elongated filaments. The longer requirement for AG activity than for AP3 activity in stamens could mean that the late functions of AG in stamens may be independent of B-class gene activity, or it could indicate that the AP3 protein is stable and persists after temperature shifts in the ap3-1 mutant (which affects RNA splicing) (Sablowski and Meyerowitz, 1998; Yi and Jack, 1998).
AG Coordinates the Maturation of Stamens by Inducing the JA Biosynthetic Pathway
One of the intriguing questions in developmental biology is how homeotic proteins orchestrate the expression of thousands of genes to form organs with proper shape and functions. For efficient fertilization in flowering plants, efficient formation of pollen grains is important, as is the temporal coordination of the elongation of filaments and the maturation of pollen and dehiscence of anthers. JA controls all three critical steps in stamen maturation in Arabidopsis (Feys et al., 1994; Xie et al., 1998; Sanders et al., 2000; Stintzi and Browse, 2000; Ishiguro et al., 2001; Park et al., 2002; von Malek et al., 2002). Here, we show that the homeotic protein AG acts to coordinate stamen maturation by controlling the initial enzymatic step in JA synthesis. Ovule identity is also controlled by AG and AG subfamily genes in a redundant manner (Pinyopich et al., 2003). It would be of interest to see if there is an AG-dependent mechanism to coordinate ovule maturation along with stamen development to allow Arabidopsis plants to achieve efficient fertilization.
Evolutionary Conservation of AG Function in Late Stamen Development
The Antirrhinum AG ortholog FARINELLI (FAR) is continuously expressed in developing stamens (Davies et al., 1999). far mutants are male-sterile with defects in anther and microspore development (Davies et al., 1999). One of the rice (Oryza sativa) C-class genes, OSMADS58, is continuously expressed in developing stamens. RNA-silenced lines of osmads58-1s1 showed white and shrunken anthers (Yamaguchi et al., 2006). This suggests that the late-stage continuous expression of AG subfamily genes in developing stamens might have similar and evolutionarily conserved functions in widely diverged flowering plants. However, it is not yet clear whether JA plays similar functions in stamen maturation in species other than Arabidopsis. Tomato (Solanum lycopersicum) mutants defective in JA biosynthesis are not male-sterile (Li et al., 2001). In tobacco (Nicotiana tabacum) mutants insensitive to ethylene, anther dehiscence is delayed (Rieu et al., 2003). Flavonoids play an important function in the maturation of pollen in maize (Zea mays) and petunia (Petunia hybrida) but not Arabidopsis (Coe et al., 1981; Mo et al., 1992; van der Meer et al., 1992; Burbulis et al., 1996; Ylstra et al., 1996; Napoli et al., 1999). Thus, in other species than Arabidopsis, other phytohormone/signaling pathways might be activated to regulate the stamen maturation downstream of the AG subfamily genes.
The stamens of basal ANITA grade plants (the earliest branching lineages of the angiosperms) have thick connectives, and some of them are leaf-shaped (Endress, 2001). In one of the ANITA plants with laminar stamens, Magnolia, the AG ortholog is expressed throughout stamen primordia at early stages, and in later stages, its expression is localized in sporogenous cells and tapetum but not maintained in the laminar region (M. Ito and M. Hasebe, personal communication). This permits the speculation that the morphological change of stamens from laminar to filamentous structures might be dependent on the duration or the expression domain of AG (perhaps along with other MADS box floral homeotic proteins). Morphological changes of laminar stamens to stamens with filaments, and vice versa, are suggested to have occurred independently several times during angiosperm evolution (Hufford, 1996). This simple possible mechanism (e.g., changes in cis-elements of the homeotic genes) might have facilitated the laminar-filamentous transformations. Multiple evolutionary origins and reversals might also explain why the function of JA in stamen maturation does not appear to be highly conserved.
