Abstract
Transcription and splicing of messenger RNAs are temporally and spatially coordinated through the recruitment by RNA polymerase II of processing factors. We questioned whether RNA polymerase I plays a role in the recruitment of the ribosomal RNA (rRNA) processing machinery. During Xenopus laevis embryogenesis, recruitment of the rRNA processing machinery to the nucleolar domain occurs in two steps: two types of precursor structures called prenucleolar bodies (PNBs) form independently throughout the nucleoplasm; and components of PNBs I (fibrillarin, nucleolin, and the U3 and U8 small nucleolar RNAs) fuse to the nucleolar domain before components of PNBs II (B23/NO38). This fusion process is independent of RNA polymerase I activity, as shown by actinomycin D treatment of embryos and by the lack of detectable RNA polymerase I at ribosomal gene loci during fusion. Instead, this process is concomitant with the targeting of maternally derived pre-rRNAs to the nucleolar domain. Absence of fusion was correlated with absence of these pre-rRNAs in nuclei where RNA polymerase II and III are inhibited. Therefore, during X. laevis embryogenesis, the recruitment of the rRNA processing machinery to the nucleolar domain could be dependent on the presence of pre-rRNAs, but is independent of either zygotic RNA polymerase I transcription or the presence of RNA polymerase I itself.
Keywords: prenucleolar body, nucleologenesis, pre-rRNA, RNA polymerase I transcription, Xenopus laevis development
Introduction
Defining the coordinating events during RNA transcription and processing is becoming increasingly important in the context of their regulatory role in gene expression and nuclear organization (for reviews see Lamond and Earnshaw 1998; Misteli and Spector 1998). It has been reported that messenger RNA (mRNA) transcription and processing is coordinated by the recruitment of processing factors to transcription sites by RNA polymerase II (RNA pol II; Jiménez-Garcia and Spector 1993; Misteli et al. 1997; Bentley 1999; Misteli and Spector 1999). Remarkably, the activation of ribosomal gene (rDNA) transcription at the end of mitosis is also accompanied by the recruitment of processing complexes (Scheer and Benavente 1990; Thiry and Goessens 1996). This therefore raises the issue of whether there is a link between active transcription and processing for ribosomal RNA (rRNA).
Processing of rRNAs involves cleavage, methylation, and pseudouridylation of the primary rRNAs (Hadjiolov 1985; Smith and Steitz 1997). Cleavage is controlled by several ribonucleoprotein (RNP) complexes that act in an ordered manner to remove the external transcribed spacers (5′ETS and 3′ETS) and the internal transcribed spacers (ITS1 and ITS2). Fibrillarin (Ochs et al. 1985b) and nucleolin (Ginisty et al. 1998) associated with several small nucleolar RNAs (snoRNAs), including U3, could play a role during the first steps of rRNA processing (for a review see Tollervey 1996). Subsequent cleavages involve endoribonuclease activities such as the MRP RNase complex (Lygerou et al. 1996a,Lygerou et al. 1996b; Dichtl and Tollervey 1997; Pluk et al. 1999; Van Eenennaam et al. 1999) for the ITS1, and protein B23 (Savkur and Olson 1998) and U8 (Michot et al. 1999) for the ITS2. In Xenopus laevis, U8 was shown to be involved both in early 3′ETS and late ITS2 cleavages (Peculis and Steitz 1993). Exoribonuclease activity is required for complete processing of 5.8 S rRNAs by the exosome complex (Mitchell et al. 1997; Briggs et al. 1998; Allmang et al. 1999).
Several proteins of the rRNA processing machinery are detected in prenucleolar bodies (PNBs; Ochs et al. 1985a), which are scattered throughout the nucleus in early G1 (Benavente et al. 1987; Jiménez-Garcia et al. 1989; Ochs and Smetana 1991; Azum-Gélade et al. 1994; Scheer and Weisenberger 1994; Beven et al. 1996; Dundr et al. 1997; Zatsepina et al. 1997). Fibrillarin, nucleolin, Nop52, PM-Scl 100/exosome, and protein B23 are found within these PNBs, as are the U3 and U14 snoRNAs (Azum-Gélade et al. 1994; Gautier et al. 1994; Jiménez-Garcia et al. 1994; Beven et al. 1996; Mitchell et al. 1997; Fomproix and Hernandez-Verdun 1999; Savino et al. 1999). Therefore, the PNBs appear to contain preassembled nucleolar complexes mainly involved in processing steps of the pre-rRNAs (Scheer and Weisenberger 1994). Distinct PNBs are involved in the delivery of specific processing complexes to the nucleolar domain (Fomproix and Hernandez-Verdun 1999; Savino et al. 1999). Since the delivery of the different PNBs follows a temporal order, it has been proposed that reassembly into nucleoli could proceed by a stepwise mechanism that reflects the role of these complexes (Savino et al. 1999). The temporally regulated targeting of PNBs during the cell cycle thus appears dependent on the constituents of these PNBs. However, the involvement of the transcription and/or the transcripts in this process is unknown.
The PNBs fuse at the nucleolar organizer regions (NORs) at the end of mitosis, when rRNA synthesis resumes (Jiménez-Garcia et al. 1994; Fomproix et al. 1998). This fusion is inhibited by microinjection into mitotic cells of antibodies directed against RNA polymerase I (RNA pol I; Benavente et al. 1987) and against DNA topoisomerase I (Weisenberger et al. 1993). Therefore, it has been proposed that fusion of PNBs into nucleoli is dependent on the resumption of rDNA transcription (Scheer et al. 1993).
Remarkably, during early X. laevis embryogenesis, a unique situation was revealed in which regroupment of fibrillarin and nucleolin around the rDNA occurred before the apparent activation of RNA pol I–dependent transcription (Verheggen et al. 1998). The first cell cycles of X. laevis embryogenesis provide an interesting biological situation since transcription is established de novo after 12 synchronized cell cycles devoid of transcription (Brown and Littna 1964; Newport and Kirschner 1982). At the midblastula transition (MBT), RNA pol II– and III–dependent transcription is activated, whereas RNA pol I transcription is initiated later (Shiokawa et al. 1981a,Shiokawa et al. 1981b; Newport and Kirschner 1982). This biological situation makes it possible to study PNB assembly and delivery in the context of active or inactive RNA pol I transcription. Before MBT, scattered PNBs containing fibrillarin exhibit similar ultrastructural features to postmitotic PNBs and at MBT fibrillarin regroups around the rDNA with maternal pre-rRNAs (Verheggen et al. 1998). At MBT, the association of rDNAs with UBF was demonstrated (Bell et al. 1997; Verheggen et al. 1998), but the presence of other partners of the transcription machinery and, in particular, the RNA pol I complex is not yet established. Indeed, at MBT it was reported that RNA pol I accumulated in nucleoplasmic structures different from PNBs (Bell and Scheer 1999), without information on its association with rDNA.
Nuclei assembled in X. laevis egg extracts contain PNB-like structures with fibrillarin, nucleolin, Nopp180, protein B23 (NO38 in X. laevis), U3, and U8 (Bell et al. 1992; Bauer et al. 1994; Bell and Scheer 1997). Since these PNBs assembled in vitro were not observed to fuse into a nucleolus, a reasonable hypothesis was proposed that this lack of nucleolar assembly is due to the absence of transcription in this system (Bell et al. 1992).
In the present study, we demonstrate that during both X. laevis embryogenesis and in nuclei assembled in vitro, two types of PNBs containing components of the rRNA processing machinery exist. During X. laevis embryogenesis, the recruitment of both types of preassembled complexes to the nucleolar domain occurs at a time when the RNA pol I complex is not detected in the nucleolar domain. Furthermore, this recruitment is not dependent on RNA pol I activity, but correlates precisely with the presence of pre-rRNAs of maternal origin. Pre-rRNAs are absent from nuclei in which RNA pol II and III transcription was inactive and, in this case, recruitment of the rRNA processing machinery does not occur.
Materials and Methods
Primary Antibodies and Probes
Antibodies with the following specificities were used: a human autoimmune serum directed against fibrillarin (Gautier et al. 1994); a rabbit polyclonal serum directed against human nucleolin (a kind gift of C. Faucher, LBME, CNRS, Toulouse, France); a monoclonal culture supernatant recognizing the X. laevis nucleolar protein NO38, a homologue of the mammalian nucleolar protein B23 (No-63; Schmidt-Zachmann et al. 1987); and a mouse monoclonal ascites fluid recognizing the X. laevis RNA pol I complex (a kind gift of M. Schmidt-Zachmann, DKFZ, Heidelberg, Germany).
The rDNA probe for rRNA detection corresponds to the entire X. laevis ribosomal transcription unit inserted into pBR322 (clone pXcr7, kindly provided by F. Amaldi, Universito Tor Vergata, Roma, Italy). Double-stranded DNA probes were labeled with the nick-translation kit (GIBCO BRL) according to the manufacturer's instructions using biotin-14-dCTP. Probes for the detection of U3 and U8 are oligonucleotides complementary to three regions (positions 9-31; 63-85; and 101-122 for U3; and 33-60; 72-93; and 107-136 for U8). They were labeled with biotin-14-dCTP using 3′ terminal transferase (Boehringer Mannheim).
Assembly of Nuclei in X. laevis Egg Extract
Eggs were obtained from female X. laevis and interphasic low-speed egg extracts were prepared as previously described (Almouzni 1998). This crude extract can be maintained on ice for 4 h without appreciable decay of nuclear assembly activity. Demembraned X. laevis sperm nuclei were prepared and permeabilized with lysolecithin (Almouzni 1998). For nuclear assembly, 105 sperm heads were added to 100 μl of egg extract and maintained at 23°C. After 45 min, when maximal decondensation of the nuclei occurred, nuclei were processed for photonic and electron microscopies.
