Abstract
Protein phosphatase type I (PP1), encoded by the single essential gene GLC7 in Saccharomyces cerevisiae, functions in diverse cellular processes. To identify in vivo subcellular location(s) where these processes take place, we used a functional green fluorescent protein (GFP)–Glc7p fusion protein. Time-lapse fluorescence microscopy revealed GFP–Glc7p localizes predominantly in the nucleus throughout the mitotic cell cycle, with the highest concentrations in the nucleolus. GFP–Glc7p was also observed in a ring at the bud neck, which was dependent upon functional septins. Supporting a role for Glc7p in bud site selection, a glc7-129 mutant displayed a random budding pattern. In α-factor treated cells, GFP–Glc7p was located at the base of mating projections, again in a septin-dependent manner. At the start of anaphase, GFP–Glc7p accumulated at the spindle pole bodies and remained there until cytokinesis. After anaphase, GFP–Glc7p became concentrated in a ring that colocalized with the actomyosin ring. A GFP–Glc7-129 fusion was defective in localizing to the bud neck and SPBs. Together, these results identify sites of Glc7p function and suggest Glc7p activity is regulated through dynamic changes in its location.
Keywords: protein phosphatase type I, yeast, GLC7, cell cycle, green fluorescent protein
Introduction
Protein phosphatase type I (PP1) is a phosphoserine/phosphothreonine-specific phosphatase that exhibits >80% sequence identity from yeast to mammals. It has been implicated in the regulation of a variety of cellular processes including cell cycle progression, glycogen metabolism, glucose repression, sporulation, RNA splicing, and muscle contraction (Mumby and Walter 1993; DePaoli-Roach et al. 1994; Shenolikar 1994). Several of these processes also appear to be conserved through evolution. One illustration of this conservation is in the regulation of mitosis. In Saccharomyces cerevisiae, conditional PP1 mutants, in the single essential gene GLC7, arrest in G2/M at the nonpermissive temperature (Hisamoto et al. 1994; MacKelvie et al. 1995; Baker et al. 1997). The G2/M arrest is due to activation of the spindle/kinetochore checkpoint, likely due to defective PP1 regulation of spindle attachment to kinetochores (Bloecher and Tatchell 1999; Sassoon et al. 1999). PP1 mutants have also been identified that have defects in progression through late mitosis or cytokinesis (Stark 1996; Bloecher and Tatchell 1999). Similarly, in mammalian cells, microinjection of neutralizing anti-PP1 antibodies into late G2 cells causes a metaphase arrest, whereas microinjection of these antibodies into cells in anaphase B causes failure to complete mitosis and cytokinesis (Fernandez et al. 1992).
In many cases the substrate specificity of PP1 appears to be regulated by targeting/regulatory subunits that localize PP1 to putative substrates and/or alter the catalytic activity of PP1 towards these substrates (Hubbard and Cohen 1993; Faux and Scott 1996). For example, PP1 in skeletal muscle associates with RGL, a targeting/regulatory subunit that targets PP1 to glycogen particles, where it dephosphorylates glycogen synthase (Hubbard and Cohen 1989; Tang et al. 1991). In S. cerevisiae, a homologous mechanism utilizes the targeting/regulatory subunit Gac1p to target PP1 to glycogen synthase (François et al. 1992).
Several targeting/regulatory subunits have been identified in S. cerevisiae. These subunits are involved in a wide variety of cellular processes, many of which are consistent with known PP1 functions (Stark 1996; Tu et al. 1996). For example, gac1, reg1, and gip1 mutants are deficient in glycogen accumulation, glucose repression, and sporulation, respectively (Stuart et al. 1994; Tu and Carlson 1995). Consistent with the regulatory roles for these subunits, the products of GLC7 mutants with defects in glycogen accumulation, glucose repression, and sporulation do not interact with Gac1p, Reg1p, or Gip1p, respectively. Additional potential regulatory/targeting subunits have been identified using two-hybrid screens (Tu et al. 1996; Uetz et al. 2000). These include Sla1p, a protein involved in cortical actin formation (Holtzman et al. 1993); Red1p, necessary for synaptonemal complex formation (Rockmill and Roeder 1990); Scd5p, involved in vesicular trafficking (Nelson et al. 1996); Bni4p, a bud neck protein required for proper chitin deposition (DeMarini et al. 1997); and several products of open reading frames of unknown function. The diversity of these potential targeting/regulatory subunits further supports the involvement of PP1 in a broad range of cellular functions.
The targeting hypothesis for PPI leads to the prediction that the phosphatase should accumulate at those subcellular locations where its activity is required. Indeed, studies in mammalian cells support this prediction. In G0 fibroblasts, indirect immunofluorescence with anti-PP1 antibodies revealed that PPI was predominantly located in the cytoplasm (Fernandez et al. 1992). Upon serum addition, PP1 translocates to the nucleus and then becomes associated with chromosomes in mitosis. The transit from the cytoplasm to the nucleus appears to be a function of growth phase rather than cell cycle phase since PP1 is largely nuclear in growing cells, irrespective of the phase of the cell cycle. In HeLa cells, antibodies specific to the three major isoforms of PP1 reveal distinct isoform-specific locations in the mitotic cell cycle (Andreassen et al. 1998) for PP1 isoforms. In interphase, PP1α is located in the nuclear matrix, PP1γ1 is concentrated in the nucleoli, and PP1δ is chromatin associated. In mitosis, PP1α localizes to centrosomes, PP1γ1 is associated with the mitotic spindle, and PP1δ is bound to chromosomes. Inhibitor-2, a PP1-specific inhibitory or regulatory subunit, also exhibits cell cycle–dependent shuttling between the cytoplasm and the nucleus (Kakinoki et al. 1997).
In contrast to the mammalian studies, little is known about the location of PPI in yeast or fungi. In S. pombe, Dis2p, a PPI gene product, is located primarily in the nucleus (Ohkura et al. 1989). A similar location has been reported for Glc7p in S. cerevisiae (Zhang et al. 1995). In this study, we have further investigated the location of PPI in S. cerevisiae using a functional myc-tagged Glc7p for indirect immunofluorescence and a functional green fluorescent protein (GFP)–Glc7p for time-lapse fluorescence and DIC microscopy. We present data showing dynamic and diverse localization of GFP–Glc7p throughout the mitotic cell cycle. Novel locations for the phosphatase at spindle pole bodies (SPBs), the mother/bud neck, the actomyosin ring, and in the nucleolus reflect known, as well as potential new roles for Glc7p.
Materials and Methods
Yeast Strains, Media, and General Methods
The yeast strains listed in Table are congenic to strain KT1112 (MAT a ura3-52 leu2 his3; Stuart et al. 1994). Yeast strains were grown in YPD media (Guthrie and Fink 1991) at 30°C, except where noted. Synthetic complete and dropout media lacking specific amino acids were made as described (Guthrie and Fink 1991). Strains were sporulated on media containing Bacto-Peptone (20 g/liter; Difco), yeast extract (10 g/liter; Difco), and potassium acetate (20 g/liter; Sigma Chemical Co.). Yeast transformation was performed using LiOAc, as previously described (Kaiser et al. 1994). Escherichia coli general methods were performed and media prepared as described (Maniatis et al. 1989).
Table 1.
S. cerevisiae Strains and Plasmids Used
| Strain | Genotype | Source or reference |
|---|---|---|
| YAB439 | MAT a leu2 his3::GFP-LacI-HIS3 ura3-52 trp1::lacO-TRP1 glc7::LEU2 pAB26 | This study |
| YAB380 | MAT a leu2 his3 ura3-52 trp1 Nuf2–GFP-URA3 glc7::LEU2 pAB70 | This study |
| YAB108 | MAT a leu2 his3 ura3-52 Nuf2–GFP-URA3 | This study |
| YAB122 | MAT a leu2 his3 ura3-52 trp1 glc7::LEU2 pAB70 | This study |
| KT1210 | MATα leu2 his3 ura3-52 glc7::HIS3 pep4 Δgac1::URA3 CY973 | This study |
| KT1208 | MAT a leu2 his3 ura3-52 glc7::HIS3 pep4 CY973 | This study |
| SB90 | MAT a leu2 ura3-52 trp1 glc7::LEU2 pNC160-PP1 | Baker et al. 1997 |
| SB91 | MATα leu2 ura3-52 trp1 glc7::LEU2 pNC160-PP1 | This study |
| YAB130 | MAT a leu2 his3 ura3-52 glc7::LEU2 pAB70 pTD150-CDC12 | This study |
| YAB453 | MAT a leu2 his3 ura3-52 pTD150-CDC12 | This study |
| KT1112 | MAT a leu2 his3 ura3-52 | Stuart et al. 1994 |
| YAB107 | MAT a leu2 his3 ura3-52 MATα leu2 his3 ura3-52 | This study |
| YAB381 | MAT a leu2 his3 ura3-52 trp1 cdc16-1 glc7::LEU2 pAB70 | This study |
| KT1556 | MAT a leu2 his3 ura3-52 trp1 glc7::LEU2 pAB26 | This study |
| KT1357 | MAT a leu2 his3 ura3-52 trp1 | This study |
| KT1358 | MATα leu2 his3 ura3-52 trp1 | This study |
| YAB58 | MAT a leu2 his3 ura3-52 trp1 cdc3-1 glc7::LEU2 pAB26 | This study |
| YAB4 | MATa leu2 ura3-52 trp1 glc7::LEU2 pAB26 MATα leu2 ura3-52 trp1glc7::LEU2 | This study |
| YAB6 | MATa leu2 ura3-52 trp1 glc7::LEU2 pAB26 rho0 MATα leu2 ura3-52 trp1 glc7::LEU2 | This study |
| YAB27 | MAT a leu2 his3 ura3-52 glc7-129 | Bloecher and Tatchell 1999 |
| YAB86 | MAT a leu2 his3 ura3-52 glc7-129 MATα leu2 his3 ura3-52 glc7-129 | This study |
| YAB122 | MAT a leu2 his3 ura3-52 trp1 glc7::LEU2 pAB70 | This study |
| Plasmid | Description/markers | Source or reference |
| pAB26 | GFP–GLC7 fusion, CEN vector/TRP1 | This study |
| pAB70 | GFP(10C)-GLC7 fusion, CEN vector/TRP1 | This study |
| pTD150-CDC12 | CDC12-GFP(m2) fusion, CEN vector/URA3 | Robinson et al. 1999 |
| pNC160-PP1 | GLC7, CEN vector/TRP1 | Baker et al. 1997 |
| CY973 | myc-GLC7 fusion, CEN vector/URA3 | Sutton et al. 1991 |
| pAB18 | GFP–glc7-129 fusion, CEN vector/TRP1 | This study |
The GFP–GLC7 gene fusion, driven from the native GLC7 promoter on a low copy CEN vector (pAB26), was transformed into a diploid yeast strain homozygous for a glc7::LEU2 disruption. This strain contained a plasmid-borne copy of GLC7 (CY973). Mitotic segregants that contained pAB26, but not CY973, were then isolated, yielding the YAB4 strain. Plasmid pAFS52, containing 256 copies of lacO, was integrated into strain KT1358 at the TRP1 locus as described (Straight et al. 1998). The lacI-GFP fusion was integrated into the resulting strain at the HIS3 locus using the linearized plasmid pAFS144 as described (Straight et al. 1998). The resulting strain (YAB404) was then crossed to YAB122. The resulting diploid strain was sporulated, tetrads were dissected, and haploid strain YAB441 was recovered. Induction of the lacI-GFP fusion was performed as described (Straight et al. 1997). The Nuf2–GFP gene fusion carried on plasmid pJK67 (Kahana et al. 1995) was linearized with BstE1 and integrated into the chromosomal NUF2 locus of strains KT1112 and YAB122 to yield strains YAB108 and YAB380, respectively.
