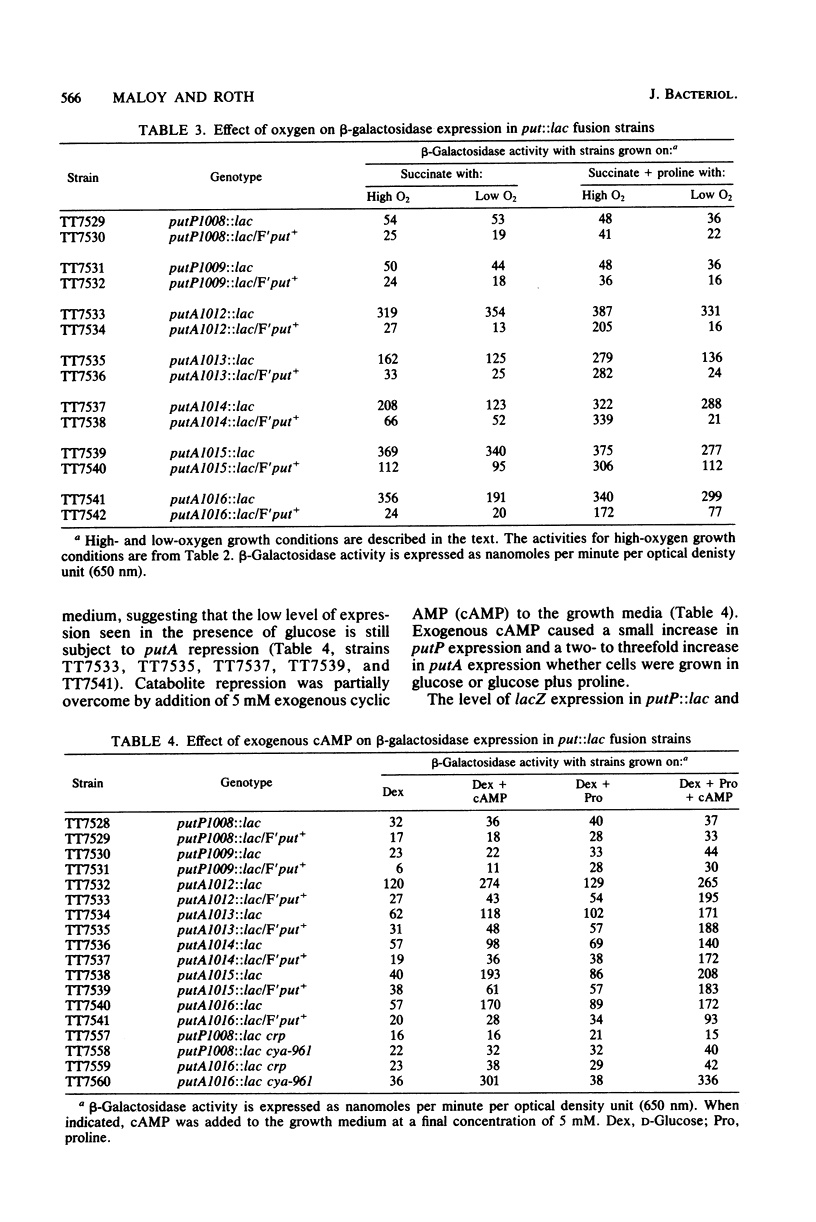
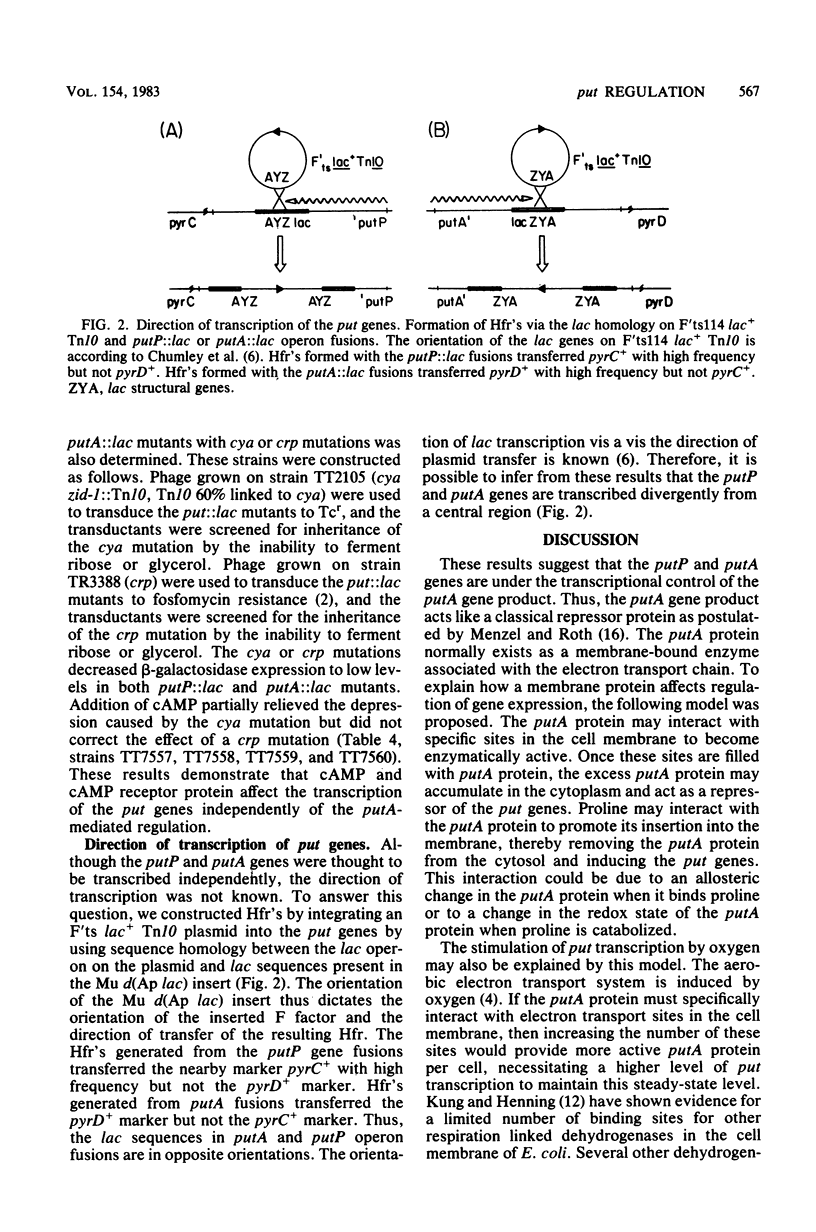
Abstract
The genes for proline utilization were fused to the structural genes of the lac operon by use of the hybrid Mu phage derivative Mu d(Ap lac). Stable deletion derivatives of these fusions were selected and used to study the transcriptional regulation of the put genes. Analysis of these fusions showed that the putA gene product, a bifunctional oxidase-dehydrogenase, also serves to negatively control transcription of the putA and putP genes. Transcription of the put genes is repressed only in putA+ strains; this repression is lifted when exogenous proline is supplied. Transcription of the put genes is stimulated by cyclic AMP in putA+ and putA strains. Maximal induction of the put genes in putA+ strains requires oxygen or an alternative electron acceptor. This oxygen effect is mediated by the putA protein since putA mutants show maximal transcription even without an electron acceptor. The orientation of the Mu d(Ap lac) insertions was determined by formation of Hfr's via the lac homology on F'ts114 lac+. The direction of chromosome mobilization by these Mu d(Ap lac)-directed Hfr's demonstrated that the putP and putA genes are divergently transcribed from a central regulatory region lying between them.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Garges S. How cyclic AMP and its receptor protein act in Escherichia coli. Cell. 1982 Jun;29(2):287–289. doi: 10.1016/0092-8674(82)90145-3. [DOI] [PubMed] [Google Scholar]
- Alper M. D., Ames B. N. Transport of antibiotics and metabolite analogs by systems under cyclic AMP control: positive selection of Salmonella typhimurium cya and crp mutants. J Bacteriol. 1978 Jan;133(1):149–157. doi: 10.1128/jb.133.1.149-157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachhuber M., Brill W. J., Howe M. M. Use of bacteriophage Mu to isolate deletions in the his-nif region of Klebsiella pneumoniae. J Bacteriol. 1976 Dec;128(3):749–753. doi: 10.1128/jb.128.3.749-753.