METHODS
Plant Materials and Chemical Treatments
All plants used in this study are in the Landsberg erecta background and were grown at 22°C under continuous light. Paraffin sections were obtained according to a published protocol (Ito et al., 2003). Plant photographs were taken by the Zeiss Stemi SV11 stereomicroscope attached to a Zeiss Axiocam. DEX treatment was done in submerging the inflorescences in solution containing either 1 or 10 μM DEX together with 0.015% Silwet L-77 for ∼1 min. The initial DEX treatment was counted as done in day 0. Exogenous phytohormone or lipid treatments were done in days 9 and 10 by submerging inflorescences in solutions containing 0.05% Tween 20 and various phytohormones or lipids: 500 or 50 μM methyl jasmonate, 100 μM GA (GA3), 0.1% (v/v) LA, and/or 0.1% (v/v) LA twice, once in day 9 and once in day 10.
Protein Gel Blot Analysis Using Crude Nuclear Extracts
The inflorescences from ag-1 35S:AG-GR mutant plants were treated with 10 μM DEX and 0.015% Silwet once or four times and the floral buds cluster, with each bud < 0.5 mm in diameter or at stage 10 and younger were harvested after 0.5, 1, 2, 3, 5, 7, and 9 d. Nuclear extraction was performed according to a previously published protocol (Ito et al., 1997). The proteins were measured by the Bradford method and suspended in 2× SDS sample buffer. Approximately 50 μg of crude nuclear extract was loaded onto a 7.5% Ready polyacrylamide gel (Bio-Rad), and the gel was blotted onto a PVDF nylon membrane (Bio-Rad). The AG-GR fusion protein was detected with AG-specific antibody (Ito et al., 1997) using SuperSignal West Dura extended duration substrate (Pierce). The band intensity was measured using NIH ImageJ 1.62f. The amount of nuclear AG-GR protein was normalized with band intensities of small nuclear proteins stained by the Memcode reversible protein stain kit for PVDF membranes (Pierce).
Expression Analysis of JA-Related Genes
Total RNA was isolated from floral bud clusters, with each bud <0.5 mm in diameter or at stage 10 and younger by the RNeasy plant mini kit (Qiagen) and reverse-transcribed by the ThermoScript RT-PCR system (Invitrogen). Four sets of independent RNA samples were prepared. Especially because floral bud at stage 8, 9, or 10 is ∼40, 200, or 400 times larger than floral buds at stage 3, respectively (T. Ito, unpublished data), RT-PCR results were presumed to mostly represent the expression patterns in stages 9 and 10 floral buds. For semiquantitative RT-PCR, the expression of DAD1, COI1, OPDA-REDUCTASE3 (OPR3), and the control lipase gene (At1g10740) were analyzed using primers shown in Supplemental Table 1 online. Each primer set was tested at 25, 30, 35 and 40 cycles, and quantitative condition was determined to be 42 cycles for DAD1, 28 cycles for COI1, 29 cycles for OPR3, and 24 cycles for control lipase. Quantitative real-time PCR assays were performed in duplicates on a 7900HT fast real-time PCR system (Applied Biosystems) using SYBR Green PCR master mix (Applied Biosystems) with the DAD1, COI1, and OPR3 primer sets shown in Supplemental Table 1 online and tubulin as an internal standard as described previously (Liu et al., 2007).
DAD1 Promoter Analysis
To produce the pDAD1:GUS construct, 3.5 kb of genomic sequence comprising the DAD1 upstream region, including 268 bp of 5′ coding region, was first amplified using wild-type Columbia genomic DNA as a template with primers DAD1-5′A2 and DAD1-5′B (see Supplemental Table 1 online) by UltraPfu High-Fidelity DNA polymerase (Stratagene) at the extension time of 4 min and then cloned into pENTR directional topo cloning vector (Invitrogen) and subsequently into pBGWFS7 binary vector by the Gateway cloning method. Transgenic plants for this construct did not show any GUS reporter expression (data not shown), and we found that a different initiation codon that is located more 5′ than the one annotated in The Arabidopsis Information Resource website was used. Thus, we digested the construct with EcoRI and AscI and inserted a short linker made by annealing two oligos, 5′-AATTCCCCGGGG-3′ and 5′-CGCGCCCCGGGG-5′, to adjust the protein reading frame. The final construct contains 3.4 kb upstream including 5′ untranslated region and 225 bp coding region of DAD1 translationally fused to green fluorescent protein–GUS reporter genes. The construct was mutated by the primers DAD1-5′MA2 and DAD1-5′MB2 using the QuikChange II XL site-directed mutagenesis kit (Stratagene). Whole inflorescences were rinsed and stained to determine GUS activity for GUS expression analysis as previously described (Ito et al., 2003).
