Abstract
Gene expression is controlled by RNA-binding proteins that modulate the synthesis, processing, transport and stability of various classes of RNA. Some RNA-binding proteins shuttle between the nucleus and cytoplasm and are thought to bind to RNA transcripts in the nucleus and remain bound during translocation to the cytoplasm. One RNA-binding protein that has been hypothesized to function in this manner is the Saccharomyces cerevisiae Scp160 protein. Although the steady-state localization of Scp160 is cytoplasmic, previous studies have identified putative nuclear localization (NLS) and nuclear export (NES) signals. The goal of this study was to test the hypothesis that Scp160 is a nucleocytoplasmic shuttling protein. We exploited a variety of yeast export mutants to capture any potential nuclear accumulation of Scp160 and found no evidence that Scp160 enters the nucleus. These localization studies were complemented by a mutational analysis of the predicted NLS. Results indicate that key basic residues within the predicted NLS of Scp160 can be altered without severely affecting Scp160 function. This finding has important implications for understanding the function of Scp160, which is likely limited to the cytoplasm. Additionally, our results provide strong evidence that the presence of a predicted nuclear localization signal within the sequence of a protein should not lead to the assumption that the protein enters the nucleus in the absence of additional experimental evidence.
INTRODUCTION
Eukaryotic cells are characterized by the separation of the genome and transcriptional machinery in the nucleus from the translational machinery in the cytoplasm. This separation is mediated by the double membrane of the nuclear envelope, which necessitates a system for transport between these discrete compartments. All macromolecular transport between the nucleus and the cytoplasm occurs via nuclear pore complexes (NPCs) embedded in the nuclear envelope (1,2). NPCs are massive multi-protein structures each composed of ∼30 proteins, which are collectively referred to as nucleoporins (3). Although passive diffusion of molecules ⩽40 kDa through NPCs can occur, most macromolecules require specific transport signal sequences to enter and exit the nucleus via the NPC (4).
Nuclear targeting signals mark proteins for transport into and out of the nucleus (4). The best-characterized nuclear targeting signal is the classical nuclear localization signal (cNLS), which consists of a monopartite or bipartite signal (5,6). The monopartite NLS contains a single cluster of four to six basic amino acids, while a bipartite NLS contains two stretches of basic amino acids separated by a stretch of 8–10 spacer residues (6,7). Nuclear export signals (NES) have also been identified. These targeting signals consist of a weak consensus of three to four hydrophobic residues (8). These classical targeting sequences can be, and often are, identified by computer searches; however, in many instances the function of these sequences is not experimentally verified.
Targeting signals are recognized by soluble nuclear transport receptors that shepherd cargo proteins through the nuclear pore complex (9). These receptors are termed karyopherins or importins/exportins. Many transport pathways involve direct cargo binding to receptors of the importin/karyopherin-β family. However, import of cargoes that contain a cNLS involves an adaptor protein, importin-α, that serves as an NLS receptor to recognize these cargos in the cytoplasm (4). Importin-α and cNLS cargo interact with importin-β to form a trimeric import complex, which then translocates through the NPC (10). Once in the nucleus, a small GTPase, Ran-GTP (11,12), binds to importin-β causing a conformational change that results in dissociation of the import complex (13). The importin receptors are then recycled back to the cytoplasm to participate in subsequent rounds of import (14). In a similar manner, NES-dependent export involves the transport receptor, Crm1 (8,15). Like other export receptors of the importin-β family (16), Crm1 forms a trimeric complex with Ran-GTP and an export cargo (17). The trimeric complex is translocated through the NPC and once it reaches the cytoplasmic face of the NPC, Ran-GTP is hydrolyzed to Ran-GDP resulting in dissociation of the export complex (18).
Many proteins contain predicted NLS and NES sequences required for entering and exiting of the nucleus. Often, the function of these predicted sequences is assumed in the absence of supporting experimental data. One example of such a case is the coding sequence of the Saccharomyces cerevisiae control of ploidy protein of 160 kDa, Scp160. Although the exact function of Scp160 is unknown, it is thought to modulate gene expression through interacting with RNA substrates (19). Scp160 contains 14 predicted K-homology (KH) RNA-binding domains and also sequence motifs that have been hypothesized to function as both a cNLS and a classical NES (20,21). Since Scp160 associates with specific mRNA transcripts (19), it has been hypothesized to shuttle into the nucleus and then exit the nucleus in complexes with mRNA.