Plasmid Construction
pAB26 was constructed by inserting an EcoRI site directly after the first codon of GLC7 in the plasmid pNC160-GLC7 (Baker et al. 1997). The EcoRI site was inserted using PCR with the primers AB1 and AB3 (AB3- cgcgaattccatttctttaatttgaat; AB1-cgcgaattcgactcacaaccagttgac). An EcoRI fragment containing the coding sequence of the F64A, S65T GFP mutant (Cormack et al. 1996; kindly provided by Dr. Lucy Robinson, LSU Medical Center, Shreveport, LA) was then inserted into the EcoRI site of pNC160-GLC7. Sequence analysis of the GFP–GLC7 fusion junction revealed an in-frame insertion after the first codon in the wild-type GLC7 gene. pAB18 was constructed by inserting the GFP–containing HindIII/SalI fragment from pAB26 into the HindIII/SalI sites of pNC160-glc7-129 (Baker et al. 1997). pAB70 was constructed by replacing a NcoI/PvuII fragment of GFP(10C) from pRSETB-GFP(10C) (Ormö et al. 1996) with a NcoI/PvuII fragment of GFP from pAB26. pTD150-CDC12 is described in Robinson et al. 1999. CY973 was a gift from Kim Arndt (Wyeth-Ayerst Research). GFP fused to Glc7p lacking the COOH-terminal eight amino acids (Δ305–312) was constructed by inserting the GFP–containing HindIII/SalI fragment from pAB26 into the HindIII/SalI sites of pNC160-glc7-305° (Baker et al. 1997).
Immunofluorescence, Rhodamine-Phalloidin, Calcofluor, and DAPI Staining
Indirect immunofluorescence against myc-tagged Glc7p was performed as described in Pringle et al. 1991 using the anti-myc antibody 9E10 (Evan et al. 1985) and a TRITC-labeled goat anti–mouse IgG secondary antibody (Sigma Chemical Co.). For nucleolar visualization, cells were treated for indirect immunofluorescence as described in Pringle et al. 1991 using the mouse anti–Nop1p primary antibody kindly provided by John Aris (University of Florida, Gainesville, FL) (Aris and Blobel 1988) and a TRITC-labeled goat anti–mouse IgG secondary antibody (Sigma Chemical Co.).
To visualize the actomyosin ring, cells were fixed in 3.5% formaldehyde for 10 min and then stained with 0.5 U/ml rhodamine-conjugated phalloidin (Molecular Probes) as described (Bi et al. 1998). Bud scars were visualized as described (Robinson et al. 1999). DNA was visualized in strain YAB6 by adding 4′,6-diamidino-2-phenylindole (DAPI; Sigma Chemical Co.) directly to the media at a final concentration of 0.2 μg/ml and incubating at room temperature for 30 min. The cells were then washed once with water and observed with fluorescence microscopy. The rho0 strain, YAB6, was generated by treating strain YAB4 with ethidium bromide as described (Guthrie and Fink 1991).
Microscopic Analysis
Time-lapse imaging of cells (see Fig. 2) was performed as described (Bloecher and Tatchell 1999). Time-lapse images, as in Fig. 8 and Fig. 9, were acquired using the Princeton Instruments Micro Max CCD camera and a custom script written for IPLab Spectrum Software similar to that described (Shaw et al. 1997). At each time interval, six fluorescent images in different Z-axis planes (∼0.5–1 μm apart) were acquired and flattened into a two-dimensional (2D) projection. A separate DIC image was captured from a central Z-axis plane.
Figure 2.
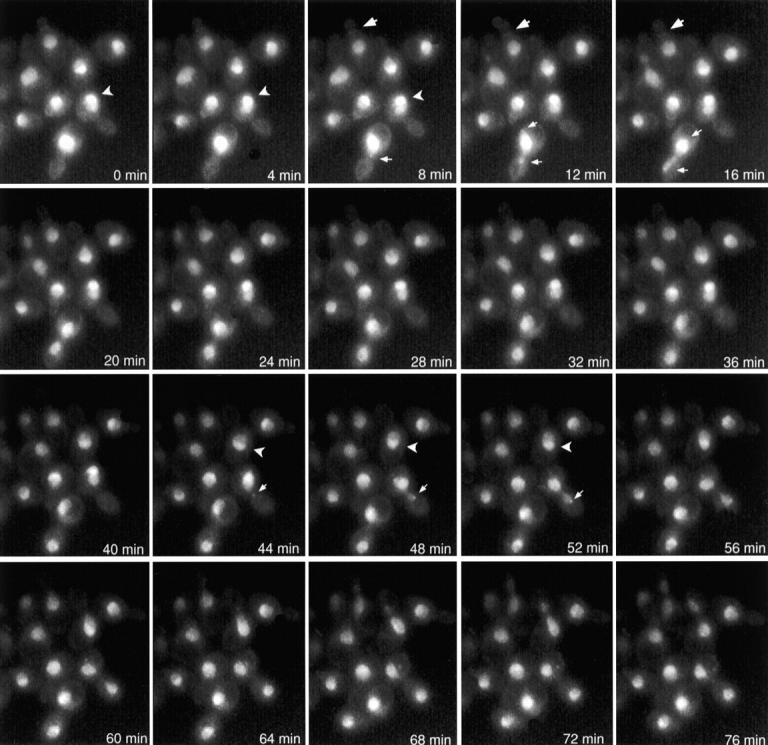
Localization of GFP–Glc7p throughout the mitotic cell cycle. YAB4 cells were placed on an agarose pad containing synthetic complete medium and visualized by fluorescence microscopy as described (Waddle et al. 1996). Each image is a 2D projection from 10 z-axis planes, each offset by 0.5 μm. Images are shown at 4-min intervals. Large arrows denote bud neck fluorescence. Carets indicate subnuclear fluorescence. Small arrows denote spots observed at nuclear periphery of cells in anaphase.
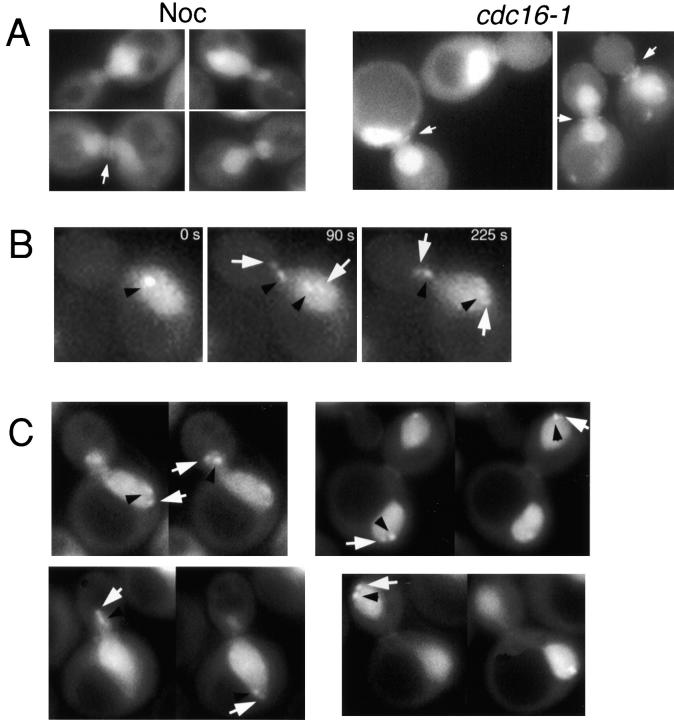
Figure 8.
GFP–Glc7p accumulates at the SPBs in early anaphase. A, Images of nocodazole (Noc)-arrested GFP–Glc7p cells (YAB4) or GFP–Glc7p cells containing cdc16-1 (YAB381) after incubation at the nonpermissive temperature for 3 h. Arrows indicate the double ring of GFP–Glc7p often observed in these arrested cells. B, Logarithmically growing GFP–Glc7p cells containing lacI-GFP/lacO marking centromeres (YAB439) were placed on an agarose pad containing synthetic complete media and imaged using a custom script written for IPLab Spectrum software (see Materials and Methods). The black arrowheads denote the lacI-GFP spots (marking the centromeres), whereas the white arrows mark the GFP–Glc7p spots at SPBs. C, Serial z-sections were taken through YAB439 cells in various stages of anaphase. Black arrowheads denote centromeres, white arrows denote GFP–Glc7p at SPBs.
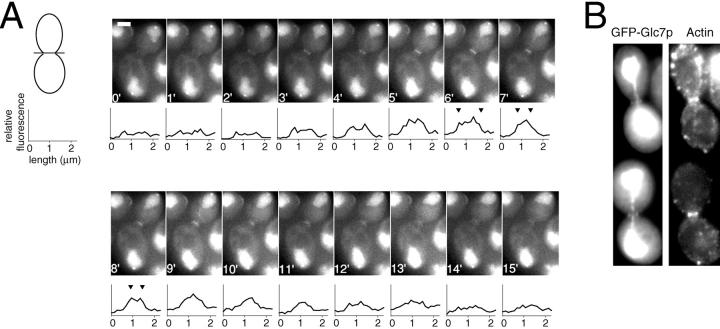
Figure 9.
GFP–Glc7p colocalizes with the actomyosin ring in late mitosis. A, Logarithmically growing GFP–Glc7p cells (YAB4) in YPD media were imaged at 1-min intervals as described in Materials and Methods. The schematic of a yeast cell is drawn to demonstrate how the line was drawn across the bud neck to quantify fluorescence. The size bar in the 0′ image represents the actual length of the line drawn across the bud neck (2.25 μm). Carets in the graphs at 6′, 7′, and 8′ mark outer edges of the GFP–Glc7p fluorescence at the bud neck. B, GFP–Glc7p cells (YAB4) were fixed and stained with rhodamine-phalloidin to visualize the actomyosin ring. Shown are two cells displaying colocalization of the GFP–Glc7p ring with the actomyosin ring.