ChIP
ChIP assays were performed as described previously (Ito et al., 1997; Liu et al., 2007) with some modifications. Inflorescences were ground and post-fixed with 1% formaldehyde for 10 min. Chromatin was isolated and solubilized by sonication with average DNA length of 400 bp. The solubilized chromatin was precleared by incubating with salmon sperm DNA/protein A agarose beads (Upstate). After centrifugation, the supernatant was incubated overnight with anti-AG serum or normal rabbit IgG as a negative control. The DNA-protein complex was precipitated by adding protein A-agarose beads, and the purified DNA samples were used for enrichment tests by real-time PCR assays. We measured the ratio in quantity between the input DNA before IP and bound DNA after IP for each primer set. The enrichment rates are the ratio of the value between each primer set and the control primer set of a Mu-like transposon. We performed two sets of fully independent ChIP experiments using samples collected separately. Both showed the same trends, and one representative data set was shown.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers of Arabidopsis genes used in this article are as follows: AG (At4G18960), DAD1 (At2g44810), COI1 (At2g39940), OPR3 (At2g06050), a lipase (At1g10740), and PFK (At4g04040).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Timed Induction of AG Activity.
Supplemental Figure 2. Effects of Four DEX Treatments to Floral Buds at Earlier Stages Than Stage 3.
Supplemental Figure 3. ChIP Assay Using ag-1 35S:AG-GR Inflorescences in Day 3 after DEX Treatments.
Supplemental Figure 4. ChIP Assay of AG Binding to DAD1 Promoter in ap1 cal 35:AP1-GR.
Supplemental Table 1. Primer Sequences Used in This Study.
Supplementary Material
Acknowledgments
We thank E.M.M.'s lab members Nicole Kubat, Jerry Guo, and Ransom Poythress and T.I.'s lab members Zhan Dan and Natsuko Ito for their diligent help with the experiments. We also thank Motomi Ito (University of Tokyo, Japan) and Mitsuyasu Hasebe (National Institute for Basic Biology, Japan) for personal communication, Pernille Rorth (Temasek Life Sciences Laboratory), former Meyerowitz lab members M. Frohlich (The Natural History Museum, London, UK), P. Kumar (National University of Singapore), G. Venugopala Reddy (University of California, Riverside), and current lab members Adrienne Roeder, Elizabeth Haswell, and Carolyn Ohno for valuable comments on the manuscript. This work was partially supported by Grant GM45697 from the National Institutes of Health to E.M.M. and by a research grant to the Temasek Life Sciences Laboratory to T.I.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Toshiro Ito (itot@tll.org.sg).
Online version contains Web-only data.