Preparation of Embryonic Nuclei from X. laevis
Embryos were produced by in vitro fertilization (Almouzni and Wolffe 1995) and allowed to develop at 23°C in 0.1× modified Barth solution (Gurdon and Wickens 1983) for different times after fertilization. At this temperature, embryos were collected at the following stages: early blastula, 6 h after fertilization (stage 8); midblastula, 7 h after fertilization (stage 8.5); late blastula, 9 h after fertilization (stage 9); early gastrula, 10 h after fertilization (stage 10); gastrula, 11 h after fertilization (stage 10.5); and late gastrula, 13 h after fertilization (stage 12); as specified by Nieuwkoop and Faber 1994. In some cases, actinomycin D was added to inhibit the onset of transcription during development. As dissociation of the blastomeres was required for drug accessibility, embryos 3 h 30 min after fertilization were transferred to Ca2+- and Mg2+-free medium containing 88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, and 7.5 mM Tris HCl, pH 7.6, as previously described (Duval et al. 1990). Actinomycin D (Sigma Chemical Co.) was added to the incubation medium at the desired concentration from a stock solution at 5 mg/ml. As a control, embryos were incubated in Ca2+- and Mg2+-free medium without actinomycin D. Other drugs, such as α-amanitin and 5,6-dichloro-β-d-ribofuranosylbenzimidazole (DRB; Sigma Chemical Co.) could not be used as inhibitors of transcription in X. laevis embryos. When α-amanitin was added to the incubation medium, it did not penetrate into the dissociated blastomeres and DRB had a deleterious effect on the division of the embryos. Suspensions of embryonic nuclei were prepared from embryos taken at precise times after fertilization as described (Verheggen et al. 1998).
Immunofluorescence and In Situ Hybridization
In vitro reconstituted nuclei and suspensions of embryonic nuclei were fixed with 1 vol 4% paraformaldehyde in PBS. The suspensions of fixed nuclei could be stored several weeks at 4°C. For immunofluorescence and in situ hybridization studies, nuclei were centrifuged onto a coverslip. After washing, the different coverslips were postfixed in methanol, permeabilized in 0.1% Triton X-100 (IBI) in PBS, and rinsed.
For immunofluorescence labeling, the coverslips were incubated with primary antibodies, followed by FITC- or TRITC-conjugated secondary antibodies (anti-human, anti-mouse, or anti-rabbit IgG; Jackson ImmunoResearch Laboratories) or cy3-conjugated anti-mouse antibodies (Sigma Chemical Co.), rinsed, counterstained with DAPI (4′-6-diamidino-2-phenylindole dihydrochloride; Polysciences, Inc.) and mounted with an antifading solution (Citifluor).
In situ hybridization of rRNAs was performed after immunolabeling as previously described (Verheggen et al. 1998). The rRNA hybridization mixture contained 40% formamide (GIBCO BRL), 10% (wt/vol) dextran sulfate (Sigma Chemical Co.), 50 ng/μl sonicated salmon testes DNA (Sigma Chemical Co.), and the biotinylated rDNA probe diluted to a final concentration of 1 ng/μl in 2× SSC. For the detection of U3 and U8, the hybridization mixture contained 30% formamide, 10% (wt/vol) dextran sulfate, 50 ng/μl sonicated salmon testes DNA, and the 3′ end biotinylated oligonucleotides complementary to U3 and U8 diluted to a final concentration of 2 ng/μl in 2× SSC. As a control for the detection of RNA, hybridization was preceded by RNase digestion as described (Highett et al. 1993).
Electron Microscopy
In vitro reconstituted nuclei were fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, at 4°C. They were washed in cacodylate buffer, postfixed in 1% OsO4 for 1 h at 4°C, dehydrated in alcohol, and embedded in Epon 812. Ultrathin sections were contrasted with uranyl acetate for 1 h and lead citrate for 2 min, and examined in a Philips EM412 electron microscope.
In Situ Transcription Assay
To localize transcription sites, 5-bromouridine-5′-triphosphate (Br-UTP) incorporation into embryonic nuclei was performed as previously described (Verheggen et al. 1998). In brief, nonfixed nuclei were permeabilized and incubated in transcription buffer in the presence of Br-UTP for 20 min at room temperature (Masson et al. 1996). Before immunolabeling, the nuclei were fixed with 2% paraformaldehyde in PBS and permeabilized with 0.1% Triton X-100. A monoclonal anti–Br-deoxyuridine antibody which also recognizes Br-UTP (Boehringer Mannheim) was used. Human antifibrillarin antibodies were used as a nucleolar marker. Transcription and fibrillarin signals were obtained using FITC-conjugated goat anti–mouse and TRITC-conjugated goat anti–human antibodies (Jackson ImmunoResearch Laboratories), respectively.
Optical Microscopy
Images were taken with a Leica epifluorescence microscope equipped with a thermoelectronically cooled charge-coupled device (CCD) camera (Leica). Grayscale images were collected separately with filter sets for FITC and rhodamine/TRITC using an oil immersion lens (63×, NA I.4 plan Apochromat). Grayscale images were pseudocolored and merged using the Adobe Photoshop 5.0 software.
Results
Components of the rRNA Processing Machinery Are Located in Two Types of PNBs in Early Embryonic and Reconstituted Nuclei
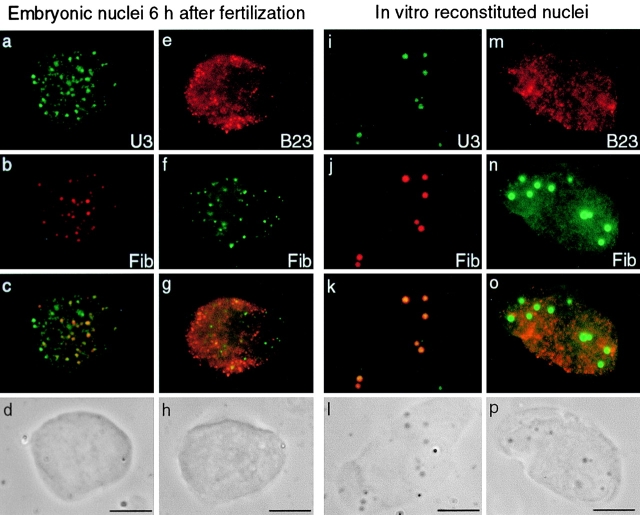
During the building process of the nucleolus in X. laevis embryonic nuclei, components of rRNA processing machinery are associated in bodies called PNBs dispersed in the nucleoplasm. A first class of PNBs (PNBs I) containing fibrillarin and nucleolin, is observed 6 h after fertilization. A maternal pool of U3 is maintained during early development of X. laevis (Xia et al. 1995) and we compared its distribution to fibrillarin in embryonic nuclei 6 h after fertilization. Fluorescent in situ hybridization (FISH) of U3 snoRNAs was performed after immunolabeling of fibrillarin. U3 snoRNAs were colocalized with fibrillarin in all PNBs I and were also present on some other dots in the nucleoplasm (Fig. 1, a–d). Moreover, U8 snoRNAs, implicated both in early and late steps of rRNA processing in X. laevis, were also detected in PNBs I by FISH (see Fig. 3). On the contrary, B23/NO38, a nucleolar protein involved in a late step of pre-rRNA processing, did not colocalize with fibrillarin when both proteins were revealed on embryonic nuclei isolated 6 h after fertilization (Fig. 1, e–h). Instead, B23/NO38 appeared diffuse in the nucleoplasm and some proteins were distributed in numerous small dots throughout the nucleoplasm, and were termed PNBs II. This could be an intermediate step in the recruitment of B23/NO38 to PNBs II since later, B23/NO38 becomes distributed in PNBs II of large size (see Fig. 4). Consequently, the formation of PNBs II appears to be delayed, compared with PNBs I.
Figure 1.
Components of rRNA processing machinery were regrouped in two types of PNBs both in embryonic and reconstituted nuclei. FISH of U3 (a; FITC) was performed after immunolabeling of fibrillarin (b; Texas red) on embryonic nuclei isolated 6 h after fertilization. U3 was distributed in numerous dots dispersed in the nucleoplasm. A large number of these dots colocalized with fibrillarin (c; yellow dots on the merged image). They were called PNBs I. Some other dots of U3 were also seen in the nucleoplasm (c; green dots on the merged image). In contrast, B23/NO38 (e; cy3) was not colocalized with fibrillarin (f, FITC) in a double immunolabeling experiment on embryonic nuclei isolated 6 h after fertilization. Labeling of B23/NO38 was diffuse and partially clustered in small dotted structures. These structures were called PNBs II. Distribution of the rRNA processing machineries was similar in in vitro reconstituted nuclei. Permeabilized sperm nuclei were incubated in low-speed X. laevis egg extract and further treated for in situ hybridization of U3 (i; FITC) after immunolabeling of fibrillarin (j; Texas red). U3 was completely colocalized with fibrillarin (k; yellow dots on the merged image). PNBs I formed in these nuclei were larger and easily distinguished by phase-contrast microscopy (l) in contrast to PNBs I in embryonic nuclei (d and h). Immunolabeling of B23/NO38 on reconstituted nuclei (m) showed the same distribution as in embryonic nuclei 6 h after fertilization. B23/NO38 (m; cy3) was not accumulated on the large PNBs I containing fibrillarin (n; FITC). Bars, 10 μm.
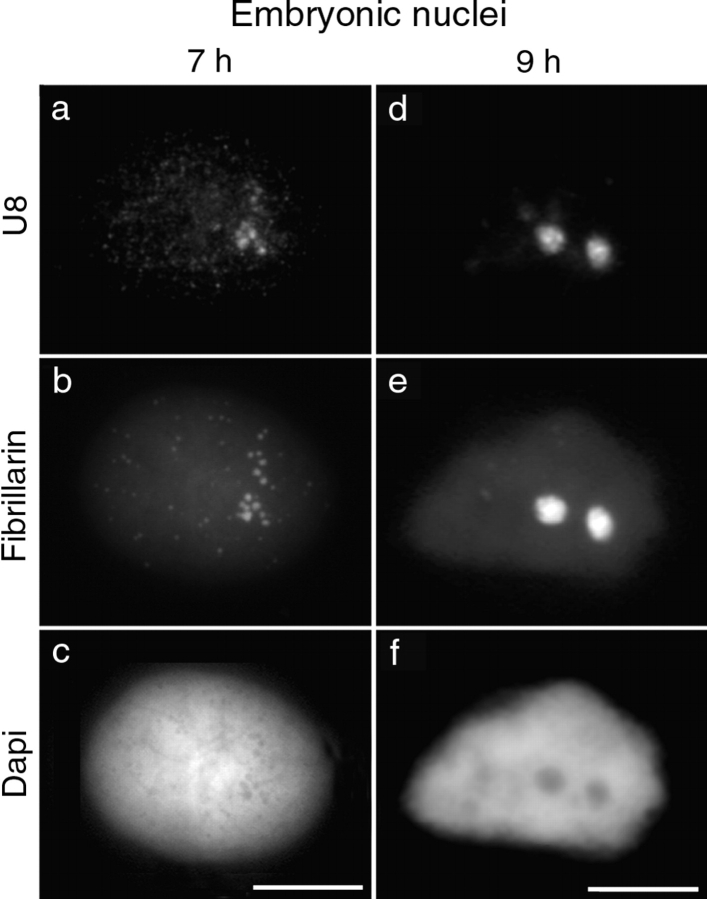
Figure 3.