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley F. G., Menzel R., Roth J. R. Hfr formation directed by tn10. Genetics. 1979 Apr;91(4):639–655. doi: 10.1093/genetics/91.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka L. N., Howe M. M., Ingraham J. L., Pierson L. S., 3rd, Turnbough C. L., Jr Infection of Salmonella typhimurium with coliphage Mu d1 (Apr lac): construction of pyr::lac gene fusions. J Bacteriol. 1981 Jan;145(1):299–305. doi: 10.1128/jb.145.1.299-305.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendinger S., Brill W. J. Regulation of proline degradation in Salmonella typhimurium. J Bacteriol. 1970 Jul;103(1):144–152. doi: 10.1128/jb.103.1.144-152.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedberg W. B., Lin E. C. Three kinds of controls affecting the expression of the glp regulon in Escherichia coli. J Bacteriol. 1973 Sep;115(3):816–823. doi: 10.1128/jb.115.3.816-823.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M. M., Bade E. G. Molecular biology of bacteriophage mu. Science. 1975 Nov 14;190(4215):624–632. doi: 10.1126/science.1103291. [DOI] [PubMed] [Google Scholar]
- Kung H. F., Henning U. Limiting availability of binding sites for dehydrogenases on the cell membrane of Escherichia coli. Proc Natl Acad Sci U S A. 1972 Apr;69(4):925–929. doi: 10.1073/pnas.69.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Al-Zarban S., Wilcox G. Genetic characterization of the araE gene in Salmonella typhimurium lt2. J Bacteriol. 1981 Apr;146(1):298–304. doi: 10.1128/jb.146.1.298-304.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Heffernan L., Wilcox G. Isolation of ara-lac gene fusions in Salmonella typhimurium LT2 by using transducing bacteriophage Mu d (Apr lac). J Bacteriol. 1980 Sep;143(3):1325–1331. doi: 10.1128/jb.143.3.1325-1331.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R., Roth J. Purification of the putA gene product. A bifunctional membrane-bound protein from Salmonella typhimurium responsible for the two-step oxidation of proline to glutamate. J Biol Chem. 1981 Sep 25;256(18):9755–9761. [PubMed] [Google Scholar]