References
- Bey, M., Stuber, K., Fellenberg, K., Schwarz-Sommer, Z., Sommer, H., Saedler, H., and Zachgo, S. (2004). Characterization of Antirrhinum petal development and identification of target genes of the class B MADS box gene DEFICIENS. Plant Cell 16 3197–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Drews, G.N., and Meyerowitz, E.M. (1991). Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 3 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulis, I.E., Iacobucci, M., and Shirley, B.W. (1996). A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. Plant Cell 8 1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capovilla, M., and Botas, J. (1998). Functional dominance among Hox genes: Repression dominates activation in the regulation of Dpp. Development 125 4949–4957. [DOI] [PubMed] [Google Scholar]
- Coe, E.H., McCormick, S.M., and Modena, S.A. (1981). White pollen in maize. J. Hered. 72 318–320. [Google Scholar]
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353 31–37. [DOI] [PubMed] [Google Scholar]
- Crickmore, M.A., and Mann, R.S. (2006). Hox control of organ size by regulation of morphogen production and mobility. Science 313 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, B., Motte, P., Keck, E., Saedler, H., Sommer, H., and Schwarz-Sommer, Z. (1999). PLENA and FARINELLI: Redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 18 4023–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, G.N., Bowman, J.L., and Meyerowitz, E.M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65 991–1002. [DOI] [PubMed] [Google Scholar]
- Endress, P.K. (2001). The flowers in extant basal angiosperms and inferences on ancestral flowers. Int. J. Plant Sci. 162 1111–1140. [Google Scholar]
- Feys, B., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido, A. (1975). Genetic control of wing disc development in Drosophila. Ciba Found. Symp. 1 161–182. [DOI] [PubMed] [Google Scholar]
- Gomez-Mena, C., de Folter, S., Costa, M.M., Angenent, G.C., and Sablowski, R. (2005). Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132 429–438. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 525–529. [DOI] [PubMed] [Google Scholar]
- Huang, H., Mizukami, Y., Hu, Y., and Ma, H. (1993). Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 21 4769–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford, L. (1996). The Origin and Early Evolution of Angiosperm Stamens. (New York: Cambridge University Press).
- Ishiguro, S., Kawai-Oda, A., Ueda, J., Nishida, I., and Okada, K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Sakai, H., and Meyerowitz, E.M. (2003). Whorl-specific expression of the SUPERMAN gene of Arabidopsis is mediated by cis elements in the transcribed region. Curr. Biol. 13 1524–1530. [DOI] [PubMed] [Google Scholar]
- Ito, T., Takahashi, N., Shimura, Y., and Okada, K. (1997). A serine/threonine protein kinase gene isolated by an in vivo binding procedure using the Arabidopsis floral homeotic gene product, AGAMOUS. Plant Cell Physiol. 38 248–258. [DOI] [PubMed] [Google Scholar]
- Ito, T., Wellmer, F., Yu, H., Das, P., Ito, N., Alves-Ferreira, M., Riechmann, J.L., and Meyerowitz, E.M. (2004). The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430 356–360. [DOI] [PubMed] [Google Scholar]
- Li, L., Li, C., and Howe, G.A. (2001). Genetic analysis of wound signaling in tomato. Evidence for a dual role of jasmonic acid in defense and female fertility. Plant Physiol. 127 1414–1417. [PMC free article] [PubMed] [Google Scholar]
- Liu, C., Zhou, J., Bracha-Drori, K., Yalovsky, S., Ito, T., and Yu, H. (2007). Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134 1901–1910. [DOI] [PubMed] [Google Scholar]
- Lloyd, A.M., Schena, M., Walbot, V., and Davis, R.W. (1994). Epidermal cell fate determination in Arabidopsis: Patterns defined by a steroid-inducible regulator. Science 266 436–439. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., Bowman, J.L., Kempin, S.A., Ma, H., Meyerowitz, E.M., and Yanofsky, M.F. (1992). Manipulation of flower structure in transgenic tobacco. Cell 71 133–143. [DOI] [PubMed] [Google Scholar]
- Meyerowitz, E.M. (2002). Plants compared to animals: The broadest comparative study of development. Science 295 1482–1485. [DOI] [PubMed] [Google Scholar]
- Meyerowitz, E.M., Bowman, J.L., Brockman, L.L., Drews, G.N., Jack, T., Sieburth, L.E., and Weigel, D. (1991). A genetic and molecular model for flower development in Arabidopsis thaliana. Dev. Suppl. 1 157–167. [PubMed] [Google Scholar]