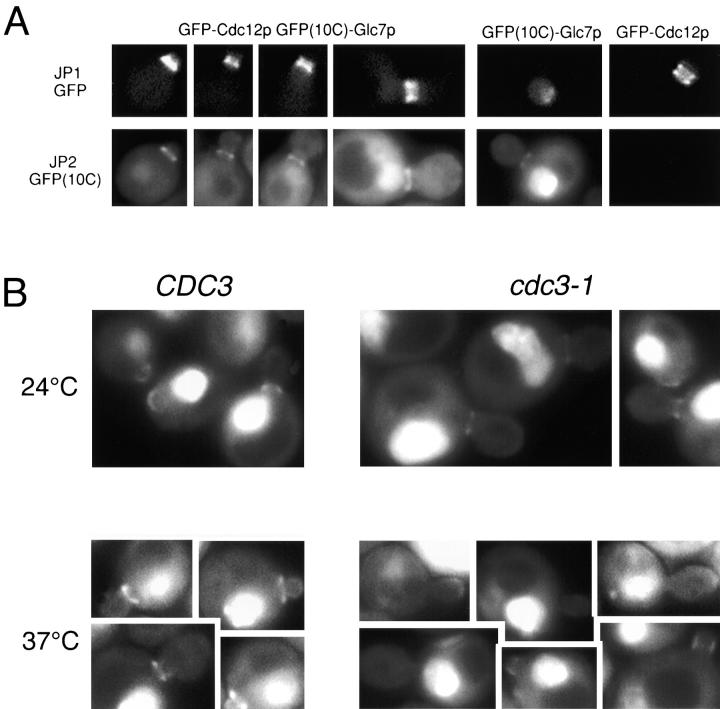
For the data presented in Fig. 7 B, Nuf2–GFP and GFP(10C)–Glc7p were imaged in the same cells using the JP1 (D470/20 exciter, D510/20 emitter, 490DCLP dichroic) and JP2 (D510/20 exciter, D560/40 emitter, 530DCLP dichroic) filter sets from Chroma Technology Corporation, respectively. The GFP variant fused to Nuf2p contains the mutations S65T, V163A (Kahana et al. 1998), whereas the GFP(10C) variant fused to Glc7p contains the mutations S65G, V68L, S72A, and T203Y (Ormö et al. 1996). Exposure times for the JP2 and JP1 filter sets were 0.4 and 1.0 s, respectively. Bleed-through was assessed for each GFP variant with images of cells containing each GFP variant (GFP(10C)–Glc7p or Nuf2–GFP) alone and found to be negligible. The merged images presented in Fig. 7 B were made by combining a red pseudocolored image from the JP2 filter set with a green pseudocolored image of the JP1 filter set using the 3D extension module from IPLab Spectrum software. In Fig. 4 A, GFP(m2)-Cdc12p and GFP(10C)–Glc7p were imaged in the same cells using the same method as above. The GFP(m2) variant fused to Cdc12p contain the mutations S65A, V68L, and S72A (Robinson et al. 1999). In situations where only a single GFP variant was imaged, the Chroma 41001 (HQ 480/40 exciter, HQ 535/50 emitter, Q 505 LP dichroic) filter set was used.
Figure 7.
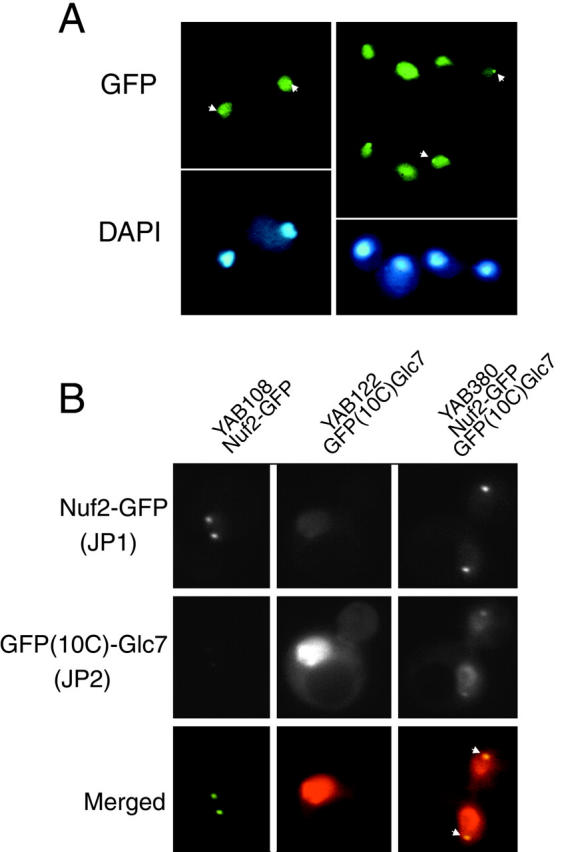
GFP–Glc7p localizes to SPBs in late mitosis. A, GFP–Glc7p and DAPI fluorescence of live, logarithmically growing cells of the rho0strain, YAB6. B, Images of logarithmically growing cells containing Nuf2–GFP (YAB108), GFP(10C)–Glc7p (YAB122), or both Nuf2–GFP and GFP(10C)–Glc7p (YAB380) captured with JP1 and JP2 filters as described in Materials and Methods. The top row shows images of Nuf2–GFP fluorescence, the middle row shows images of GFP(10C)–Glc7p fluorescence, and in the bottom row are merged images of Nuf2–GFP and GFP(10C)–Glc7p fluorescence. Areas of colocalization are yellow (arrows) in these pseudocolored images.
Figure 4.
The GFP–Glc7p ring at the mother/bud neck is dependent upon septins. A, Images of cells containing Cdc12-GFPm2 (YAB453), GFP(10C)–Glc7p (YAB122), or Cdc12-GFPm2 and GFP (10C)–Glc7p (YAB441) were captured using the JP1 or JP2 filter sets as described in Materials and Methods. The top row shows images of Cdc12-GFP fluorescence and the bottom row shows images of GFP(10C)–Glc7p fluorescence. The first four columns show images of Cdc12-GFP, GFP(10C)–Glc7p cells in different stages of the cell cycle. The fifth and sixth columns include control images of a GFP(10C)–Glc7p cell and a Cdc12-GFP cell showing lack of significant bleed-through into the JP1 and JP2 filter sets, respectively. B, Images of wild-type GFP–GLC7 (KT1556) or cdc3-1 GFP–GLC7 (YAB58) cells growing logarithmically at the permissive temperature (24°C) or after a shift to the nonpermissive temperature (37°C) for 50 min.
Quantitation of GFP–Glc7p fluorescence was performed on images of an asynchronous population of live cells in which 10 z-sections (∼0.5 μm apart) through the cells were captured using the Princeton Instruments Micro Max CCD camera. To determine fluorescence values at SPBs, the brightest pixel value (arbitrary units) in the z-section in which the SPB was in focus was determined using IPLab software. For quantitation of fluorescence across the bud neck, a 2.25-μm line (segment) was drawn across the GFP–Glc7p ring at the bud neck. The intensity of the pixels on the segment were then graphically represented using the row/col plot command. The fluorescence values were changed from 12-bit to 8-bit data when graphed using this method.
Online Supplemental Material
The online version of this article includes videos that accompany the figures presented here. Videos are available at http://www.jcb.org/cgi/ content/full/149/1/125/DC1.
Video 1.
Depicts Fig. 2. Time-lapse imaging of cells was performed as described (Bloecher and Tatchell 1999). The video contains images of GFP–Glc7p cells taken every 2 min for 3 h and 46 min.
Video 2.
Depicts Fig. 9 A. The video contains images of a GFP–Glc7p cell taken every minute for 41 min (see Microscopic Analysis for a description of the time-lapse imaging).
Results
Myc-tagged Glc7 Localizes Predominantly to the Nucleus
To investigate the subcellular distribution of Glc7p, we used indirect immunofluorescence microscopy to visualize a myc-tagged variant of Glc7p. This myc-Glc7p fusion protein was previously shown to be fully functional (Sutton et al. 1991; Hisamoto et al. 1995; Baker et al. 1997). Logarithmically growing cells of the diploid strain (KT1208/KT1210), homozygous for a glc7::LEU2 disruption and carrying myc-GLC7 expressed from the CEN vector YCp50 (Sutton et al. 1991) were fixed in 5% formaldehyde and stained with the anti-myc mAb 9E10 (Evan et al. 1985). As shown in Fig. 1, myc-Glc7p was located predominantly in the nucleus at all stages of the cell cycle. These results are consistent with the predominant nuclear location of PP1 reported previously in yeast, as well as other eukaryotes (Fernandez et al. 1992; Zhang et al. 1995). Although the staining was mostly nuclear, the anti-myc fluorescence did not strictly correlate with bulk DNA, as determined by DAPI staining. The highest concentration of myc-Glc7p was located in a domain that was separate from the bulk of the chromatin (Fig. 1, large arrow). myc-Glc7p was also observed at the bud neck (Fig. 1, small arrow), a novel location for the phosphatase. Together, these results suggest that Glc7p distribution in the cell is more diverse than previously reported (Zhang et al. 1995).
Figure 1.
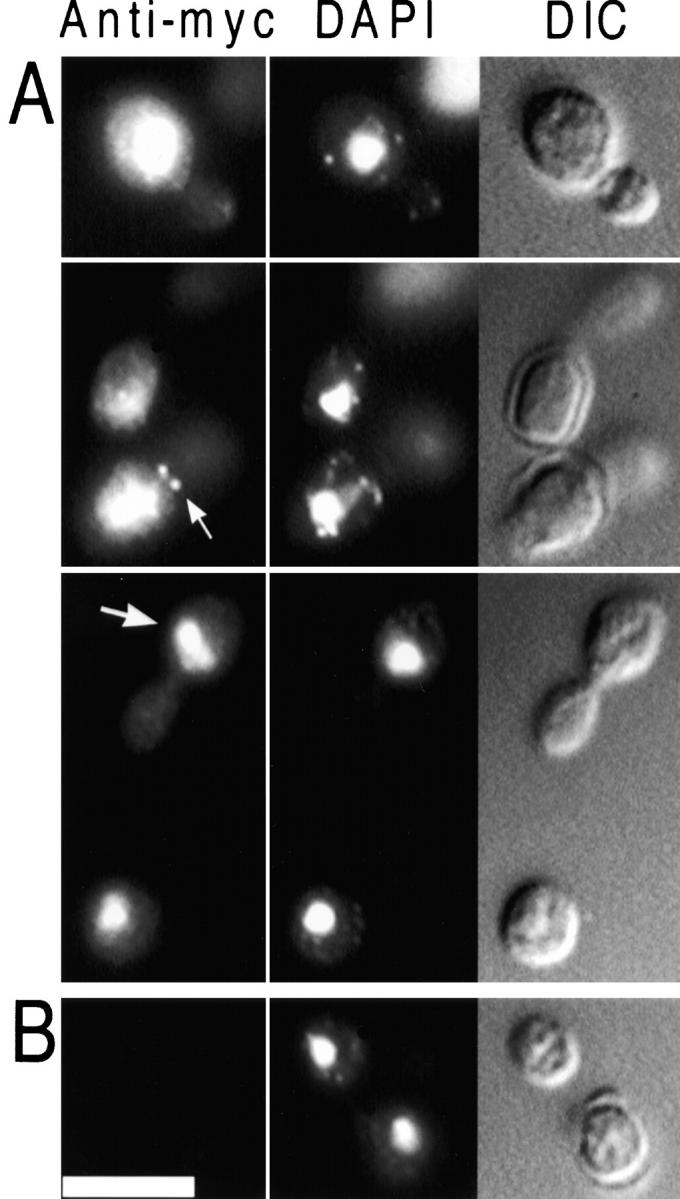
Localization of myc-Glc7p. Diploid strain KT1208/1210 (A), which contains myc-GLC7, and the control strain SB90/91 (B), which lacks myc-GLC7, were harvested from log phase in YPD medium, fixed, and prepared for indirect immunofluorescence microscopy as described in the Materials and Methods. myc-Glc7p was visualized using 9E10 primary antibody (left), DNA was visualized with DAPI (middle) and cells were visualized with DIC optics (right). Bar, 5 μm.
GFP–Glc7p Localizes to Diverse Subcellular Sites throughout the Mitotic Cell Cycle
Indirect immunofluorescence microscopy has several limitations. Accurate detection of the protein is dependent upon the sensitivity of the antibody to that protein. Cells must be chemically fixed, which can disrupt or alter the location of proteins. Fixation also prevents investigation of dynamic aspects of protein localization in vivo. To bypass these limitations, we constructed and expressed a GFP–Glc7p fusion in yeast. The diploid strain (YAB4), whose only GLC7 gene was GFP–GLC7 expressed from a CEN-vector, grew at wild-type rates at 22, 30, and 37°C (data not shown).