Despite the presence of a sequence noted as an NLS, the steady-state localization of Scp160 is clearly cytoplasmic (22–25). This cytoplasmic localization is consistent with at least two models: (i) Scp160 is solely restricted to the cytoplasm; or (ii) Scp160 shuttles into and out of the nucleus but the rate of export exceeds the rate of import. To distinguish between these two possibilities, we exploited yeast mutants that block different nuclear export pathways and assessed the localization of Scp160 in these mutant cells under conditions where these specific nuclear export pathways are blocked. If Scp160 does, in fact, shuttle between the nucleus and the cytoplasm, blocking its export would cause accumulation of the protein within the nucleus. However, if Scp160 does not shuttle, we would expect the localization to remain cytoplasmic despite blocked export pathways.
This general approach has been used previously to uncover the nucleocytoplasmic shuttling properties of other proteins. For example, the yeast poly(A)-binding protein 1, Pab1, has a well-characterized function in the cytoplasm where it binds to the poly(A) tail of mRNA transcripts and modulates their stability and translation (26). Consistent with these functions, Pab1 is localized to the cytoplasm at steady state (26). However, recent studies show that Pab1 accumulates within the nucleus of cells that express a conditional mutant of the NES receptor, crm1-1, revealing the surprising finding that this protein with a well-characterized cytoplasmic function can enter the nucleus (27,28). These studies validate the use of export mutants to test whether Scp160 is also a shuttling protein.
The goal of this study was to carry out a functional analysis of the nuclear targeting signals present in the Scp160 protein. To accomplish this goal, we first exploited a number of conditional yeast mutants that impact nuclear export. We found that mutations that rendered each of the major export pathways nonfunctional did not cause accumulation of Scp160 within the nucleus, suggesting that Scp160 may not enter the nucleus. Second, we generated amino acid substitutions predicted to disrupt the function of the putative NLS of Scp160 and found that these mutations did not significantly impair Scp160 function, indicating that the putative NLS is not critical for the key cellular function of Scp160. These data challenge the model that Scp160 performs critical functions in the nucleus, and suggest that Scp160 functions are carried out exclusively in the cytoplasm.
MATERIALS AND METHODS
Plasmids and yeast strains
All yeast manipulations were performed according to standard protocols (29). All yeast strains used in this study are listed and described in Table 1. The wild-type yeast strain used for microscopy, JFY4493, was derived from the haploid parent strain W303 (MAT a ura3-1 leu2-3 his3-11, 15 trp1-1 ade2-1 can1-100 RAD5+, a gift from Dr R. Rothstein, Columbia University, NY, USA) by integration of a FLAG epitope onto the N-terminus of the Scp160 open reading frame. All recombinant DNA manipulations were performed according to standard procedures (30) utilizing the XL10-gold (Stratagene) strain of Escherichia coli. All mutagenesis was done using a QuikChange Site-Directed Mutagenesis Kit (Stratagene). Experiments were carried out at 34°C unless otherwise noted.
Table 1.
Strains and plasmids used in this study
| Strains/plasmids | Description | Reference |
|---|---|---|
| ACY191 | mat α rpb1-1, ura2-52, his4-53 | (41) |
| ACY194 | mat α rat7-1, ura3-52, leu2Δ1, his3Δ200 | (46) |
| ACY372 | mat a crm1-3, ade2-1, ura3, leu2Δ1, his3, trp1 | (38) |
| ACY545 | mat α MEX67::KanMX4, ura3, leu2Δ1, his3Δ200, [+mex67-5 LEU2] | (42) |
| ACY1508 | mat α MLP1-RFP::KanMX4, ura3, leu2Δ1, his3Δ200, lys2 | (56) |
| JFY4247 | mat a SCP160::HIS3, EAP1::HIS3, ade2-1, ura3, leu2-3, his3-11, trp1-1, can1-100, [+SCP160-URA3] | (36) |
| JFY4493 | mat a FLAG-SCP160::SCP160, ade2-1, ura3-1, leu2-3, his3-11, trp1-1, can1-100 | (57) |
| pRS312 | TRP1, CEN, AMP | (58) |
| pRS315 | LEU2, CEN, AMP | (58) |
| pAC213 | SV40 NLS-PK1 NES-GFP, URA3, 2 μM, AMP | (39) |
| pAC980 | ΔRGG Nab2-GFP, URA3, CEN, AMP | (32) |
| JF4470 | SCP160-GFP, TRP1, CEN, AMP | This study |
| JF4823 | FLAG-SCP160, LEU2, CEN, AMP | This study |
| JF4843 | FLAG-SCP160-NLSmut, LEU2, CEN, AMP | This study |
Lysate preparation and immunoblot analysis
Cells were grown to mid-log phase and harvested by centrifugation (5 min, 4000g at 4°C). Following two washes with ddH2O, cells were lyzed by vortex agitation with an equal volume of glass beads in 0.5 ml lysis buffer (25 mM Tris pH 7.2, 50 mM KCl, 30 mM MgCl2, 1.5% Triton). Lysates were then transferred to a clean microfuge tube and centrifuged for 5 min at 3000 g at 4°C. The supernatant was again transferred to a clean microfuge tube and centrifuged at 12 000 g for 10 min at 4°C.