- Mizukami, Y., and Ma, H. (1992). Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71 119–131. [DOI] [PubMed] [Google Scholar]
- Mo, Y., Nagel, C., and Taylor, L.P. (1992). Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc. Natl. Acad. Sci. USA 89 7213–7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal, P., Ellis, C.M., Weber, H., Ploense, S.E., Barkawi, L.S., Guilfoyle, T.J., Hagen, G., Alonso, J.M., Cohen, J.D., Farmer, E.E., Ecker, J.R., and Reed, J.W. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132 4107–4118. [DOI] [PubMed] [Google Scholar]
- Napoli, C.A., Fahy, D., Wang, H.Y., and Taylor, L.P. (1999). white anther: A petunia mutant that abolishes pollen flavonol accumulation, induces male sterility, and is complemented by a chalcone synthase transgene. Plant Physiol. 120 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.H., Halitschke, R., Kim, H.B., Baldwin, I.T., Feldmann, K.A., and Feyereisen, R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31 1–12. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 200–203. [DOI] [PubMed] [Google Scholar]
- Pinyopich, A., Ditta, G.S., Savidge, B., Liljegren, S.J., Baumann, E., Wisman, E., and Yanofsky, M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424 85–88. [DOI] [PubMed] [Google Scholar]
- Riechmann, J., Wang, M., and Meyerowitz, E. (1996). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24 3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu, I., Wolters-Arts, M., Derksen, J., Mariani, C., and Weterings, K. (2003). Ethylene regulates the timing of anther dehiscence in tobacco. Planta 217 131–137. [DOI] [PubMed] [Google Scholar]
- Sablowski, R.W., and Meyerowitz, E.M. (1998). Temperature-sensitive splicing in the floral homeotic mutant apetala3–1. Plant Cell 10 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, P.M., Lee, P.Y., Biesgen, C., Boone, J.D., Beals, T.P., Weiler, E.W., and Goldberg, R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Sekimoto, Y., et al. (2005). Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J. 44 653–668. [DOI] [PubMed] [Google Scholar]
- Shiraishi, H., Okada, K., and Shimura, Y. (1993). Nucleotide sequences recognized by the AGAMOUS MADS domain of Arabidopsis thaliana in vitro. Plant J. 4 385–398. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E., and Meyerowitz, E.M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer, I.M., Stam, M.E., van Tunen, A.J., Mol, J.N., and Stuitje, A.R. (1992). Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell 4 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Malek, B., van der Graaff, E., Schneitz, K., and Keller, B. (2002). The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216 187–192. [DOI] [PubMed] [Google Scholar]
- Weatherbee, S.D., Halder, G., Kim, J., Hudson, A., and Carroll, S. (1998). Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev. 12 1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer, F., Alves-Ferreira, M., Dubois, A., Riechmann, J.L., and Meyerowitz, E.M. (2006). Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet 2 e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer, F., Riechmann, J.L., Alves-Ferreira, M., and Meyerowitz, E.M. (2004). Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16 1314–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, T., Lee, D.Y., Miyao, A., Hirochika, H., An, G., and Hirano, H.Y. (2006). Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W.C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldmann, K.A., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346 35–39. [DOI] [PubMed] [Google Scholar]
- Yi, Y., and Jack, T. (1998). An intragenic suppressor of the Arabidopsis floral organ identity mutant apetala3-1 functions by suppressing defects in splicing. Plant Cell 10 1465–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylstra, B., Muskens, M., and Van Tunen, A.J. (1996). Flavonols are not essential for fertilization in Arabidopsis thaliana. Plant Mol. Biol. 32 1155–1158. [DOI] [PubMed] [Google Scholar]
- Zachgo, S., Silva Ede, A., Motte, P., Trobner, W., Saedler, H., and Schwarz-Sommer, Z. (1995). Functional analysis of the Antirrhinum floral homeotic DEFICIENS gene in vivo and in vitro by using a temperature-sensitive mutant. Development 121 2861–2875. [DOI] [PubMed] [Google Scholar]
- Zik, M., and Irish, V.F. (2003). Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell 15 207–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.