To investigate the localization of Glc7p during the mitotic cell cycle, time-lapse microscopy was performed on GFP–Glc7p containing cells (YAB4). Cells were grown on synthetic complete medium under a coverslip and 2D projections of images acquired from different focal planes were collected every two minutes. A montage of selected images from such an experiment is presented in Fig. 2 (see Video 1 available at http://www.jcb.org/cgi/content/full/149/1/125/DC1). GFP–Glc7p was located predominantly in the nucleus throughout all stages of the mitotic cell cycle, consistent with the indirect immunofluorescence results shown in Fig. 1. Several additional sites of GFP–Glc7p concentration were revealed in the time-lapse images. First, GFP–Glc7p localized to two spots at the periphery of the nucleus in anaphase, similar to the location of SPBs or centromeres (Fig. 2, small arrows). Similar to immunofluorescence results, GFP–Glc7p was unevenly distributed throughout the nucleus (Fig. 2, carets). Finally, a ring of GFP–Glc7p at the bud neck was observed (Fig. 2, large arrows). Each of these areas of localization will be presented in more detail below.
GFP–Glc7p Is Highly Concentrated in the Nucleolus
As shown in Fig. 1, myc-Glc7p was highly concentrated in a region of the nucleus (large arrow) that contained relatively little DAPI fluorescence, similar to the staining pattern of nucleolar antigens (de Beus et al. 1994). In time-lapse experiments, the brighter fluorescing portion of the nucleus was observed in all phases of the cell cycle except anaphase, and was consistently oriented opposite the bud, similar to the location of the nucleolus (Yang et al. 1989). To determine if the intranuclear regions of high GFP–Glc7p concentration coincide with the nucleolus, Nop1p, a nucleolar protein, was localized in GFP–GLC7 cells (YAB4) using an anti-Nop1p mAb. NOP1 encodes yeast fibrillarin, an abundant nucleolar protein required for rRNA processing (Aris and Blobel 1988). As shown in Fig. 3 A, the regions of high GFP–Glc7p concentration exactly overlapped the Nop1p staining. As expected for nucleolar staining, strong DNA staining did not colocalize with nucleolar staining. These results demonstrate that GFP–Glc7p is highly concentrated in the nucleolus.
Figure 3.
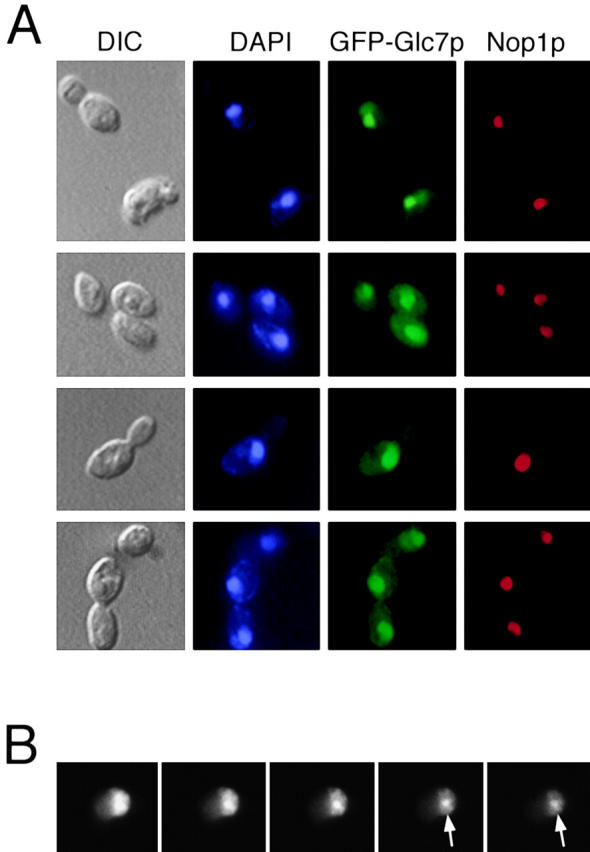
GFP–Glc7p is highly concentrated in the nucleolus. A, GFP–Glc7p cells (YAB4) were fixed and stained for DNA (DAPI) and Nop1p (a known nucleolar protein) as described in Materials and Methods. GFP–Glc7p was visualized by its native fluorescence. B, Serial z-sections through a live cell (YAB4) reveal the nonuniform nucleolar fluorescence pattern of GFP–Glc7p. Arrows indicate a bright spot commonly associated with the nucleus/nucleolus.
High resolution images of GFP–Glc7p indicated that nucleolar fluorescence was not uniform. In Fig. 3 B, a montage of serial z-sections through the nucleus reveals a nonuniform nucleolar distribution of GFP–Glc7p. The nucleolar fluorescence pattern is reminiscent of the morphological characteristics of the yeast nucleolus, recently shown to be analogous to those of higher eukaryotic nucleoli (Leger-Silvestre et al. 1999). Specifically, GFP–Glc7p fluorescence is patchy in appearance, most resembling the dense fibrillar component and fibrillar centers of the nucleolus. We have also noted that 29% of the cells (n = 415) contained a bright GFP–Glc7p spot that was commonly associated with the nucleolus and less often in the nucleus (Fig. 3 B, arrows). The identity of this spot is not known; however, in time-lapse experiments, it did not segregate in mitosis.
GFP–Glc7p Localization to the Mother/Bud Neck Is Dependent Upon Septins
Analysis of time-lapse data similar to that presented in Fig. 2 revealed that GFP–Glc7p localized to the presumptive bud site 15 ± 3 min before bud emergence (n = 4). GFP–Glc7p accumulation in a ring or spot on the cell cortex was never observed in a cdc28-4 mutant (Reed 1980) arrested at the nonpermissive temperature (data not shown), indicating that GFP–Glc7p moves to the presumptive bud neck after START. GFP–Glc7p remains at the bud neck until anaphase, when the concentration was markedly reduced.
The integrity of the bud neck is controlled by septin proteins, encoded by the genes CDC3, CDC10, CDC11, and CDC12, which make up the 10-nm neck filaments and are required for bud morphogenesis and cell division (Longtine et al. 1996). To compare the location of GFP–Glc7p with respect to septins, a functional CDC12-GFPm2 gene fusion (Robinson et al. 1999) was introduced into strain YAB122 containing the GFP(10C)-GLC7 fusion. The GFP proteins, fused to Glc7p and Cdc12p, were fluorescent variants that could be distinguished with appropriate filters (see Materials and Methods). Under the exposure conditions and relative levels of expression we used in these experiments, the Cdc12-GFPm2 and GFP(10C)–Glc7p fusions were imaged independently and with minimal bleed-through with the JP1 and JP2 filter sets, respectively. As shown in Fig. 4 A, strain YAB453, which contains the CDC12-GFPm2 gene fusion, exhibited fluorescence only with the JP1 filter set and not with the JP2 filter set. In contrast, strain YAB122, which contains only the GFP(10C)-GLC7 gene fusion, exhibited strong fluorescence with the JP2 filter set and minimal bleed-through in the JP1 set. Strain YAB130, which contains both GFP(10C)-GLC7 and CDC12-GFPm2 exhibited fluorescence with both the JP1 and JP2 filter sets.
Before anaphase, Cdc12-GFPm2 was readily observed on both sides of the bud neck as predicted (Haarer and Pringle 1987; Lippincott and Li 1998a). In contrast, GFP(10C)–Glc7p was located only on the mother side of the bud neck (Fig. 4 A). Assessment of the location of the mother/bud neck using the chitin-specific stain calcofluor also placed GFP–Glc7p on the mother side of the neck (data not shown). In large-budded cells before anaphase, GFP–Glc7p was occasionally observed as two rings, one in the mother and one in the bud. The ring in the bud was always much fainter compared with the mother ring. In cells arrested before anaphase with nocodazole or in a cdc16 mutant, the GFP–Glc7p double ring was more evident (see Fig. 8 A, arrows). Temperature-sensitive alleles of any of the four septins result in disappearance of 10-nm neck filaments (Ford and Pringle 1991). To determine if the localization of GFP–Glc7p at the bud neck is dependent upon septins, GFP–Glc7p was visualized in temperature-sensitive septin mutants. As shown in Fig. 4 B, GFP–Glc7p was present at the bud neck in wild-type cells, but not in cdc3-1 cells, after 50 min at the nonpermissive temperature (37°C). GFP–Glc7p was present at the permissive temperature in the cdc3-1 strain (Fig. 4 B). As a control, Cdc12-GFPm2 was not detected at the mother/bud neck in cdc3-1 cells after 50 min at 37°C. Similar results were obtained using a temperature-sensitive allele of another septin gene, cdc10-1 (data not shown). These results indicate that GFP–Glc7p is dependent upon a functional septin ring structure for accumulation at the bud neck.
GFP–Glc7p also faintly stained the tips and sides of small and medium-sized buds. As is apparent in Fig. 4 B, this location of GFP–Glc7p is not dependent on septins since GFP–Glc7p remains at this location in the cdc3-1 mutant incubated at 37°C. Although not apparent in the data presented in Fig. 4 B, small GFP–Glc7p spots were commonly seen in the nucleolus and less often in the nucleus and the cytoplasm when cells were shifted from 24 to 37°C.
GFP–Glc7p Localizes to the Base of the Mating Projection
Septins are located in a diffuse band at the base of mating projections in pheromone-treated cells (Ford and Pringle 1991; Kim et al. 1991). Because GFP–Glc7p colocalized with septins during vegetative growth, we examined the location of GFP–Glc7p in cells treated with the mating pheromone α-factor. Haploid strain KT1556 was treated with α-factor (8 μg/ml) and images were taken after three hours. As shown in Fig. 5 A, GFP–Glc7p localized to a diffuse band at the base of the mating projection similar to that observed for septins (Fig. 5 A, top, arrows). This band was also observed in a cdc3-1 mutant (YAB58) grown at the permissive temperature (Fig. 5 B), but not after incubation at the nonpermissive temperature (37°C) for 40 min (Fig. 5 D). Similar results were obtained using a cdc10-1 mutant (data not shown). Wild-type strain KT1556 maintained normal GFP–Glc7p localization at 37°C (Fig. 5 C).
Figure 5.
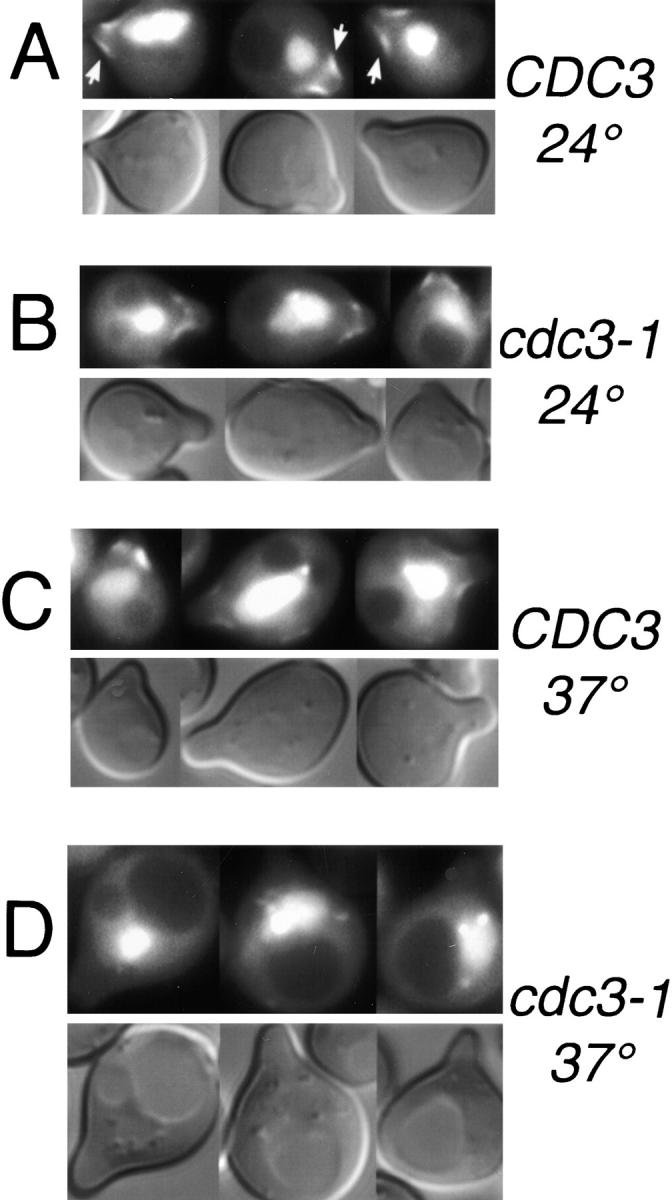
GFP–Glc7p localizes to the base of the mating projection. Images of GFP–Glc7p or DIC in wild-type (YAB1556; A and C) or cdc3-1 cells (YAB58; B and D) that were arrested in α-factor (8 μg/ml) for 3 h at the permissive temperature (24°C; A and B), or after a 40-min shift to the nonpermissive temperature (37°C; C and D). Arrows in A indicate GFP–Glc7p fluorescence at the base of the mating projection.