Immunoblot analysis was performed essentially as described previously (22). Briefly, samples to be analyzed were mixed with sample buffer, boiled and run through a 10% SDS–polyacrylamide gel. The gel was then electroblotted onto a nitrocellulose membrane (Bio-Rad) and GFP-fusion proteins were detected by incubation of the membrane with a 1:10 000 dilution of an α-GFP polyclonal antibody (31).
Direct fluorescence microscopy
Imaging was performed as described previously (32). In brief, yeast cells transformed with plasmids expressing GFP-fusion proteins were grown to mid-log phase in media lacking uracil and supplemented with adenine. Cultures were split and half was left at room temperature while the other half was shifted to 37°C for 2 h. Direct fluorescence and DIC images were collected using an Olympus BX epifluorescence microscope equipped with a Photometrics Quantix digital camera. IP Lab Spectrum software was used to capture images of cells. Prior to imaging, cells were incubated with 5 µM Hoechst dye to visualize the chromatin within the nucleus.
Functional analysis of Scp160 variants in vivo
To assess the function of an Scp160 NLS variant (Scp160-NLSmut) four lysine residues (321, 322, 331 and 333) within the predicted NLS were changed to alanines. We then tested whether this variant of Scp160 could rescue the synthetic lethality of an Δscp160Δeap1 mutant using a standard plasmid shuffle assay (33). Briefly, Scp160-NLSmut (LEU2) was transformed into an Δscp160Δeap1 double mutant containing a wild-type URA3 maintenance plasmid. Cells were inoculated into liquid medium and grown to saturation for 2 days. Cell numbers were equalized, serially diluted 1:10, and then spotted onto control plates of synthetic medium lacking leucine and test plates of synthetic medium lacking leucine and containing the drug 5-fluororotic acid (5-FOA). 5-FOA is toxic to cells containing the URA3 gene product and, therefore, the only cells that grow on 5-FOA plates have lost the URA3 plasmid and retain only the LEU2 test plasmid (34). Test plasmids encoding a second wild-type copy of SCP160 and empty vector (pRS315) were analyzed in parallel as positive and negative controls, respectively. Plates were incubated for two days at 34°C.
RESULTS
Scp160-GFP is localized to the cytoplasm at steady state
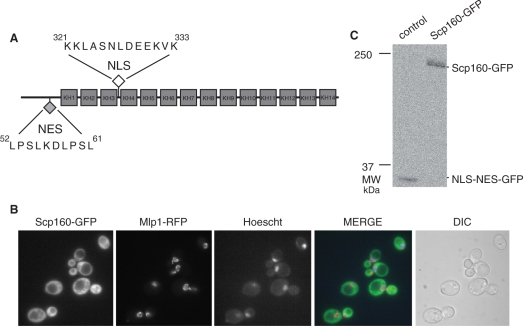
Figure 1A shows a schematic representation of the domain structure and postulated sequence motifs of Scp160. The predicted NES sequence is contained within the N-terminal portion of the protein, while the predicted NLS is located between KH domains three and four (KH3 and KH4), spanning a few residues into both KH3 and KH4. The amino acid sequence for each predicted transport signal is shown.
Figure 1.