Targeting of PNB I components to the nucleolar domain. Embryonic nuclei isolated 7 and 9 h after fertilization were labeled with antibodies to fibrillarin (b and e) before performing in situ hybridization of U8 (a and d). In nuclei 7 h after fertilization, U8 (a) and fibrillarin (b) were colocalized on PNBs I and gathered in two areas of the nucleoplasm as a first step through the recruitment process. In most nuclei isolated 9 h after fertilization, fibrillarin (e) and U8 (d) were colocalized in two nucleolar domains. The corresponding DNA staining with DAPI is shown (c and f). Bars, 10 μm.
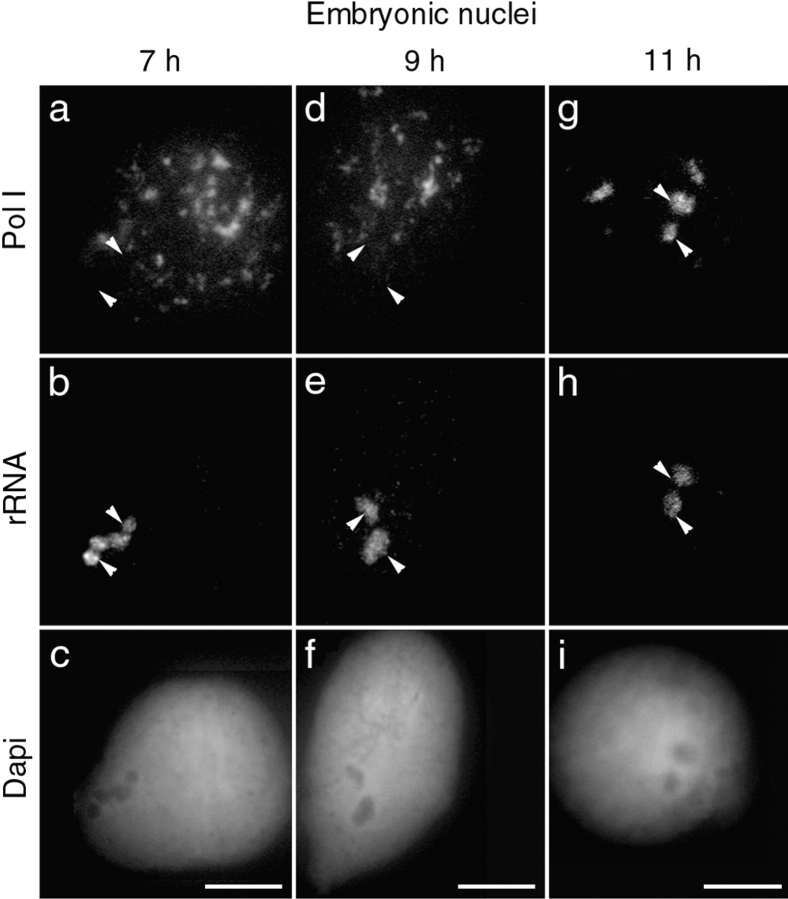
Figure 4.
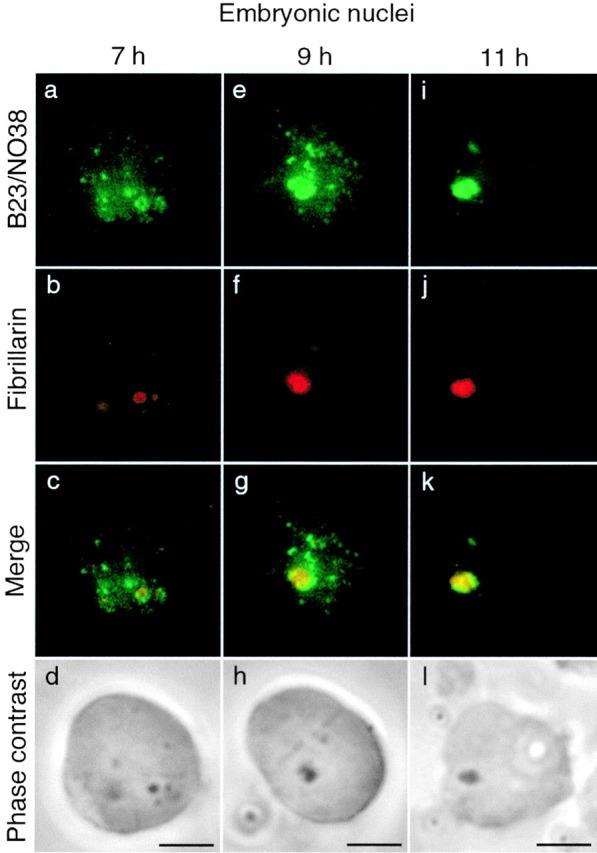
Recruitment of PNB I and PNB II components to the nucleolar domain follows different kinetics. Double immunolabeling of B23/NO38 (FITC) and fibrillarin (Texas red) was performed on embryonic nuclei isolated 7, 9, and 11 h after fertilization. In a fraction of nuclei 7 h after fertilization, B23/NO38 (a) was found colocalized with fibrillarin (b) in large clusters that began to form at the nucleolar domain (c; yellow dots on the merged image). These clusters were seen by phase-contrast microscopy (d; dense area). Nucleolar delivery of PNBs I (fibrillarin) and PNBs II (B23/NO38) thus appears to be initiated at the same time. In nuclei 9 h after fertilization, B23/NO38 (e) was not completely relocalized to the nucleolar domain in contrast to fibrillarin (f). Large dots of B23/NO38 (g; green dots on the merged image) were seen outside of the nucleolar domain (g; yellow area on the merged image). In nuclei 11 h after fertilization, B23/NO38 (i) and fibrillarin (j) were regrouped and colocalized to the nucleolar domain (k). The nucleolar domain appears dense by phase-contrast microscopy (h and l). Bars, 10 μm.
The recruitment of nucleolar proteins and snoRNAs of the rRNA processing machinery to fibrillar PNB-like structures has been described using in vitro reconstituted nuclei (Bell et al. 1992; Bauer et al. 1994). We performed double immunolabeling experiments on reconstituted nuclei to see whether the components of the processing machinery were in the same structures or in different classes of PNBs as in embryonic nuclei. Permeabilized sperm nuclei incubated in a low-speed X. laevis egg extract formed largely decondensed nuclei. After immunolabeling of fibrillarin, U3 and U8 snoRNAs were detected by FISH in these reconstituted nuclei. U3 was colocalized with fibrillarin in large round-shaped structures (Fig. 1, i–l). U8 displayed a similar distribution (not shown). B23/NO38 did not colocalize with fibrillarin (Fig. 1, m–p), but showed a distribution similar to that of the embryonic nuclei 6 h after fertilization. In particular, B23/NO38 accumulated in small and numerous dots. In EM, two types of nucleolar bodies were observed (Fig. 2). In reconstituted nuclei, the general view of sections makes it possible to discriminate few dense structures and numerous small nuclear bodies (Fig. 2 a). The dense bodies (mean diameter, 0.5 μm) exhibited a fibrillar matrix containing densely packed granules of 10–15 nm (Fig. 2 b), structures also observed at preMBT in embryonic PNBs (data not shown). Their size, distribution, and morphological features are compatible with the identification of these bodies as PNBs I. On the contrary, the small nuclear bodies (mean diameter, 0.1 μm) were gray without visible dense granules (Fig. 2 c). Based on their size, distribution (Fig. 2 a), and number, we postulated that these nuclear bodies might correspond to PNBs II. Small nuclear bodies with low contrast were also observed in embryonic nuclei (data not shown). Immunolabeling using B23/NO38 antibody was attempted, but without success, most probably because for EM, the antibody has too low a titer. Thus, morphologically different nuclear bodies were formed in reconstituted nuclei. We propose that they correspond to PNBs I and PNBs II.
Figure 2.