Glc7p Is Required for Proper Bud-site Selection
GFP–Glc7p localized to the presumptive bud site 15 ± 3 min before bud emergence (n = 4). Using the temperature-sensitive allele cdc28-4 (Reed 1980) to arrest cells at START, the GFP–Glc7p bud neck ring was never observed (data not shown). Septins also localize to the presumptive bud site ∼15 min before bud emergence (Longtine et al. 1996). Additionally, septin mutants have been shown to have defects in bud site selection (Flescher et al. 1993; Chant et al. 1995). To determine if Glc7p has a role in bud site selection, wild-type and glc7 mutants were grown at 30°C in YPD and stained with the chitin-specific dye, calcofluor, to stain their bud scars (see Materials and Methods). As shown in Fig. 6 A, wild-type diploid cells display the typical bipolar budding pattern, whereas glc7-129 cells display a more random pattern. glc7-129 cells also display an approximately two- to threefold increase in chitin staining compared with wild-type cells (Fig. 6 A). Quantitative analysis of the budding pattern in haploids and diploids for the wild-type, glc7-129, and glc7-133 mutants revealed a clear randomization of the budding pattern in glc7-129 cells (Fig. 6B and Fig. C). This random budding pattern was allele-specific as the glc7-133 mutant displayed wild-type budding patterns in haploids and diploids. These results suggest that Glc7p may have a role at the bud neck to control the site of bud emergence and chitin deposition.
Figure 6.
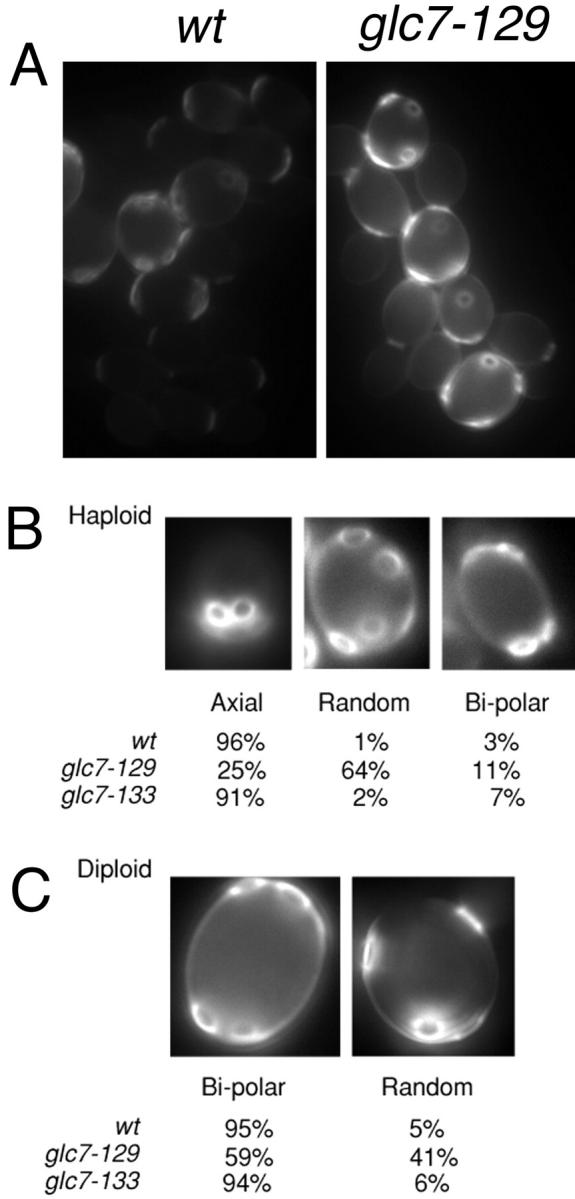
glc7-129 mutants exhibits a random budding pattern. A, Images of wild-type and glc7-129/glc7-129 diploid cells stained with calcofluor. Note that the normalization values for these two images are identical, illustrating the increased level of calcofluor staining in glc7-129 mutants. B, Haploid wild-type (KT1112) and glc7-129 strains (YAB305) were scored for their budding pattern. n = 210 for wild-type cells, n = 211 for glc7-129 cells. C, Diploid wild-type (YAB107) and glc7-129/glc7-129 strains (YAB86) were scored as in A. n = 198 for wild-type cells, n = 186 for glc7-129 cells.
GFP–Glc7p Localizes to Spindle Pole Bodies in Anaphase
In the time-lapse images of YAB4, cells in late mitosis contained two GFP–Glc7p spots at opposite ends of the presumed nucleus (Fig. 2, 12 min, small arrows). In an asynchronous population of YAB4 cells, two GFP–Glcp7p spots were always observed in cells in anaphase (n = 34). To more precisely locate these GFP–Glc7p spots in late mitosis with respect to DNA, a rho0strain containing GFP–Glc7p (YAB6) was stained with the DNA-specific stain DAPI. In Fig. 7 A, three cells in anaphase/telophase are shown that have GFP–Glc7p spots at both ends of the nucleus. The level of fluorescence of the spots in the mother cell (299 ± 131, arbitrary units) and the bud (263 ± 128, arbitrary units) were found to be equal (n = 32 cells).
The location of the GFP–Glc7p spots late in mitosis resembles the pattern observed for both SPBs and centromeres. To test whether the GFP–Glc7p spots colocalize with SPBs, we constructed a yeast strain containing Nuf2–GFP and GFP(10C)–Glc7p. Using immunofluorescence confocal microscopy, Nuf2p was located at the nuclear face of SPBs (Osborne et al. 1994) and a functional Nuf2–GFP fusion was located at SPBs throughout the cell cycle (Kahana et al. 1995). As in the case of the data presented in Fig. 4 A, the GFP proteins fused to Glc7p and Nuf2p were variants that could be distinguished with the JP1 and JP2 filter sets (see Materials and Methods). When Nuf2–GFP and GFP(10C)–Glc7p images were merged, colocalization of Nuf2–GFP and GFP(10C)–Glc7p was evident as the yellow SPBs in the merged images in Fig. 7 B (arrows). Therefore, we conclude that GFP–Glc7p localizes to SPBs, not centromeres (Fig. 8 B), in late mitosis.
To determine when GFP–Glc7p associates with SPBs in mitosis, GFP–Glc7p cells were arrested before anaphase with the microtubule depolymerizing drug, nocodazole. Cells were also arrested before anaphase using a temperature-sensitive cdc16-1 mutant whose product is a component of the anaphase promoting complex. Arrest was confirmed in both cases as >77% large budded cells (n >200). Images of nocodazole-arrested and cdc16-1-arrested cells revealed GFP–Glc7p was concentrated in the nucleus, but GFP–Glc7p spots corresponding to presumed SPBs were never observed (Fig. 8 A). These results indicate that GFP–Glc7p does not localize to SPBs until after anaphase onset.
To more precisely define when GFP–Glc7p localizes to SPBs in anaphase, time-lapse microscopy was performed on a strain (YAB439) that contains GFP–Glc7p and the lacI-GFP/lacO system to visualize kinetochores (Straight et al. 1997). In this system, many copies of lacO are integrated at the TRP1 locus (∼10 kb from CEN4). LacI-GFP that is expressed in these cells binds to lacO, thus marking the kinetochore of centromere IV. Using this system, it was observed that kinetochores move close to their respective SPBs during anaphase A, but can clearly be distinguished from SPBs at the level of light microscopy throughout anaphase (Straight et al. 1997). A montage of a YAB439 cell undergoing anaphase is presented in Fig. 8 B. Before anaphase and sister chromatid separation, when one lacI-GFP spot is observed (Fig. 8 B, black arrowhead, 0 s), GFP–Glc7p spots at SPBs are not seen. At anaphase B onset, defined as the first time point at which the nucleus begins to elongate (Fig. 8 B, 90 s), four spots were visible, two corresponding to the GFP–Glc7p spots (white arrows) at the SPBs and two kinetochore spots trailing the SPBs (black arrowheads). To further confirm localization of GFP–Glc7p to SPBs in anaphase, serial z-sections were captured of YAB439 cells in early and late anaphase. In Fig. 8 C, images of several anaphase cells are shown. Cells in early and late anaphase have GFP–Glc7p concentrated at SPBs. From these and other examples, we conclude that Glc7p mobilizes to the SPB early in anaphase, within 90 s after the initiation of anaphase. GFP–Glc7p remains at the SPB throughout anaphase, but was not observed at SPBs after cytokinesis or cell separation. We compared the time of disappearance of Glc7p from the SPB with the disappearance of Glc7p from the actomyosin ring (see below) and found that the SPB localization of Glc7p became undetectable 2 ± 3 min (n = 6) after disappearance of GFP–Glc7p from the actomyosin ring. Together, these results indicate that GFP–Glc7p becomes concentrated at both SPBs at the start of anaphase and remains there until just before cell separation.
GFP–Glc7p Localizes to a Ring at the Mother/Bud Neck Late in Mitosis
Although not visible in the time-lapse images presented in Fig. 2, higher resolution images of cells in late mitosis (postanaphase) reveal that GFP–Glc7p accumulates in a ring or bar at the mother/bud neck. This structure was observed in all postanaphase cells examined. Time-lapse experiments reveal the dynamic properties of this ring. GFP fluorescence at the bud neck in a representative cell in Fig. 9 A (see Video 2 available at http://www.jcb.org/cgi/content/ full/149/1/125/DC1) is first apparent at the two- or three-minute time point. The band becomes more intense and narrower for another five minutes and then fades in intensity. Fluorescence intensity was measured through the bud neck along a line perpendicular to the elongated spindle at each time point. The fluorescence intensity for each pixel on the line was then plotted below each image. The y-axis represents arbitrary fluorescence intensity and the x-axis represents the position on the 2.5-μm line (from left to right) across the bud neck. The images, as well as the graphs, showed that the GFP–Glc7p ring fluorescence at the bud neck increased near the completion of anaphase. Initially, the ring appeared to undergo contraction-like movement (see carets) without a significant fluorescence increase, and then fluorescence gradually diffused until it was barely detectable by the 15-min time point. Cell separation (defined by the rotation of the daughter cell with respect to the mother cell) occurred 13 ± 2 min later (n = 3). In an asynchronous population of cells, 8% of the cells contained bud neck fluorescence similar to that seen in Fig. 9 A (n = 277). This percentage is consistent with the 8–10 min duration of the ring observed in the time-lapse experiments, assuming a 90-min doubling time at 30°C.