Scp160 is localized to the cytoplasm at steady state. (A) Schematic representation of Scp160 domain structure and location of putative nuclear targeting motifs. The predicted NLS is indicated by the filled diamond and the predicted NES is indicated by the open diamond. The sequences for the predicted NLS and NES motifs as well as their positions within the open reading frame are indicated. Numbered boxes represent the 14 KH domains. (B) The Scp160-GFP protein was expressed in wild-type cells that express a red-fluorescent protein-tagged nuclear rim protein, Mlp1 (56) to mark the position of the nucleus. Fluorescent protein localization was examined by direct fluorescence microscopy. Cells were also stained with Hoechst dye to mark the position of chromatin within the nucleus. A merged fluorescence image is shown as well as the corresponding DIC image. (C) Immunoblot analysis of GFP in protein lysate from wild-type cells transformed with plasmids encoding FLAG-Scp160-GFP (right lane) or a control NLS-NES-GFP (left lane). Migration of protein standards is indicated to the left of the image. The position of the bands corresponding to FLAG-Scp160-GFP and NLS-NES-GFP is indicated.
Figure 1B shows that, as previously reported (23–25), Scp160-GFP is localized to the cytoplasm and excluded from the nucleus. To confirm that the GFP signal reflects the localization of full-length Scp160, soluble lysates of wild-type cells expressing Scp160-GFP were subjected to immunoblot analysis with an α-GFP antibody (Figure 1C). Lysate from cells expressing NLS-NES-GFP served as a control for the specificity of the antibody. FLAG-Scp160-GFP migrated as a single band at ∼200 kDa while NLS-NES-GFP migrated at 35 kDa.
As an additional control, we tested whether the GFP tag impacts the function of Scp160 by assaying the ability of Scp160-GFP to replace wild-type Scp160 in vivo using a plasmid shuffle assay. A TRP1 plasmid containing SCP160-GFP was transformed into Δscp160Δeap1 yeast containing a wild-type URA3 maintenance plasmid of SCP160. Although Scp160 is not essential for cell viability (23), Scp160 becomes essential when the spindle pole body protein, Eap1 (35), is also deleted (36). Thus, we can employ this double mutant background where Scp160 is essential to rapidly assess Scp160 function. Transformants containing both the wild-type URA3 plasmid and the TRP1 test plasmid were streaked onto synthetic complete plates lacking uracil and tryptophan (control) and 5-FOA plates lacking tryptophan. No significant growth defect was observed in the Scp160-GFP versus the untagged Scp160 (data not shown). As expected, cells containing vector alone failed to grow, confirming the requirement for Scp160 in this genetic background.
Scp160 does not accumulate in the nucleus in a NES export mutant
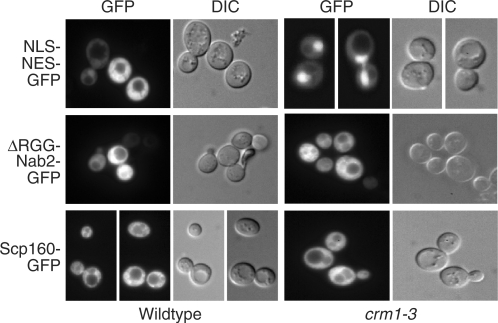
To begin to assess whether Scp160 can enter the nucleus, we took advantage of the observation that Scp160 contains a predicted classical NES (20). If this NES mediates nuclear export of Scp160, then blocking the NES-dependent export pathway should cause nuclear accumulation of Scp160. To block this pathway, we utilized a conditional mutant of the NES receptor, Crm1 (37). crm1-3 cells show defects in export of NES-cargo (38). Plasmids encoding Scp160-GFP or control cargoes either containing a classical NES, NLS-NES-GFP (39), or lacking an NES, ΔRGG-Nab2-GFP (32), were transformed into wild type and crm1-3 cells. Protein localization was assessed by direct fluorescence microscopy. Scp160 remained strictly localized to the cytoplasm in crm1-3 cells at the non-permissive temperature with no apparent nuclear concentration (Figure 2). In contrast, the control NLS-NES-GFP accumulated in the nucleus of crm1-3 cells. The localization of a control non-NES cargo protein, ΔRGG-Nab2-GFP, was not altered in the crm1-3 cells compared to wild-type cells. These results indicate that either Scp160 does not enter the nucleus or its export is not dependent on the Crm1 export pathway.
Figure 2.