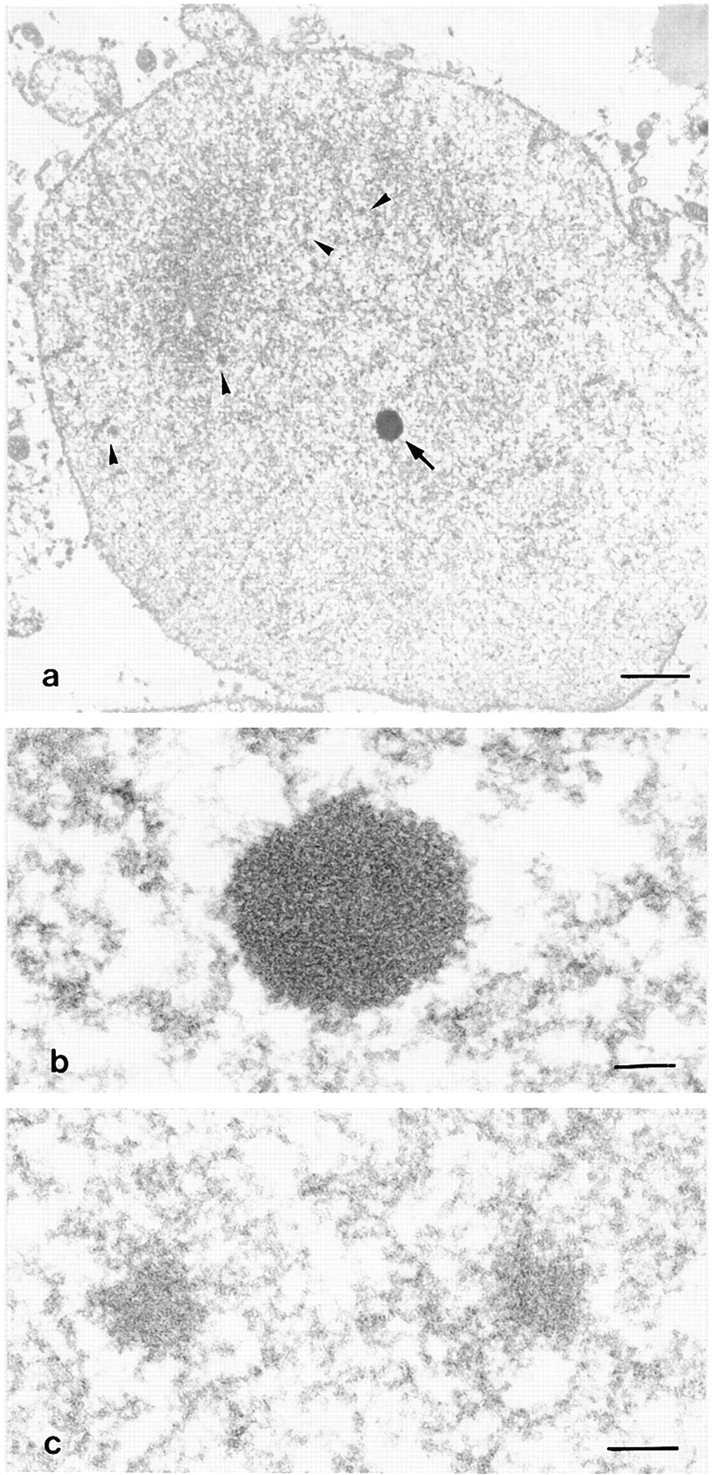
In vitro reconstituted nuclei observed by EM. a, General view of the nucleus surrounded by the nuclear envelope. The chromatin is decondensed and two types of nuclear bodies are visible: a dense body (arrow) and several gray bodies (arrow heads). Bar, 1 μm. b, High magnification of a dense nuclear body. This body corresponds to PNB I. Granules are densely packed inside. Bar, 0.1 μm. c, High magnification of two gray bodies exhibiting a fibrillar feature. These bodies correspond to PNB II. Bar, 0.1 μm.
PNBs I and PNBs II Are Targeted to the Nucleolar Domain with Different Kinetics during Development
We next defined the kinetics of recruitment of PNBs I and PNBs II to the nucleolar domain. Embryonic nuclei were isolated at various times during development, between 7 and 9 h after fertilization. Labeling of PNB I components (fibrillarin, nucleolin, U3, and U8) revealed a progression towards regrouping in nucleolar domain. PNBs I were first seen gathered in two restricted areas of the nucleoplasm in some nuclei 7 h after fertilization (Fig. 3, a–c). By 9 h after fertilization, they had fused in two large nucleolar domains in most nuclei (Fig. 3, d–f). Intermediate structures larger than PNBs I and smaller than the nucleolar domain were formed (Fig. 4 b) and were visible by phase-contrast microscopy (Fig. 4 d) in nuclei between 7 and 9 h after fertilization. B23/NO38 colocalized with fibrillarin in these structures (Fig. 4 c). Thus, fusion of the PNB I and PNB II components to the nucleolar domain was initiated nearly at the same time. In contrast, large amounts of B23/NO38 remained in PNBs II in the nucleoplasm, even when all PNBs I had completely fused (Fig. 4 g). PNBs II in nuclei 7 (Fig. 4 a) and 9 h (Fig. 4 e) after fertilization were larger than in nuclei 6 h after fertilization (Fig. 1). Complete targeting of B23/NO38 to the nucleolar domain was only observed in nuclei 11 h after fertilization (Fig. 4 i). Targeting of B23/NO38 to the nucleolar domain was thus achieved at a later time, compared with the other rRNA processing components found in PNBs I. Similar delay between B23 and fibrillarin targeting was also observed in early G1 X. laevis A6 cells (data not shown).
Fusion of PNB components to the nucleolar domain does not occur in in vitro reconstituted nuclei after several hours of incubation in the egg extract. B23/NO38 distribution remained diffuse and in small PNBs, as in embryonic nuclei 6 h after fertilization. Therefore, we attempted to identify the events that occur in embryonic nuclei and not in reconstituted nuclei that could be responsible for targeting of the rRNA processing machinery to the nucleolar domain.
RNA Polymerase I Was Excluded from the Nucleolar Domain during the Recruitment of Maternally Derived Pre-rRNAs and their Processing Machinery
It is generally accepted that the onset of rDNA transcription in cultured cells at the end of mitosis is an event required for relocalization of the rRNA processing machinery to the nucleolar domain (Scheer et al. 1993). RNA pol I could play a role in this recruitment process. This prompted us to investigate the distribution of RNA pol I at the time of nucleolar assembly in X. laevis embryos.
Immunolabeling of nuclei between 7 h and 11 h after fertilization with an mAb that immunoprecipitates the X. laevis RNA pol I complex (M. Schmidt-Zachmann, personal communication) revealed a speckled distribution (Fig. 5), as recently described (Bell and Scheer 1999). rRNAs were previously detected in the nucleolar domain at the time of PNB gathering and were identified as 40S transcripts, probably derived from a maternal pool (Verheggen et al. 1998). We performed in situ hybridization of rRNAs on nuclei previously labeled with the antibody against RNA pol I, to simultaneously visualize the RNA pol I complex and the pre-rRNAs on the nucleolar domain in nuclei isolated between 7 (Fig. 5, a–c) and 9 h after fertilization (Fig. 5, d–f). Speckle-like structures containing RNA pol I were far from nucleolar domains and consequently from rDNAs. This indicated that pre-rRNAs were maternally derived. A fraction of RNA pol I was found associated with the nucleolar domain only in nuclei 11 h after fertilization (Fig. 5, g–i). This corresponded precisely with the time when RNA pol I transcription began to be observed by the in situ transcription assay (see below).
Figure 5.
RNA polymerase I is not associated with the newly formed nucleolar domain. Embryonic nuclei isolated 7, 9, and 11 h after fertilization were labeled with an antibody to RNA polymerase I (a, d, and g) before performing in situ hybridization of the rRNAs (b, e, and h). When the nucleolar domain began to assemble in nuclei 7 h after fertilization, maternally derived rRNAs were detected (b). No RNA polymerase I was seen colocalized to the FISH signal at this time (a; arrowheads). Instead, RNA polymerase I appeared as speckles dispersed in the nucleoplasm. This distribution was maintained in nuclei 9 h after fertilization (d). RNA polymerase I was not yet colocalized with rRNA (e). Only after 11 h of development, RNA polymerase I (g) was seen partially colocalized with rRNAs (h). The corresponding DNA staining with DAPI is shown (c, f, and i). Bars, 10 μm.
Zygotic Pre-rRNA Is Not Required for the Recruitment of the Maternal Pre-rRNAs and Processing Machinery to the Nucleolar Domain
To confirm that zygotic nascent pre-RNAs, even in undetectable amounts, were not present in the newly formed nucleolar domain between 7 h and 9 h after fertilization, embryos were treated with low doses of actinomycin D to inhibit RNA pol I transcription. To allow the drug to diffuse in all the embryonic cells, actinomycin D was diluted in Ca2+- and Mg2+-free medium, conditions that dissociate blastomeres while they remain within the vitellin membrane (Duval et al. 1990). It is of note that neither the dissociation of blastomeres nor the presence of actinomycin D appeared to block cell division. Nuclei, isolated from dissociated embryos that were developed in 0.2 and 10 μg/ml of actinomycin D, were comparable in size to nuclei from control embryos. Embryos were maintained at 23°C until 13 h after fertilization. At this time, divisions still occurred, but gastrulation movements were abolished.
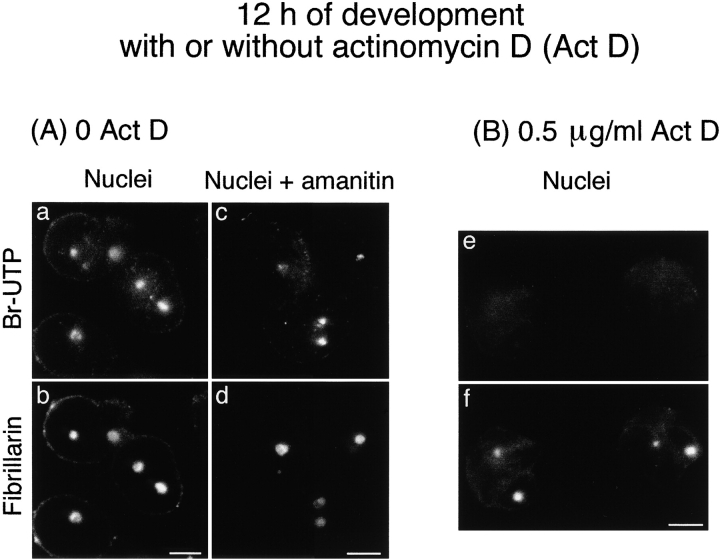
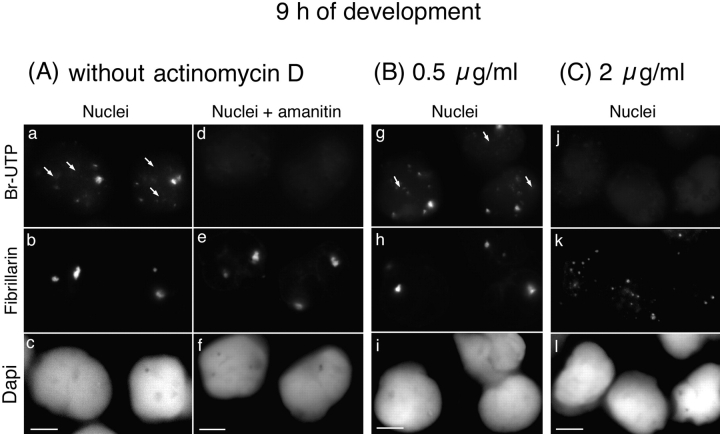
It was first necessary to check the ability of actinomycin D when applied to whole embryos to selectively block RNA pol I transcription. This prompted us to use embryos 12 h after fertilization and in situ transcription assays in conditions favoring the detection of RNA pol I transcription (see Materials and Methods). Transcription, revealed by Br-UTP incorporation in isolated nuclei from control and actinomycin D–treated embryos, was characterized by its sensitivity to α-amanitin. In control embryos, the transcription detected in nuclei isolated 12 h after fertilization (Fig. 6 a) was RNA pol I–dependent (Fig. 6 c). Indeed, transcription was not sensitive to 100 μg/ml of α-amanitin and consequently was not RNA pol II– and III–dependent. Accordingly, with RNA pol I transcription the transcripts were colocalized with fibrillarin (Fig. 6 b and d). In contrast, RNA pol I transcription was not detectable in 95% of the nuclei isolated from embryos treated with 0.5 μg/ml of actinomycin D and collected 12 h after fertilization (Fig. 6 e). This demonstrates inhibition of RNA pol I in actinomycin D–treated (0.5 μg/ml) whole embryos. Interestingly, the recruitment of fibrillarin occurred even when onset of RNA pol I transcription was abolished (Fig. 6 f).