The actomyosin ring has been shown previously to form between two septin rings before cell division and to undergo contraction-like movement before cell separation (Lippincott and Li 1998a). The dynamic nature of the GFP–Glc7p ring in late mitosis suggests that the phosphatase could associate with the actomyosin ring. To determine whether GFP–Glc7p colocalizes with the actomyosin ring late in mitosis, the GFP–Glc7p containing strain, YAB4, was fixed and stained with rhodamine-phalloidin for visualization of F-actin. In Fig. 9 B, images of cells in late mitosis show that the GFP–Glc7p ring colocalizes with the actomyosin ring. The brief accumulation of Glc7p at the site of cytokinesis is consistent with a role for GLC7 in this process, as documented by a glc7 mutant with a defect in cytokinesis (Andrews and Stark 2000).
GFP–Glc7-129p Is Defective in Localization to the Bud Neck and SPBs
The sites of Glc7p accumulation may reflect locations where the phosphatase acts on key substrates. If accumulation at the bud neck, SPBs and actomyosin ring reflect sites of phosphatase activity, the products of GLC7 mutations with defects in the cell cycle and bud emergence/morphogenesis may not localize to some or all of these sites. Therefore, we fused GFP to glc7-129 and expressed the fusion protein from the CEN vector pNC160 in wild-type cells (KT1357). glc7-129 mutants have a mitotic delay caused by activation of the mitotic checkpoint (Bloecher and Tatchell 1999) and as shown in Fig. 6, display defects in bud site selection and chitin synthesis. The overall fluorescence levels in these transformants were less than half that of the wild-type GFP–Glc7p fusion, which correlates with the reduced steady state levels of GFP–Glc7-129p observed using immunoblot analysis (data not shown). GFP–Glc7-129p retains its nuclear localization. However, as shown in Fig. 10 A, GFP–Glc7-129p fails to localize to the bud neck of cells with small and medium-sized buds (arrows). In contrast, GFP–Glc7-129p did accumulate at the bud neck in large-budded cells that had completed anaphase (asterisks).
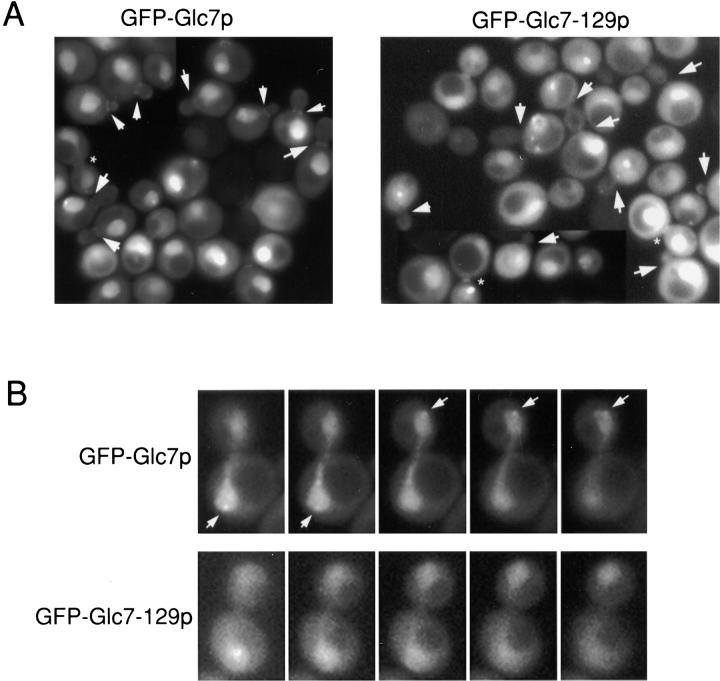
Figure 10.
GFP–Glc7-129p is defective in localization to the bud neck and SPBs. A, Images of wild-type strain KT1357 transformed with either pAB26 (GFP–GLC7) or pAB18 (GFP–glc7-129) after inoculation into YPD media for 4 h at 30°C. Arrows indicate bud neck localization of GFP–Glc7p and the lack of this localization for GFP–Glc7-129p. Asterisks denote GFP–Glc7p or GFP–Glc7-129p at the presumed actomyosin ring. For ease of comparison, grayscale values for GFP–Glc7-129p were increased to approximately the same levels as those of GFP–Glc7p. B, Serial z-sections are shown through anaphase KT1357 cells containing pAB26 or pAB18. Arrows indicate GFP–Glc7p spots at SPBs. This localization is not observed for GFP–Glc7-129p.
Notably, GFP–Glc7-129p was also reduced in concentration at the SPBs of anaphase cells. Serial z-sections of representative cells in Fig. 10 B illustrate the loss of GFP–Glc7-129p from the SPB. To quantitate SPB localization, anaphase cells were examined in five z-sections and scored for the presence of GFP spots at the presumptive SPBs. GFP–Glc7p was identified in 87% (59/68) of the SPBs (34 anaphase cells), whereas GFP–Glc7-129p was only found at 19% (13/70) of the SPBs (35 anaphase cells). We also constructed a gene fusion to glc7-127, an allele with defects in glucose repression and sporulation (Baker et al. 1997), but with normal bud site selection, chitin deposition, and cell cycle regulation (data not shown). GFP–Glc7-127p localized normally to the bud neck and to SPBs (data not shown), strengthening the argument that Glc7p at the bud neck, and possibly the SPB, is required for activities involved in bud morphogenesis and cell cycle regulation, respectively.
Discussion
We have shown dynamic changes in the location of Glc7p throughout the cell cycle. Glc7p appears at the site of bud initiation ∼15 min before bud formation and remains there until mitosis. At the end of mitosis, Glc7p reappears at the bud neck, this time overlapping directly with the actomyosin ring that forms during cytokinesis. At the start of anaphase, Glc7p appears at the SPBs and remains there until cytokinesis. More static sites of Glc7p accumulation occur in the nucleolus, where it accumulates in the highest concentration, in the nucleus, and in the cytoplasm. The plethora of locations of Glc7p accumulation is in good agreement with the many roles that this phosphatase plays in cellular processes. It is worth noting that we may have identified only the sites of highest Glc7p concentration; physiologically important sites may have been missed if the concentration of Glc7p is below the limit of detection at these sites. The dynamic changes in GFP–Glc7p subcellular localization are consistent with redistribution of the phosphatase rather than changes in expression. GLC7 transcript levels are constant during the cell cycle, as judged using DNA microarrays in synchronized cultures (Spellman et al. 1998) and GFP–Glc7p fluorescence levels do not undergo dramatic changes during the cell cycle.
Glc7p accumulates to its highest levels in the nucleus and nucleolus throughout the cell cycle. PP1 has also been reported to be predominantly nuclear in mammals (Fernandez et al. 1992), A. nidulans (Doonan and Morris 1989), and fission yeast (Ohkura et al. 1989). A nuclear location for Glc7p is consistent with its proposed role in regulating microtubule/kinetochore attachment in mitosis (Bloecher and Tatchell 1999; Sassoon et al. 1999) and the expression of CUP1 (Lin and Lis 1999). How does GFP–Glc7p localize to the nucleus and nucleolus? Using the PSORT II program (Leger-Silvestre et al. 1999) for predicting subcellular localization, Glc7p contains a putative nuclear localization signal, but was predicted to be cytoplasmic, based on overall amino acid composition. The predicted nuclear localization signal, RKKK, is located at the extreme COOH terminus (amino acids 309–312). This sequence is missing from the product of GLC7-305°, a fully functional nonsense mutant that lacks the COOH-terminal eight amino acid residues of Glc7p (Baker et al. 1997). A GFP–Glc7-305°p fusion also localizes normally to the nucleus, nucleolus, SPBs, bud neck, and the actomyosin ring in a manner identical to wild-type GFP–Glc7p (Bloecher, A., and K. Tatchell, unpublished data). This indicates that Glc7p does not have a conventional nuclear localization signal and may depend upon binding to another protein(s) for nuclear targeting. Glc7p is located primarily in the nucleus during logarithmic growth, but is found dispersed uniformly throughout the cell in stationary phase (Bloecher, A., and K. Tatchell, unpublished data). A similar distribution pattern has been reported for PP1 in mammalian tissue culture cells, where PP1 is located primarily in the cytoplasm in quiescent cells, but accumulates in the nucleus upon serum addition and entry into the mitotic cell cycle (Fernandez et al. 1992).
The isoform-specific patterns of PP1 localization in mammalian cells have been proposed to be due to the unique COOH-terminal extensions on PP1 subtypes (Andreassen et al. 1998). These extensions, which vary in length from 20–30 amino acid residues, could potentially bind targeting factors and direct PP1 isoforms to their specified locations. Glc7p is directed to some of the same locations that are occupied by single mammalian PP1 isoforms: Glc7p and PP1γ are directed to nucleoli, and Glc7p and PP1α are directed to SPBs and centrosomes, respectively. However, the COOH terminus of Glc7p is not required for proper localization or function, indicating that targeting subunits may interact productively with only the catalytic domain of Glc7p. It has been observed that a conserved hydrophobic pocket on the catalytic domain is important for binding many PP1 targeting subunits (Egloff et al. 1997).
The phenotypes of glc7 mutants and the functions of Glc7p-interacting proteins do not readily reveal a role for Glc7p in the nucleolus. In A. nidulans, a PP1 mutant has hyperphosphorylated nucleolar proteins, supporting the idea that substrates for PP1 are found in the nucleolus (Doonan and Morris 1989). Mammalian PP1γ localizes to the nucleolus during interphase (Andreassen et al. 1998). Mammalian ribosomal protein L5, the homologue to the yeast ribosomal protein Rpl5p, associates with PP1 in the yeast two hybrid system (Hirano et al. 1995), but the possibility that Glc7p has a role in ribosome assembly or some other nucleolar function has not been examined. Another possibility is that Glc7p is sequestered in the nucleolus similar to the dual-specificity phosphatase, Cdc14p. Cdc14p is retained in the nucleolus during most of the cell cycle, but is released during anaphase when it plays a key role in the exit from mitosis (Shou et al. 1999; Straight et al. 1999; Visintin et al. 1999). Time-lapse data showed that GFP–Glc7p was concentrated in the nucleolus throughout the mitotic cell cycle, indicating nucleolar sequestration and release is not a regulatory mechanism for Glc7p. In a fraction of anaphase cells, nucleolar fluorescence of GFP–Glc7p was not apparent. Because GFP–Glc7p was observed in a clearly defined nucleolus in early and late anaphase cells, loss of GFP–Glc7p fluorescence in the nucleolus may be a consequence of a morphological change in the nucleolus during chromosome segregation.