- Menzel R., Roth J. Regulation of the genes for proline utilization in Salmonella typhimurium: autogenous repression by the putA gene product. J Mol Biol. 1981 May 5;148(1):21–44. doi: 10.1016/0022-2836(81)90233-3. [DOI] [PubMed] [Google Scholar]
- Mitchell C. G., Dawes E. A. The role of oxygen in the regulation of glucose metabolism, transport and the tricarboxylic acid cycle in Pseudomonas aeruginosa. J Gen Microbiol. 1982 Jan;128(1):49–59. doi: 10.1099/00221287-128-1-49. [DOI] [PubMed] [Google Scholar]
- Mulligan J. T., Margolin W., Krueger J. H., Walker G. C. Mutations affecting regulation of methionine biosynthetic genes isolated by use of met-lac fusions. J Bacteriol. 1982 Aug;151(2):609–619. doi: 10.1128/jb.151.2.609-619.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzkin B., Grabnar M., Roth J. Regulation of the major proline permease gene of Salmonella typhimurium. J Bacteriol. 1978 Feb;133(2):737–743. doi: 10.1128/jb.133.2.737-743.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzkin B., Roth J. Cluster of genes controlling proline degradation in Salmonella typhimurium. J Bacteriol. 1978 Feb;133(2):744–754. doi: 10.1128/jb.133.2.744-754.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld S. A., Brenchley J. E. Bacteriophage P1 as a vehicle for Mu mutagenesis of Salmonella typhimurium. J Bacteriol. 1980 Nov;144(2):848–851. doi: 10.1128/jb.144.2.848-851.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119(1):75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- Scott J. F., Roth J. R., Artz S. W. Regulation of histidine operon does not require hisG enzyme. Proc Natl Acad Sci U S A. 1975 Dec;72(12):5021–5025. doi: 10.1073/pnas.72.12.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. M. Genetics of L-proline utilization in Escherichia coli. J Bacteriol. 1981 Jun;146(3):895–901. doi: 10.1128/jb.146.3.895-901.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]