Figure 6.
Fibrillarin is targeted to the nucleolar domain even in the absence of zygotic RNA pol I transcription. The fluorescent in situ transcription assay by immunolabeling of incorporated Br-UTP coupled to immunolabeling of fibrillarin was performed on nuclei isolated 12 h after fertilization from control embryos (A) and embryos treated with 0.5 μg/ml of actinomycin D (B). A, Sites of transcription were labeled in nuclei isolated from control embryos (a) and colocalized with fibrillarin in the nucleolar domain (b). This transcription was RNA pol I transcription because it was not inhibited by incubation of nuclei in 100 μg/ml of α-amanitin before and during incorporation of Br-UTP (c). B, When whole embryos were treated with 0.5 μg/ml of actinomycin D during development, no RNA pol I transcription was detected (e). Although RNA pol I activity was inhibited, fibrillarin was clustered in the nucleolar domain (f) as in control nuclei. Bars, 10 μm.
Thus, the treatment of whole embryos by low doses of actinomycin D provides a means to follow the changes in the distribution of the rRNA processing components in embryos without activation of RNA pol I transcription. From the same group of actinomycin D–treated (0.5 μg/ml) embryos, the nuclei were isolated 9 h after fertilization. Fibrillarin was already recruited to the nucleolar domain at this time (Fig. 7 e). Maternally derived pre-rRNAs were also detected by FISH in the nucleolar domain (Fig. 7 d). Indeed, formation of the nucleolar domain was initiated normally at the MBT, even if the onset of RNA pol I transcription was blocked. Based on these results, it appears that neither zygotic RNA pol I transcription nor the catalytic enzyme RNA polymerase I itself, are responsible for the recruitment of processing machinery.
Figure 7.
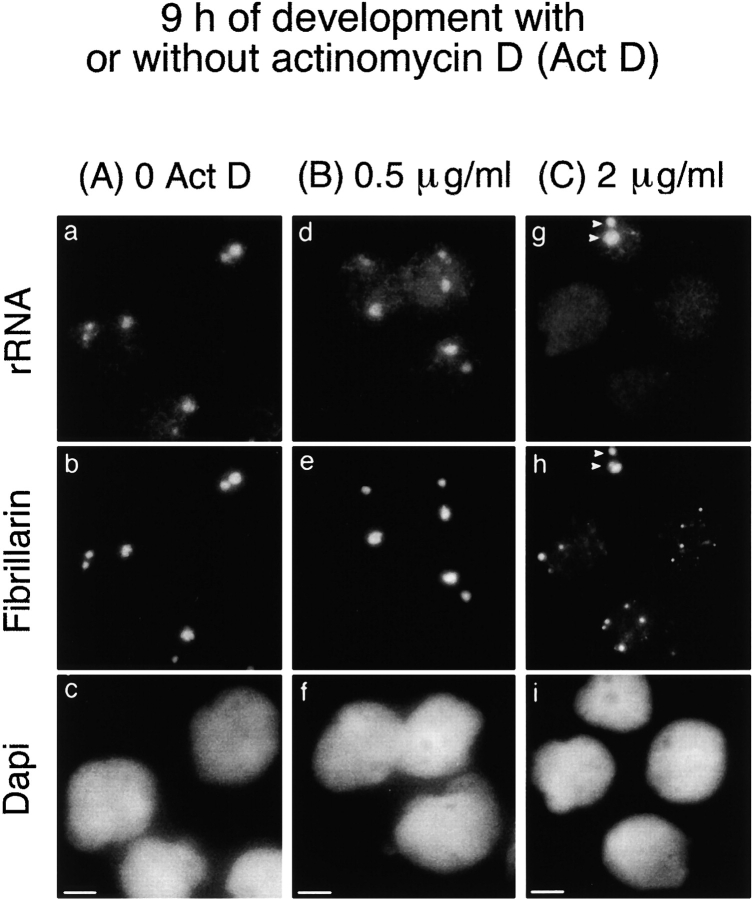
Presence of maternally derived pre-rRNAs for the recruitment of fibrillarin to the nucleolar domain. In situ hybridization of rRNA was performed after immunolabeling of fibrillarin on nuclei isolated 9 h after fertilization from control embryos (a–c) and embryos treated with 0.5 μg/ml (inhibition of RNA pol I; d–f) or 2 μg/ml of actinomycin D (inhibition of RNA pol I, II, and III; g–i). The nucleolar domain was formed in control and 0.5 μg/ml actinomycin D–treated nuclei. rRNAs (a and d) were colocalized with fibrillarin (b and e) in this domain. rRNA was not detected in most nuclei from embryos treated with 2 μg/ml of actinomycin D (g). In this case, fibrillarin was not recruited to the nucleolar domain, but was maintained in large PNBs in the nucleoplasm (h). Only a few nuclei formed a nucleolar domain containing rRNA and fibrillarin (arrowheads in g and h, respectively). The corresponding DNA staining with DAPI is shown (c, f, and i). Bars, 10 μm.
Recruitment of the rRNA Processing Machinery Does Not Occur in the Absence of Maternal Pre-rRNAs in the Nucleolar Domain
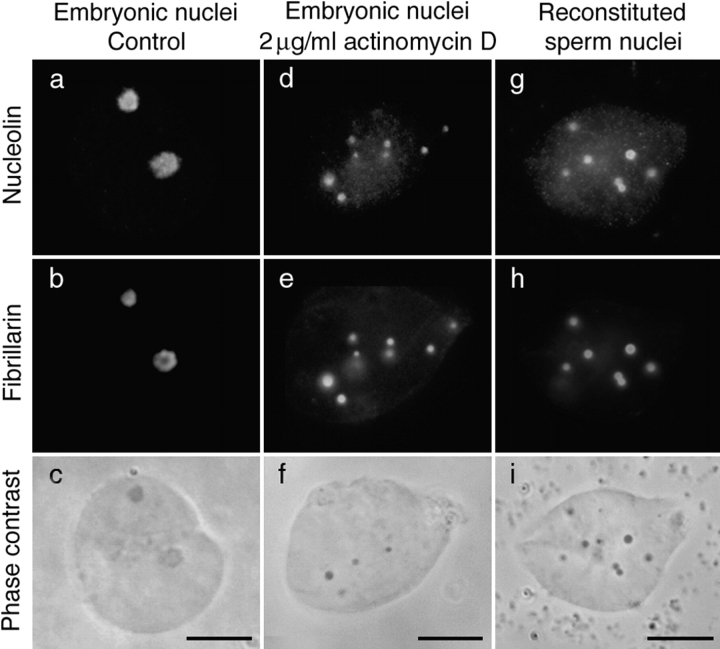
As maternally derived pre-rRNAs were the only pre-rRNAs in embryonic nuclei at MBT, we wondered if they could play a role in recruiting the rRNA processing machinery to the nucleolar domain. A correlation could be established between the presence of pre-rRNAs in the nucleolar domain and the ability of the rRNA processing machinery to be recruited to this domain. Two types of nuclei were experimentally induced in which pre-rRNAs were not detected by FISH, and in both cases the rRNA processing machinery was maintained in large PNBs. The first type of nuclei was isolated from embryos treated with 2 μg/ml of actinomycin D and collected 9 h after fertilization (Fig. 7, g–i). At this time, Br-UTP was incorporated into large foci (Fig. 8 a) not colocalized with fibrillarin (Fig. 8 a, b arrows). These foci were not further detected, probably due to changes in the transcription pattern during development. Transcription observed 9 h after fertilization was inhibited by α-amanitin, indicating that it corresponds to RNA pol II and/or III activities (Fig. 8 d). RNA pol II and/or III activities were also detected in most nuclei isolated from embryos treated with 0.5 μg/ml of actinomycin D (Fig. 8 g). In contrast, the onset of RNA pol II and III activities was inhibited in 86% of nuclei isolated from embryos treated with 2 μg/ml of actinomycin D (Fig. 8 j). In the latter nuclei, fibrillarin was maintained in large PNBs (Fig. 7 h and 8 k) and no pre-rRNA was detected by in situ hybridization (Fig. 7 g). In a few nuclei only, fibrillarin and pre-rRNAs were regrouped in the nucleolar domain (Fig. 7g and Fig. h, arrowheads). These nuclei most likely escaped inhibition of RNA pol II and III activities by actinomycin D, as indicated by the detection of fibrillarin clustered to nucleolar domain in the few nuclei in which RNA pol II and III activities were detected (data not shown). The PNBs that remained in the transcriptionally quiescent nuclei (Fig. 7 h and 8 k) were larger than the PNBs I observed in nuclei 6 h after fertilization (Fig. 1 b). Nucleolin, U3, and U8 were also found in these large structures (Fig. 9d and Fig. e). B23/NO38 was maintained in small diffuse PNBs similar to those observed in nuclei isolated 6 h after fertilization (data not shown). A second type of nuclei were in in vitro reconstituted sperm nuclei. Again, the absence of nucleolar domain formation was observed (Fig. 9, g–i). This is consistent with the transcriptional inactive state of these nuclei, which was previously reported (Bell et al. 1992).