GFP–Glc7p, Septins, and Bud Site Selection
Septins have roles in cytokinesis, morphogenesis, cell wall deposition, bud-site selection, sporulation, and mating (Longtine et al. 1996). Temperature-sensitive septin mutants form elongated buds at the restrictive temperature with actin hyperpolarized towards the bud tip (Adams and Pringle 1984). We have shown that the accumulation of GFP–Glc7p at the bud neck and at the base of mating projections is septin-dependent. In support of a functional role for this association, Andrews and Stark 2000 have noted that glc7-10 mutants have defects in bud morphology. We have also found that glc7-129 mutants frequently develop elongated buds at elevated temperatures (Bloecher, A., and K. Tatchell, unpublished data) and also show increased and delocalized chitin deposition (Fig. 6 A). The failure of GFP–Glc7-129p to localize to the bud neck is consistent with Glc7p acting at this location to regulate bud morphogenesis. We have no evidence for a direct interaction between septins and Glc7p. However, Bni4p, a septin binding protein that targets chitin synthase to the bud neck, has recently been shown to interact with Glc7p in a two hybrid screen (Uetz et al. 2000). Bni4p is also located on the mother side of the bud neck (DeMarini et al. 1997) as predicted, if Bni4p is responsible for tethering Glc7p to the bud neck. Further work will be necessary to determine if this or other proteins target Glc7p to the bud neck.
We discovered that glc7-129 mutants exhibited a random budding pattern in haploid and diploid cells. Mutants that are defective in bud site selection have been classified into three different groups based on their effect on budding pattern in haploid and diploid cells. One group, including septin mutants (Flescher et al. 1993; Longtine et al. 1996), changes the axial budding pattern to bipolar in haploid cells, but does not affect bud site selection in diploid cells. Another group affects bipolar budding in the diploid, but has no effect on haploid cells. The third group, of which glc7-129 is a member, causes random budding in both haploid and diploid cells. This group, consisting of mutant in BUD1, BUD2, and BUD5 (Chant et al. 1991; Chant and Herskowitz 1991; Powers et al. 1991; Park et al. 1993) is believed to transmit spatial cues from the presumptive bud site to proteins involved in initiating polarized growth, such as the GTPase Cdc42p and its guanine nucleotide exchange factor, Cdc24p. cdc24 mutants also exhibit a random pattern of bud site selection at the permissive temperature (Sloat et al. 1981). Interestingly, a genetic connection exists between GLC7 and CDC24/CDC42. Mutations in GLC7, including glc7-Y170 and glc7-129, can partially suppress the growth defects caused by mutations in CDC24 and CDC42 (Hisamoto et al. 1994; Bloecher, A., and K. Tatchell, unpublished data). We do not know if the bud site selection defect of the cdc24 mutant is also suppressed by glc7 mutants.
The appearance of the GFP–Glc7p ring at the bud neck prompted us to determine if Glc7p functioned in proper selection of the bud site. However, the defect we observed in the glc7-129 mutant, randomization of the bud site in both haploid and diploid cells, is contrary to what we would predict if the Glc7p holoenzyme that acts in bud site selection is located at the septin ring. This is because septin mutants have not been reported to affect the bipolar budding pattern in diploid cells. Because the bud neck association of GFP–Glc7p in diploid cells is lost in septin mutants, this would suggest that the Glc7p holoenzyme responsible for bud site selection resides at another location.
GFP–Glc7p and SPB Localization
At the onset of anaphase, we find that GFP–Glc7p accumulates at SPBs and remains there until cytokinesis. Similarly, mammalian PP1α is located at centrosomes during mitosis (Andreassen et al. 1998). In A. nidulans, the spindle plaques of a PP1 mutant contain hyperphosphorylated proteins (Doonan and Morris 1989), supporting the possibility that PP1 is located at the spindle plaques. There are several plausible functions for Glc7p at the SPBs. First, Glc7p may regulate microtubule dynamics. Although glc7-129, a GLC7 mutant known to activate the spindle/kinetochore checkpoint, is not sensitive to microtubule depolymerization, rates of anaphase spindle elongation are more rapid than wild-type (Bloecher and Tatchell 1999). Furthermore, several microtubule-dependent events, such as nuclear migration, spindle orientation, and anaphase spindle breakdown are partially defective in this glc7 mutant (Bloecher, A., and K. Tatchell, unpublished data). Regulation of microtubule dynamics by Glc7p could be through the regulation of microtubule-dependent motor proteins. Strains containing glc7-129 are inviable when they lack any one of several microtubule-dependent motor proteins or components of dynactin (Bloecher, A., and K. Tatchell, unpublished data). The loss of GFP–Glc7-129p from SPBs suggests that one or more of these functions may be carried out at the SPB.
The spindle checkpoint was recently shown to consist of two pathways: Bub1p, Bub3p, Mad1p, Mad2p, Mad3p, and Mps1p constitute components of the checkpoint that block anaphase onset, whereas Bub2p is necessary for a later block before cytokinesis (Alexandru et al. 1999). glc7-129 mutants delay anaphase due to activation of the Mad1p/Bub3p-dependent checkpoint, but also delay cytokinesis (Bloecher and Tatchell 1999), suggesting that the Bub2p-dependent checkpoint may also be activated. Like GFP–Glc7p, Bub2p has also been shown to localize to SPBs in anaphase (Fraschini et al. 1999; Li 1999). Therefore, it is possible that the SPB is the site at which the glc7-129 defect causes a delay in cytokinesis.
The relevant substrate or substrates at the SPB for Glc7p are not known, but one intriguing possibility is Spc110, an essential component of the central plaque of SPBs (Rout and Kilmartin 1990) that exists in multiple phosphorylated states (Friedman et al. 1996; Stirling and Stark 1996). A hyperphosphorylated form of the protein that migrates as a 120-kD protein by SDS-PAGE is converted to a 112-kD form during anaphase. In vitro phosphatase treatment of the 120-kD form shifts its electrophoretic mobility to that of the 112-kD form. Treatment of cells with nocodazole and alpha factor results in an accumulation of the 120-kD form and 112-kD form, respectively. These and other experiments indicate that Spc110 becomes hyperphosphorylated when the mitotic spindle forms and is rapidly dephosphorylated at the onset of anaphase (Friedman et al. 1996). Thus, Glc7p is temporally positioned to act on Spc110 during anaphase.
GFP–Glc7p at the Mother/Bud Neck before Cytokinesis
Near the end of anaphase, we observed the appearance of a GFP–Glc7p ring at the mother/bud neck that colocalizes with the ring of F-actin. The temporal appearance of this ring is very similar to that of Cyk1p, an actin-recruiting protein involved in cytokinesis (Lippincott and Li 1998b). Based on this and the dynamic nature of the GFP–Glc7p ring, we propose that Glc7p can associate with the actomyosin ring. A functional role for Glc7p at the actomyosin ring is supported by the finding that a GLC7 mutant is delayed in cytokinesis or cell separation (Andrews and Stark 2000). Substrates for Glc7p at the actomyosin ring, known to contain actin, Myo1p (type II myosin), Cyk1p, and Cyk2p (Bi et al. 1998; Lippincott and Li 1998a) are not known, but in mammalian cells, a PP1 holoenzyme composed of PP1 and the GM targeting subunit has been shown to dephosphorylate myosin II light chain (Chisholm and Cohen 1988).
Acknowledgments
The authors would like to thank Jim Waddle for technical assistance with microscopy, Aaron Straight, Andrew Murray, John Aris, Elaine Yeh, Aaron Neiman, Lucy Robinson, Jason Kahana, and Pam Silver for plasmids, strains, and antibodies; Debbie Frederick, Bill Venturi, Lucy Robinson, Eric Aamodt and Dan Burke for helpful discussions.
This work was supported by National Institutes of Health grant GM-477899.
Footnotes
The online version of this article contains supplemental material.
Abbreviations used in this paper: 2D, two-dimensional; DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; PP1, protein phosphatase type 1; SPB, spindle pole body.
References
- Adams A., Pringle J. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae . J. Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandru G., Zachariae W., Schleiffer A., Nasmyth K. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2707–2721. doi: 10.1093/emboj/18.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen P.R., Lacroix F.B., Villa-Moruzzi E., Margolis R.L. Differential subcellular localization of protein phosphatase-1 alpha, gamma1, and delta isoforms during both interphase and mitosis in mammalian cells. J. Cell Biol. 1998;141:1207–1215. doi: 10.1083/jcb.141.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P.D., Stark M.J. Type 1 protein phosphatase is required for maintenance of cell wall integrity, morphogenesis and cell cycle progression in Saccharomyces cerevisiae . J Cell Sci. 2000;113:507–520. doi: 10.1242/jcs.113.3.507. [DOI] [PubMed] [Google Scholar]
- Aris J.P., Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.H., Frederick D.L., Bloecher A., Tatchell K. Alanine scanning mutagenesis of protein phosphatase type 1 in the yeast Saccharomyces cerevisiae . Genetics. 1997;145:615–626. doi: 10.1093/genetics/145.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Maddox P., Lew D.J., Salmon E.D., McMillan J.N., Yeh E., Pringle J.R. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloecher A., Tatchell K. Defects in Saccharomyces cerevisiae protein phosphatase type I activate the spindle/kinetochore checkpoint. Genes Dev. 1999;13:517–522. doi: 10.1101/gad.13.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J., Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991;65:1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- Chant J., Corrado K., Pringle J.R., Herskowitz I. Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1 . Cell. 1991;65:1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- Chant J., Mischke M., Mitchell E., Herskowitz I., Pringle J.R. Role of Bud3p in producing the axial budding pattern of yeast. J. Cell Biol. 1995;129:767–778. doi: 10.1083/jcb.129.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm A.A.K., Cohen P. The myosin-bound form of protein phosphatase 1 (PP-1M) is the enzyme that dephosphorylates native myosin in skeletal and cardiac muscles. Biochim. Biophys. Acta. 1988;971:163–169. doi: 10.1016/0167-4889(88)90188-7. [DOI] [PubMed] [Google Scholar]
- Cormack B.P., Valdivia R.H., Falkow S. FACs-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- de Beus E., Brockenbrough J.S., Hong B., Aris J.P. Yeast NOP2 encodes an essential nucleolar protein with homology to a human proliferation marker. J. Cell Biol. 1994;127:1799–1813. doi: 10.1083/jcb.127.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini D.J., Adams A.E., Fares H., De Virgilio C., Valle G., Chuang J.S., Pringle J.R. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePaoli-Roach A.A., Park I.K., Cerovsky V., Csortos C., Durbin S.D., Kuntz M.J., Sitikov A., Tang P.M., Verin A., Zolnierowicz S. Serine/threonine protein phosphatases in the control of cell function. Adv. Enzyme Regul. 1994;34:199–224. doi: 10.1016/0065-2571(94)90017-5. [DOI] [PubMed] [Google Scholar]
- Doonan J.H., Morris N.R. The bimG gene of Aspergillus nidulans, required for completion of anaphase, encodes a homolog of mammalian phosphoprotein phosphatase 1. Cell. 1989;57:987–996. doi: 10.1016/0092-8674(89)90337-1. [DOI] [PubMed] [Google Scholar]
- Egloff M.P., Johnson D.F., Moorhead G., Cohen P.T., Cohen P., Barford D. Structural basis for the recognition of regulatory subunits by catalytic subunits of protein phosphatase 1. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:1876–1887. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G., Lewis G., Ramsay G., Bishop J.M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faux M.C., Scott J.D. More on target with protein phosphorylationconferring specificity by location. TIBS. 1996;21:312–315. [PubMed] [Google Scholar]