Figure 8.
Fibrillarin was not recruited to the nucleolar domain when the onset of nonribosomal zygotic transcription was inhibited. The fluorescent in situ transcription assay by immunolabeling of incorporated Br-UTP coupled to immunolabeling of fibrillarin was performed on nuclei isolated 9 h after fertilization from control embryos (A) and embryos treated with actinomycin D (B and C). A, At this time, incorporation of Br-UTP was detected in the nucleoplasm as punctuated signals (a) not colocalized with fibrillarin to the nucleolar domain in control nuclei (b). This transcription signal was inhibited by incubation of nuclei in 100 μg/ml of α-amanitin before and during incorporation of Br-UTP (d). B and C, A non-RNA pol I transcription signal was also detected in nuclei isolated 9 h after fertilization when embryos were treated with 0.5 μg/ml of actinomycin D (g), but not when embryos were treated with 2 μg/ml of actinomycin D (j). In this latter case, fibrillarin was maintained in large PNBs dispersed in the nucleoplasm (k). Arrows indicate the position of the nucleolar domains in a and g. DNA staining with DAPI is shown in (c, f, i, and l). Bars, 10 μm.
Figure 9.
Fibrillarin and nucleolin were maintained in large PNBs throughout development in the presence of actinomycin D or in in vitro reconstituted nuclei. In a double immunolabeling experiment, nucleolin (a) and fibrillarin (b) were colocalized in the nucleolar domain in nuclei isolated 9 h after fertilization from control embryos. This domain appeared dense by phase-contrast microscopy (c). The nucleolar domain was not formed in most nuclei isolated from embryos treated with 2 μg/ml of actinomycin D (inhibition of RNA pol I, II, and III). Instead, nucleolin (d) and fibrillarin (e) were maintained in large PNBs. A similar distribution was also observed in in vitro reconstituted sperm nuclei (g and h). PNB structures, maintained at the MBT stage in treated embryos, were easily distinguished in nuclei by phase-contrast microscopy (f) and similar to structures formed in in vitro reconstituted nuclei (i). Bars, 10 μm.
Discussion
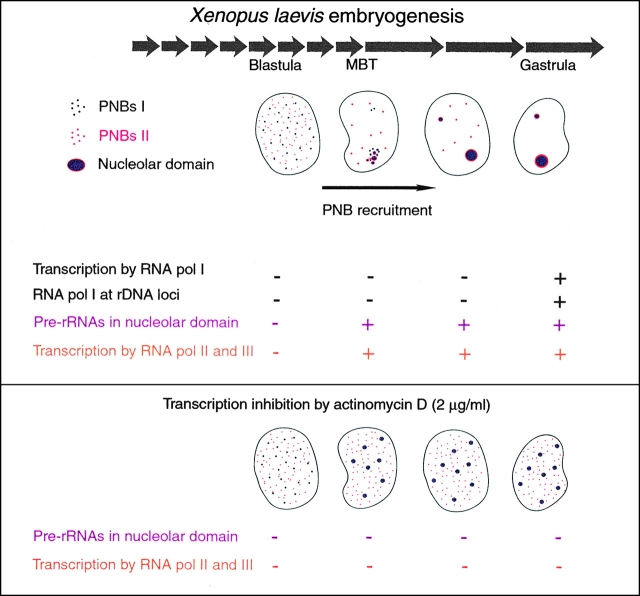
During early X. laevis embryogenesis, components of the rRNA processing machinery are first associated in bodies, known as the PNBs, which are dispersed in the nucleoplasm. These components are subsequently recruited to rDNA loci to form the nucleolar domain (Fig. 10). Both of these steps play an important part of the nucleolar assembly process. Interestingly, these phenomena also occur during nucleolar assembly at the end of mitosis (Hadjiolov 1985; Scheer and Benavente 1990; Thiry and Goessens 1996). A comparison of these biological situations reveals the general features necessary for recruitment of the rRNA processing machinery.
Figure 10.
Recruitment of rRNA processing machinery related to transcription during X. laevis development. Processing machinery components were first assembled in the nucleoplasm in two types of PNBs: PNBs I (in purple) containing fibrillarin, nucleolin, U3, and U8; and PNBs II (in red) containing B23/NO38. PNB I and PNB II components were recruited to the nucleolar domain with different kinetics. Material contained in PNBs I was regrouped in the nucleolar domain before material contained in PNBs II. At the time of the recruitment process: zygotic transcription by RNA pol II and pol III was observed (+ in orange); RNA polymerase I was found in the nucleoplasm. It was relocalized on rDNA and activated later (+ in blue); pre-rRNAs of maternal origin were detected at the time of nucleolar domain formation (+ in fuchsia). In nuclei isolated from embryos treated with 2 μg/ml of actinomycin D that inhibited RNA pol II and pol III transcriptions, pre-rRNAs were not detected by FISH and the rRNA processing machinery components were maintained in large PNBs in the nucleoplasm. A similar situation was encountered in in vitro reconstituted nuclei.
PNB Assembly as the First Step of Nucleologenesis
In cultured cells, PNBs are formed at the end of mitosis when components of the rRNA processing machinery associate (Ochs et al. 1985a; Schmidt-Zachmann et al. 1987; Jiménez-Garcia et al. 1989; Azum-Gélade et al. 1994). PNB-like structures with a similar composition also form in vitro in reconstituted interphasic nuclei (Bauer et al. 1994; Bell et al. 1992; Bell and Scheer 1997; Bauer and Gall 1997). During X. laevis embryogenesis, PNBs assemble in early blastula embryonic nuclei and remain as discrete structures throughout interphase (Verheggen et al. 1998). In this study, we demonstrate the existence of two types of PNBs in both X. laevis embryonic nuclei and permeabilized sperm nuclei reconstituted in egg extracts. The components in PNBs I are all involved in early steps of rRNA processing, whereas the components found in PNBs II are involved in a late step of processing. Distinct PNBs for proteins involved in early and late steps of rRNA processing were also described in telophase cells (Fomproix and Hernandez-Verdun 1999; Savino et al. 1999).
To date, the mechanisms that govern PNB assembly are poorly understood. By systematic depletion of the egg extract, it has been shown that fibrillarin, nucleolin, B23/NO38, and U3 are dispensable for PNB assembly in reconstituted nuclei (Bell and Scheer 1997). At the end of mitosis, PNB formation and the onset of transcription occur simultaneously, suggesting a common regulatory event (Fomproix et al. 1998). Nevertheless, PNB assembly occurs in the absence of transcription in both cultured cells (Morcillo et al. 1976; Benavente et al. 1987) and reconstituted nuclei (Bell et al. 1992). Nucleolus-like particles with features of PNBs were assembled in a soluble extract of nucleoli (Trimbur and Walsh 1993). PNBs were also induced in interphasic cells which had been released from hypotonic shock and transferred to normal isotonic medium, even in the presence of actinomycin D (Zatsepina et al. 1997). During X. laevis embryogenesis, PNBs are assembled from maternally derived components at a transcriptionally silent stage (Verheggen et al. 1998). In the present study, we observe PNBs in early and late stage nuclei from embryos treated with high concentrations of actinomycin D, confirming that PNB assembly during X. laevis embryogenesis is totally independent of any transcription.
Transcription and Processing Machineries Assemble in Bodies at Locations Distinct from RNA Synthesis and Processing Sites
Accumulation of machineries in domains distinct from their effector sites is not limited to rRNA processing machinery. We also showed that RNA pol I was not at rDNA loci at the time of nucleolar domain formation during X. laevis embryogenesis. Several extranucleolar foci containing RNA pol I distinct from PNBs were observed in both blastula nuclei (Bell and Scheer 1999) and in vitro reconstituted nuclei (Bell et al. 1997). TBP is known to accumulate in these foci at the gastrula stage (Bell and Scheer 1999). However, the significance of the presence of these complexes in nucleoplasmic foci remains to be elucidated.
Several examples of domains for accumulation of transcription and processing machineries have been reported. Stress granules, containing the transcription factor HSF1, formed in response to heat shock and independent of the sites of active transcription (Jolly et al. 1999a). Speckled distribution has been described for components of the mRNA processing machinery (for reviews see Moen et al. 1995; Huang and Spector 1996a; Lamond and Earnshaw 1998; Misteli and Spector 1998). Speckles are prominent in cells with low transcriptional activity, whereas upon activation of transcription, factors are recruited from speckles to sites of active transcription (Jiménez-Garcia and Spector 1993; Huang and Spector 1996b; Jolly et al. 1999b). In addition, coiled bodies are sites where components of the processing machinery localize, but the precise role of these bodies is not yet defined (Boudonck et al. 1999; Frey et al. 1999). Recently, a model was proposed in which the coiled body (cajal body) is the site of preassembly of RNA transcription and processing complexes before transport to the appropriate genes (Gall et al. 1999).
PNB Recruitment to the Nucleolar Domain as the Second Step of Nucleologenesis
Fusion of the PNB components to the nucleolar domain constitutes the second event of delivery of the processing machinery to the nucleolus. This stage was shown to be dependent on RNA pol I activity in cultured cells at the end of mitosis (Giménez-Martin et al. 1974; Ochs et al. 1985a; Benavente et al. 1987; Scheer and Benavente 1990). In contrast, we showed that during the de novo assembly of the nucleolus in X. laevis embryos, neither RNA pol I activity, nor the catalytic enzyme RNA pol I itself were required for this recruitment process. Formation of the nucleolar domain in the absence of RNA pol I has been observed in yeast mutant strains in which rRNA was transcribed from plasmids containing rDNA flanked by an RNA pol II–dependent promoter (Oakes et al. 1998). However, the structure and position of the nucleolar domain were abnormal, even when the plasmids were integrated in chromosomal loci. RNA pol I was therefore suggested to play a role in the organization of the nucleolar domain. However, the authors did not exclude the possibility that a transcription factor, rather than the RNA pol I itself, was involved (Oakes et al. 1998).