- Fernandez A., Brautigan D.L., Lamb N.J.C. Protein phosphatase type 1 in mammalian cell mitosischromosome localization and involvement in mitotic exit. J. Cell Biol. 1992;116:1421–1430. doi: 10.1083/jcb.116.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flescher E.G., Madden K., Snyder M. Components required for cytokinesis are important for bud site selection in yeast. J. Cell Biol. 1993;122:373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford S.K., Pringle J.R. Cellular morphogenesis in the Saccharomyces cerevisiae cell cyclelocalization of the CDC11 gene product and the timing of events at the budding site. Dev. Genet. 1991;12:281–292. doi: 10.1002/dvg.1020120405. [DOI] [PubMed] [Google Scholar]
- François J.M., Thompson-Jaeger S., Skroch J., Zellenka U., Spevak W., Tatchell K. GAC1 may encode a regulatory subunit for protein phosphatase type 1 in Saccharomyces cerevisiae . EMBO (Eur. Mol. Biol. Organ.) J. 1992;11:87–96. doi: 10.1002/j.1460-2075.1992.tb05031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., Formenti E., Lucchini G., Piatti S. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J. Cell Biol. 1999;145:979–991. doi: 10.1083/jcb.145.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D.B., Sundberg H.A., Huang E.Y., Davis T.N. The 110-kD spindle pole body component of Saccharomyces cerevisiae is a phosphoprotein that is modified in a cell cycle-dependent manner. J. Cell Biol. 1996;132:903–914. doi: 10.1083/jcb.132.5.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G.R. Fink. 1991. Guide to yeast genetics and molecular biology. In Methods Enzymol. Vol. 194. Academic Press, San Diego, CA. 933 pages. [PubMed]
- Haarer B.K., Pringle J.R. Immunofluorescence localization of the Saccharomyces cerevisiae CDC12 gene product to the vicinity of the 10-nm filaments in the mother-bud neck. Mol. Cell. Biol. 1987;7:3678–3687. doi: 10.1128/mcb.7.10.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K., Ito M., Hartshorne D.J. Interaction of the ribosomal protein, L5, with protein phosphatase type 1. J. Biol. Chem. 1995;270:19786–19790. doi: 10.1074/jbc.270.34.19786. [DOI] [PubMed] [Google Scholar]
- Hisamoto N., Sugimoto K., Matsumoto K. The Glc7 type 1 protein phosphatase of Saccharomyces cerevisiae is required for cell cycle progression in G2/M. Mol. Cell. Biol. 1994;14:3158–3165. doi: 10.1128/mcb.14.5.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamoto N., Frederick D.L., Sugimoto K., Tatchell K., Matsumoto K. The EGP1 gene may be a positive regulator of protein phosphatase type 1 in the growth control of Saccharomyces cerevisiae . Mol. Cell. Biol. 1995;15:3767–3776. doi: 10.1128/mcb.15.7.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman D.A., Yang S., Drubin D.G. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae . J. Cell Biol. 1993;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard M., Cohen P. The glycogen-binding subunit of protein phosphatase-1G from rabbit skeletal musclefurther characterization of its structure and glycogen-binding properties. Eur. J. Biochem. 1989;180:457–465. doi: 10.1111/j.1432-1033.1989.tb14668.x. [DOI] [PubMed] [Google Scholar]
- Hubbard M.J., Cohen P. On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem. Sci. 1993;18:172–177. doi: 10.1016/0968-0004(93)90109-z. [DOI] [PubMed] [Google Scholar]
- Kahana J.A., Schnapp B.J., Silver P.A. Kinetics of spindle pole body separation in budding yeast. Proc. Natl. Acad. Sci. USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana J.A., Schlenstedt G., Evanchuk D.M., Geiser J.R., Hoyt M.A., Silver P.A. The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol. Biol. Cell. 1998;9:1741–1756. doi: 10.1091/mbc.9.7.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A. Methods in Yeast Genetics 1994. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: pp. 234 [Google Scholar]
- Kakinoki Y., Somers J., Brautigan D.L. Multisite phosphorylation and the nuclear localization of phosphatase inhibitor 2-green fluorescent protein fusion protein during S phase of the cell growth cycle. J. Biol. Chem. 1997;272:32308–32314. doi: 10.1074/jbc.272.51.32308. [DOI] [PubMed] [Google Scholar]
- Kim H.B., Haarer B.K., Pringle J.R. Cellular morphogenesis in the Saccharomyces cerevisiae cell cyclelocalization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol. 1991;112:535–544. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger-Silvestre I., Trumtel S., Noaillac-Depeyre J., Gas N. Functional compartmentalization of the nucleus in the budding yeast Saccharomyces cerevisiae . Chromosoma. 1999;108:103–113. doi: 10.1007/s004120050357. [DOI] [PubMed] [Google Scholar]
- Li R. Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc. Natl. Acad. Sci. USA. 1999;96:4989–4994. doi: 10.1073/pnas.96.9.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.T., Lis J.T. Glycogen synthase phosphatase interacts with heat shock factor to activate CUP1 gene transcription in Saccharomyces cerevisiae . Mol. Cell. Biol. 1999;19:3237–3245. doi: 10.1128/mcb.19.5.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution J. Cell Biol. 143 1998. 1947 1960a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis J. Cell Biol. 140 1998. 355 366b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., DeMarini D.J., Valencik M.L., Al-Awar O.S., Fares H., De Virgilio C., Pringle J.R. The septinsroles in cytokinesis and other processes. Curr. Opin. Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- MacKelvie S.H., Andrews P.D., Stark M.J.R. The Saccharomyces cerevisiae gene SDS22 encodes a potential regulator of the mitotic function of yeast type 1 protein phosphatase. Mol. Cell. Biol. 1995;15:3777–3785. doi: 10.1128/mcb.15.7.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T.J., Sambrook, Fritsch E.F. Molecular Cloning, A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Mumby M.C., Walter G. Protein serine/threonine phosphatasesstructure, regulation, and functions in cell growth. Physiol. Rev. 1993;73:673–699. doi: 10.1152/physrev.1993.73.4.673. [DOI] [PubMed] [Google Scholar]
- Nelson K.K., Holmer M., Lemmon S.K. SCD5, a suppressor of clathrin deficiency, encodes a novel protein with a late secretory function in yeast. Mol. Biol. Cell. 1996;7:245–260. doi: 10.1091/mbc.7.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., Kinoshita N., Miyatani S., Toda T., Yanagida M. The fission yeast dis2 + gene required for chromosome disjoining encodes one of two putative type 1 protein phosphatases. Cell. 1989;57:997–1007. doi: 10.1016/0092-8674(89)90338-3. [DOI] [PubMed] [Google Scholar]
- Ormö M., Cubitt A.B., Kallio K., Gross L.A., Tsien R.Y., Remington S.J. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- Osborne M.A., Schlenstedt G., Jinks T., Silver P.A. Nuf2, a spindle pole body-associated protein required for nuclear division in yeast. J. Cell Biol. 1994;125:853–866. doi: 10.1083/jcb.125.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Chant J., Herskowitz I. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature. 1993;365:269–274. doi: 10.1038/365269a0. [DOI] [PubMed] [Google Scholar]
- Powers S., Gonzales E., Christensen T., Cubert J., Broek D. Functional cloning of BUD5, a CDC25-related gene from S. cerevisiae that can suppress a dominant-negative RAS2 mutant. Cell. 1991;65:1225–1231. doi: 10.1016/0092-8674(91)90017-s. [DOI] [PubMed] [Google Scholar]
- Pringle J.R., Adams A.E., Drubin D.G., Haarer B.K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Reed S.I. The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics. 1980;95:561–577. doi: 10.1093/genetics/95.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson L.C., Bradley C., Bryan J.D., Jerome A., Kweon Y., Panek H.R. The yck2 yeast casein kinase 1 isoform shows cell cycle-specific localization to sites of polarized growth and is required for proper septin organization. Mol. Biol. Cell. 1999;10:1077–1092. doi: 10.1091/mbc.10.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B., Roeder G.S. Meiosis in asynaptic yeast. Genetics. 1990;126:563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Kilmartin J.V. Isolation of the yeast spindle and spindle pole body. J. Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassoon I., Severin F.F., Andrews P.D., Taba M.R., Kaplan K.B., Ashford A.J., Stark M.J.R., Sorger P.K., Hyman A.A. Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 1999;13:545–555. doi: 10.1101/gad.13.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S.L., Yeh E., Bloom K., Salmon E.D. Imaging green fluorescent protein fusion proteins in Saccharomyces cerevisiae . Curr. Biol. 1997;7:701–704. doi: 10.1016/s0960-9822(06)00299-5. [DOI] [PubMed] [Google Scholar]
- Shenolikar S. Protein serine/threonine phosphatasesnew avenues for cell regulation. Annu. Rev. Cell Biol. 1994;10:55–86. doi: 10.1146/annurev.cb.10.110194.000415. [DOI] [PubMed] [Google Scholar]
- Shou W., Seol J.H., Shevchenko A., Baskerville C., Moazed D., Chen Z.W., Jang J., Charbonneau H., Deshaies R.J. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Sloat B.F., Adams A., Pringle J.R. Roles of the CDC24 gene product in cellular morphogenesis during the Saccahromyces cerevisiae cell cycle. J. Cell Biol. 1981;89:395–405. doi: 10.1083/jcb.89.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman P.T., Sherlock G., Zhang M.Q., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M.J.R. Yeast protein serine/threonine phosphatasesmultiple roles and diverse regulation. Yeast. 1996;12:1647–1675. doi: 10.1002/(SICI)1097-0061(199612)12:16%3C1647::AID-YEA71%3E3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Stirling D.A., Stark M.J. The phosphorylation state of the 110 kDa component of the yeast spindle pole body shows cell cycle dependent regulation. Biochem. Biophys. Res. Commun. 1996;222:236–242. doi: 10.1006/bbrc.1996.0728. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Marshall W.F., Sedat J.W., Murray A.W. Mitosis in living budding yeastanaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Sedat J.W., Murray A.W. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A.F., Shou W., Dowd G.J., Turck C.W., Deshaies R.J., Johnson A.D., Moazed D. Net1, a sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- Stuart J.S., Frederick D.L., Varner C.M., Tatchell K. The mutant type 1 protein phosphatase encoded by glc7-1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol. Cell. Biol. 1994;14:896–905. doi: 10.1128/mcb.14.2.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A., Lin F., Sarabia M.J.F., Arndt K.T. The SIT4 protein phosphatase is required in late G1 for progression into S phase. Cold Spring Harbor Symp. Quant. Biol. 1991;56:75–81. doi: 10.1101/sqb.1991.056.01.011. [DOI] [PubMed] [Google Scholar]
- Tang P., Bandor J., Swiderek K., DePaoli-Roach A. Molecular cloning and expression of the regulatory (RG1) subunit of the glycogen-associated protein phosphatase. J. Biol. Chem. 1991;266:15782–15879. [PubMed] [Google Scholar]
- Tu J., Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae . EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J., Song W., Carlson M. Protein phosphatase type 1 interacts with proteins required for meiosis and other cellular processes in Saccharomyces cerevisiae . Mol. Cell. Biol. 1996;16:4199–4206. doi: 10.1128/mcb.16.8.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae . Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang E.S., Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- Waddle J.A., Karpova T.S., Waterston R.H., Cooper J.A. Movement of cortical actin patches in yeast. J. Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.H., Lambie E.J., Hardin J., Craft J., Snyder M. Higher order structure is present in the yeast nucleusautoantibody probes demonstrate that the nucleolus lies opposite the spindle pole body. Chromosoma. 1989;98:123–128. doi: 10.1007/BF00291048. [DOI] [PubMed] [Google Scholar]
- Zhang S., Guha S., Volkert F.C. The Saccharomyces SHP1 gene, which encodes a regulator of phosphoprotein phosphatase 1 with differential effects on glycogen metabolism, meiotic differentiation, and mitotic cell cycle progression. Mol. Cell. Biol. 1995;15:2037–2050. doi: 10.1128/mcb.15.4.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]