Recruitment of the mRNA processing machinery has been shown to be mediated by the COOH-terminal domain (CTD) of the RNA pol II (Corden and Patturajan 1997; McCracken et al. 1997; Steinmetz 1997). Even in the presence of pre-mRNA in the transcription sites, the processing machinery is not recruited when the RNA pol II CTD is deleted (Misteli and Spector 1999). A different recruiting mechanism is used for rRNA processing machinery as RNA pol I is dispensable for nucleolar domain assembly during X. laevis embryogenesis.
Protein recruitment to a specific location can be controlled by reversible phosphorylation, which could provide an attractive mechanism for recruitment of PNB to the nucleolar domain. For example, the nuclear import of nucleolin in early blastula of X. laevis has been shown to depend on its phosphorylation (Mebmer and Dreyer 1993). The phosphorylation state also influences the subnuclear distribution of SR (Ser/Arg-rich) proteins (Misteli and Spector 1998) and their activity in splicing of mRNA during development of Ascaris lumbricoides (Sanford and Bruzik 1999). A hypothesis could be that recruitment of the rRNA processing machinery to the nucleolar domain involves changes in phosphorylation of proteins during X. laevis embryogenesis.
The major event for recruitment of the rRNA processing machinery is likely to be the presence of pre-rRNAs in the nucleolar domain. In cultured cells, the onset of rDNA transcription at the end of mitosis could provide pre-rRNAs required for recruitment of the rRNA processing machinery to the nucleolar domain (Scheer et al. 1993). This reassembly of PNBs is prevented if activation of rDNA transcription is inhibited (Benavente et al. 1987; Scheer and Benavente 1990). Remarkably, during X. laevis embryogenesis, inhibition of rDNA transcription does not prevent nucleolar assembly. Maternally derived pre-rRNAs are maintained during X. laevis embryogenesis and targeted to the nucleolar domain at MBT (Verheggen et al. 1998). Accumulation of these pre-rRNAs in the nucleolar domain could be a prerequisite for the recruitment of the processing machinery. The absence of nucleolar accumulation of maternally derived pre-rRNAs in actinomycin D–treated embryos appears to be a consequence of RNA pol II and III transcription inhibition. An event required for pre-rRNA accumulation could be promoted by zygotic transcription.
Besides the predominant role of pre-rRNAs, additional regulatory mechanisms must exist to allow the delayed translocation of PNB II components relative to PNB I components. During X. laevis embryogenesis, we observed that most of B23/NO38 was recruited later than fibrillarin and nucleolin. A comparable situation was observed for nucleolus formation in telophase cell (Fomproix and Hernandez-Verdun 1999; Savino et al. 1999). Considering that steps for nucleolar assembly are common to both embryonic and cultured cells at the end of mitosis, it is tempting to speculate that nucleolar assembly involves common regulatory pathways in both cases. The fact that components of rRNA processing machinery involved in distinct steps of processing are delayed in their recruitment could be in itself a regulatory mechanism for their stepwise involvement in rRNA processing. This is raising the question whether alteration of the timing of the recruitment process might impair processing of rRNA.
Acknowledgments
The authors are grateful to M. Schmidt-Zachmann for providing anti-pol I and anti-B23 antibodies and to D. Roche for preparing the X laevis eggs and embryos. We thank P. Ridgway, R. Bastos, and A.-L. Haenni for critical reading of the manuscript.
This work was supported in part by grants from the Centre National de la Recherche Scientifique (programme Biologie Cellulaire no. 96098) and l'Association pour la Recherche sur le Cancer (contracts nos. 9143 and 5304 to D. Hernandez-Verdun, and no. 1030 to G. Almouzni). C. Verheggen was a recipient of a fellowship from l'Association pour la Recherche sur le Cancer.
Footnotes
Abbreviations used in this paper: Br-UTP, 5-bromouridine-5′-triphosphate; DAPI, 4′-6-diamidino-2-phenylindole dihydrochloride; ETS, external transcribed spacers; FISH, fluorescent in situ hybridization; ITS, internal transcribed spacers; MBT, midblastula transition; mRNA, messenger RNA; PNB, prenucleolar bodies; rDNA, ribosomal gene; rRNA, ribosomal RNA; RNA pol I, RNA polymerase I; snoRNA, small nucleolar RNA.
References
- Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′-5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni, G. 1998. Assembly of chromatin and nuclear structures in Xenopus egg extract. In Chromatin, A Practical Approach. 195–218.
- Almouzni G., Wolffe A.P. Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:1752–1765. doi: 10.1002/j.1460-2075.1995.tb07164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azum-Gélade M.-C., Noaillac-Depeyre J., Caizergues-Ferrer M., Gas N. Cell cycle redistribution of U3 snRNA and fibrillarin. Presence in the cytoplasmic nucleolus remnant and in the prenucleolar bodies at telophase. J. Cell Sci. 1994;107:463–475. doi: 10.1242/jcs.107.2.463. [DOI] [PubMed] [Google Scholar]
- Bauer D.W., Gall J.G. Coiled bodies without coilin. Mol. Biol. Cell. 1997;8:73–82. doi: 10.1091/mbc.8.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D.W., Murphy C., Zheng'an W., Wu C.H., Gall J.G. In vitro assembly of coiled bodies in Xenopus egg extract. Mol. Biol. Cell. 1994;5:633–644. doi: 10.1091/mbc.5.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell P., Scheer U. Prenucleolar bodies contain coilin and are assembled in Xenopus egg extract depleted of specific nucleolar proteins and U3 RNA. J. Cell Sci. 1997;110:43–54. doi: 10.1242/jcs.110.1.43. [DOI] [PubMed] [Google Scholar]
- Bell P., Scheer U. Developmental changes in RNA polymerase I and TATA box-binding protein during early Xenopus embryogenesis. Exp. Cell Res. 1999;248:122–135. doi: 10.1006/excr.1999.4411. [DOI] [PubMed] [Google Scholar]
- Bell P., Dabauvalle M.C., Scheer U. In vitro assembly of prenucleolar bodies in Xenopus egg extract. J. Cell Biol. 1992;118:1297–1304. doi: 10.1083/jcb.118.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell P., Mais C., McStay B., Scheer U. Association of the nucleolar transcription factor UBF with the transcriptionally inactive rRNA genes of pronuclei and early Xenopus embryos. J. Cell Sci. 1997;110:2053–2063. doi: 10.1242/jcs.110.17.2053. [DOI] [PubMed] [Google Scholar]
- Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- Beven A.F., Lee R., Razaz M., Leader D.J., Brown J.W.S., Shaw P.J. The organization of ribosomal RNA processing correlates with the distribution of nucleolar snRNAs. J. Cell Sci. 1996;109:1241–1251. doi: 10.1242/jcs.109.6.1241. [DOI] [PubMed] [Google Scholar]
- Boudonck K., Dolan L., Shaw P.J. The movement of coiled bodies visualized in living plant cells by the green fluorescent protein. Mol. Biol. Cell. 1999;10:2297–2307. doi: 10.1091/mbc.10.7.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs M.W., Burkard K.T.D., Butler J.S. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- Brown D.D., Littna E. RNA synthesis during development of Xenopus laevis, the South African clawed toad. J. Mol. Biol. 1964;8:669–687. doi: 10.1016/s0022-2836(64)80116-9. [DOI] [PubMed] [Google Scholar]
- Corden J.L., Patturajan M. A CTD function linking transcription to splicing. Trends Biochem. Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- Dichtl B., Tollervey D. Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:417–429. doi: 10.1093/emboj/16.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Meier U.T., Lewis N., Rekosh D., Hammarskjöld M.-L., Olson M.O.J. A class of nonribosomal nucleolar components is located in chromosome periphery and in nucleolus-derived foci during anaphase and telophase. Chromosoma. 1997;105:407–417. doi: 10.1007/BF02510477. [DOI] [PubMed] [Google Scholar]
- Duval C., Bouvet P., Omilli F., Roghi C., Dorel C., LeGuellec R., Paris J., Osborne H.B. Stability of maternal mRNA in Xenopus embryosrole of transcription and translation. Mol. Cell. Biol. 1990;10:4123–4129. doi: 10.1128/mcb.10.8.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomproix N., Hernandez-Verdun Effects of anti-PM-Scl 100 (Rr6p exonuclease) antibodies on prenucleolar body dynamics at the end of mitosis. Exp. Cell Res. 1999;251:452–464. doi: 10.1006/excr.1999.4578. [DOI] [PubMed] [Google Scholar]
- Fomproix N., Gébrane-Younes J., Hernandez-Verdun D. Effects of anti-fibrillarin antibodies on building of functional nucleoli at the end of mitosis. J. Cell Sci. 1998;111:359–372. doi: 10.1242/jcs.111.3.359. [DOI] [PubMed] [Google Scholar]
- Frey M.R., Bailey A.D., Weiner A.M., Matera A.G. Association of snRNA genes with the coiled bodies is mediated by nascent snRNA transcripts. Curr. Biol. 1999;9:126–135. doi: 10.1016/s0960-9822(99)80066-9. [DOI] [PubMed] [Google Scholar]
- Gall J.G., Bellini M., Wu Z., Murphy C. Assembly of the nuclear transcription and processing machinerycajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier T., Fomproix N., Masson C., Azum-Gélade M.C., Gas N., Hernandez-Verdun D. Fate of specific nucleolar perichromosomal proteins during mitosiscellular distribution and association with U3 snoRNA. Biol. Cell. 1994;82:81–93. doi: 10.1016/s0248-4900(94)80010-3. [DOI] [PubMed] [Google Scholar]
- Giménez-Martin G.C., De la Torre C., Fernandez-Gomez M.E., Gonzalez-Fernandez A. Experimental analysis of nucleolar reorganization. J. Cell Biol. 1974;60:502–507. doi: 10.1083/jcb.60.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H., Amalric F., Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon J.B., Wickens M.P. Use of Xenopus oocytes for the expression of cloned genes. In: Wu R., Grossman L., Moldave K., editors. Methods in Enzymology. Academic Press; New York: 1983. pp. 101:370–386. [DOI] [PubMed] [Google Scholar]
- Hadjiolov, A.A. 1985. The Nucleolus and Ribosome Biogenesis. Vol. 12. M. Alfert, W. Beermann, L. Goldstein, K.R. Porter, and P. Sitte, editors. Springer-Verlag, New York. 268 pp.
- Highett M.I., Rawlins D.J., Shaw P.J. Different patterns of rDNA distribution in Pisum sativum nucleoli correlate with different levels of nucleolar activity. J. Cell Sci. 1993;104:843–852. [Google Scholar]
- Huang S., Spector D.L. Dynamic organization of pre-mRNA splicing factors J. Cell Biochem. 62 1996. 191 197a [DOI] [PubMed] [Google Scholar]
- Huang S., Spector D.L. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription J. Cell Biol. 133 1996. 719 732b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Garcia L.F., Spector D.L. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell. 1993;73:47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- Jiménez-Garcia L.F., Rothblum L.I., Busch H., Ochs R.L. Nucleologenesisuse of non-isotopic in situ hybridization and immunocytochemistry to compare the localization of rDNA and nucleolar proteins during mitosis. Biol. Cell. 1989;65:239–246. doi: 10.1111/j.1768-322x.1989.tb00795.x. [DOI] [PubMed] [Google Scholar]
- Jiménez-Garcia L.F., Segura-Valdez M.L., Ochs R.L., Rothblum L.I., Hannan R., Spector D.L. NucleologenesisU3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol. Biol. Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C., Usson Y., Morimoto R. Rapid and reversible relocalization of heat shock factor 1 within seconds to nuclear stress granules Proc. Natl. Acad. Sci. USA. 96 1999. 6769 6774a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C., Vourc'h C., Robert-Nicoud M., Morimoto R.I. Intron-independent association of splicing factors with active genes J. Cell Biol. 145 1999. 1133 1143b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A.I., Earnshaw W.C. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lygerou Z., Allmang C., Tollervey D., Séraphin B. Accurate processing of a eukaryotic pre-rRNA by RNase MRP in vitro Science. 272 1996. 268 270a [DOI] [PubMed] [Google Scholar]
- Lygerou Z., Pluk H., van Venrooij W.J., Séraphin B. hPop1an autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins EMBO (Eur. Mol. Biol. Organ.) J. 15 1996. 5936 5948b [PMC free article] [PubMed] [Google Scholar]
- Masson C., Bouniol C., Fomproix N., Szöllösi M.S., Debey P., Hernandez-Verdun D. Conditions favoring RNA polymerase I transcription in permeabilized cells. Exp. Cell Res. 1996;226:114–125. doi: 10.1006/excr.1996.0209. [DOI] [PubMed] [Google Scholar]
- McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Mebmer B., Dreyer C. Requirement for nuclear translocation and nucleolar accumulation of nucleolin of Xenopus laevis . Eur. J. Cell. Biol. 1993;61:369–382. [PubMed] [Google Scholar]
- Michot B., Joseph N., Mazan S., Bachellerie J.P. Evolutionarily conserved structural features in the ITS2 of mammalian pre-rRNAs and potential interactions with the snoRNA U8 detected by comparative analysis of new mouse sequences. Nucleic Acids Res. 1999;27:2271–2282. doi: 10.1093/nar/27.11.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T., Spector D.L. The cellular organization of gene expression. Curr. Opin. Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- Misteli T., Spector D. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- Misteli T., Caceres J.F., Spector D.L. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. The exosomea conserved eukaryotic RNA processing complex containing multiple 3′-5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Moen P.T., Smith K.P., Lawrence J.B. Compartmentalization of specific pre-mRNA metabolisman emerging view. Hum. Mol. Genet. 1995;4:1779–1789. doi: 10.1093/hmg/4.suppl_1.1779. [DOI] [PubMed] [Google Scholar]
- Morcillo G., De la Torre C., Giménez-Martin G. Nucleolar transcription during plant mitosis. Exp. Cell Res. 1976;102:311–316. doi: 10.1016/0014-4827(76)90046-x. [DOI] [PubMed] [Google Scholar]
- Newport J., Kirschner M. A major developmental transition in early Xenopus embryos. I. Characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P.D., Faber J. Normal table of Xenopus laevis (Daudin) 1994. Garland Publishing, Inc; New York: pp. 252 [Google Scholar]
- Oakes M., Aris J.P., Brockenbrough J.S., Wai H., Vu L., Nomura M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae . J. Cell Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs R.L., Smetana K. Detection of fibrillarin in nucleolar remnants and the nucleolar matrix. Exp. Cell Res. 1991;197:183–190. doi: 10.1016/0014-4827(91)90421-p. [DOI] [PubMed] [Google Scholar]
- Ochs R.L., Lischwe M.A., Shen E., Caroll R.E., Busch H. Nucleologenesiscomposition and fate of prenucleolar bodies Chromosoma. 92 1985. 330 336a [DOI] [PubMed] [Google Scholar]
- Ochs R.L., Lischwe M.A., Spohn W.H., Busch H. Fibrillarina new protein of the nucleolus identified by autoimmune sera Biol. Cell. 54 1985. 123 134b [DOI] [PubMed] [Google Scholar]
- Peculis B.A., Steitz J.A. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- Pluk H., Van Eenennaam H., Rutjes S.A., Pruijn G.J.M., Van Venrrooij W.J. RNA–protein interactions in the human RNase MRP ribonucleoprotein complex. RNA. 1999;5:512–524. doi: 10.1017/s1355838299982079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.R., Bruzik J.P. Developmental regulation of SR protein phosphorylation and activity. Genes Dev. 1999;13:1513–1518. doi: 10.1101/gad.13.12.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino T.M., Bastos R., Jansen E., Hernandez-Verdun D. The nucleolar antigen Nop 52, the human homologue of the yeast ribosomal RNA processing RRP1, is recruited at late stages of nucleologenesis. J. Cell Sci. 1999;112:1889–1900. doi: 10.1242/jcs.112.12.1889. [DOI] [PubMed] [Google Scholar]
- Savkur R.S., Olson M.O.J. Preferential cleavage in pre-ribosomal RNA by protein B23 endoribonuclease. Nucleic Acids Res. 1998;26:4508–4515. doi: 10.1093/nar/26.19.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U., Thiry M., Goessens G. Structure, function and assembly of the nucleolus. Trends Cell Biol. 1993;3:236–241. doi: 10.1016/0962-8924(93)90123-i. [DOI] [PubMed] [Google Scholar]
- Scheer U., Weisenberger D. The nucleolus. Curr. Opin. Cell Biol. 1994;6:354–359. doi: 10.1016/0955-0674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Schmidt-Zachmann M.S., Hügle-Dörr B., Franke W.W. A constitutive nucleolar protein identified as a member of the nucleoplasmin family. EMBO (Eur. Mol. Biol. Organ.) J. 1987;6:1881–1890. doi: 10.1002/j.1460-2075.1987.tb02447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiokawa K., Misumi Y., Yamana K. Demonstration of rRNA synthesis in pre-gastrular embryos of Xenopus laevis Dev. Growth Differ. 23 1981. 579 587a [DOI] [PubMed] [Google Scholar]
- Shiokawa K., Tashiro K., Misumi Y., Yamana K. Non-coordinated synthesis of RNA's in pre-gastrular embryos of Xenopus laevis Dev. Growth Differ. 23 1981. 589 597b [DOI] [PubMed] [Google Scholar]
- Smith C.M., Steitz J.A. Sno storm in the nucleolusnew roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Steinmetz E.J. Pre-mRNA processing and the CTD of RNA polymerase IIthe tail that wags the dog? Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- Thiry M., Goessens G. The nucleolus during the cell cycle 1996. Springer-Verlag; Heidelberg: pp. 146 [Google Scholar]
- Tollervey D. Trans-acting factors in ribosome synthesis. Exp. Cell Res. 1996;229:226–232. doi: 10.1006/excr.1996.0364. [DOI] [PubMed] [Google Scholar]
- Trimbur G.M., Walsh C.J. Nucleolus-like morphology produced during the in vitro reassociation of nucleolar components. J. Cell Biol. 1993;122:753–766. doi: 10.1083/jcb.122.4.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eenennaam H., Pruijn G.J.M., Van Venrooij W.J. hPop4a new protein subunit of the human RNase MRP and RNase P ribonucleoprotein complexes. Nucleic Acids Res. 1999;27:2465–2472. doi: 10.1093/nar/27.12.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C., Le Panse S., Almouzni G., Hernandez-Verdun D. Presence of pre-rRNAs before activation of polymerase I transcription in the building process of nucleoli during early development of Xenopus laevis . J. Cell Biol. 1998;142:1167–1180. doi: 10.1083/jcb.142.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger D., Scheer U., Benavente R. The DNA topoisomerase I inhibitor camptothecin blocks postmitotic reformation of nucleoli in mammalian cells. Eur. J. Cell Biol. 1993;61:189–192. [PubMed] [Google Scholar]
- Xia L., Liu J., Sage C., Trexler E.B., Andrews M.T., Maxwell E.S. Intronic U14 snoRNAs of Xenopus laevis are located in two different parent genes and can be processed from their introns during early oogenesis. Nucleic Acids Res. 1995;23:4844–4849. doi: 10.1093/nar/23.23.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatsepina O.V., Dudnic O.A., Todorov I.T., Thiry M., Spring H., Trendelenburg M.F. Experimental induction of prenucleolar bodies (PNBs) in interphase cellsinterphase PNBs show similar characteristics as those typically observed at telophase of mitosis in untreated cells. Chromosoma. 1997;105:418–430. doi: 10.1007/BF02510478. [DOI] [PubMed] [Google Scholar]