Localization of Scp160-GFP in crm1-3 and wild-type cells. The localization of Scp160-GFP was examined in crm1-3 cells following a 2 h shift to 37°C where NES-dependent nuclear export is blocked. Scp160 is localized to the cytoplasm in both wild-type and crm1-1 cells. As controls, we also localized NLS-NES-GFP and ΔRGG-Nab2-GFP. As expected, NLS-NES-GFP accumulates in the nucleus of crm1-1 cells, but ΔRGG-Nab2-GFP, which is exported via an mRNA export-dependent pathway, does not. Corresponding DIC images are shown.
Scp160 does not accumulate in the nucleus when poly(A) RNA export is blocked
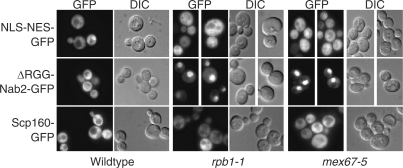
Since Scp160 associates with mRNA (19), we hypothesized that Scp160 localization could depend on ongoing mRNA synthesis and mRNA export. A number of mRNA-binding proteins that undergo nucleocytoplasmic shuttling depend both on ongoing transcription and mRNA export (32,40). Therefore, we examined the steady-state localization of Scp160 in both rpb1-1 cells where RNA polymerase II transcription is decreased (41) and mex67-5 mutant cells where poly(A) RNA export from the nucleus is blocked (42,43). As controls we also examined the localization of NLS-NES-GFP, which is not affected by mRNA export (44), and ΔRGG-Nab2-GFP, which depends on ongoing mRNA export and synthesis for export from the nucleus (44).
No nuclear accumulation of Scp160 was observed in either the mex67-5 or rpb1-1 cells (Figure 3). As expected, the control mRNA binding protein, ΔRGG-Nab2-GFP, accumulated in the nucleus of both mex67-5 and rpb1-1 cells following a shift to the non-permissive temperature. No change in the localization of NLS-NES-GFP was observed. These results indicate that either Scp160 does not enter the nucleus or its export does not depend on ongoing mRNA synthesis or mRNA export.
Figure 3.
Localization of Scp160-GFP in rpb1-1, mex67-5 and wild-type cells. The localization of Scp160-GFP was examined in wild-type, rpb1-1 and mex67-5 cells following a 1 h shift to 37°C. Results indicates that Scp160 is localized to the cytoplasm in both rpb1-1 and mex67-5 cells. As controls, we also localized NLS-NES-GFP and ΔRGG-Nab2-GFP. As expected, ΔRGG-Nab2-GFP, which is exported in an mRNA export-dependent manner, accumulates in the nuclei of both rpb1-1 and mex67-5 cells, but NLS-NES-GFP does not.
Scp160 does not accumulate in the nucleus of cells with an export-deficient NPC
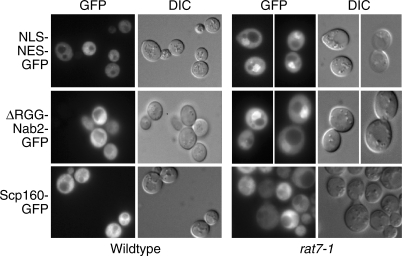
As a final test of whether Scp160 enters the nucleus, we exploited a nuclear pore mutant that blocks all known nuclear export pathways, rat7-1 (32,45–47). We hypothesized that if Scp160 enters the nucleus it should accumulate in the nucleus of rat7-1 cells. Although RAT7 (NUP159) is essential, the rat7-1 allele is a temperature-sensitive allele that can be shifted to the non-permissive temperature to inactivate Rat7/Nup159 and block export pathways (46). We examined the localization of Scp160-GFP and control proteins, NLS-NES GFP and ΔRGG-Nab2-GFP, in rat7-1 cells. Following the shift to the non-permissive temperature, Scp160-GFP remained cytoplasmic in rat7-1 cells similar to wild-type cells (Figure 4). In contrast, NLS-NES-GFP and ΔRGG-Nab2-GFP both accumulated in the nuclei of rat7-1 cells. These results taken together with localization of Scp160 in other export mutants suggest that Scp160 does not enter the nucleus, at least under normal growth conditions.
Figure 4.
Localization of Scp160-GFP in rat7-1 and wild-type cells. The localization of Scp160-GFP was examined in rat7-1 cells following a 30 min shift to 37°C where all known transport pathways are blocked. As shown, Scp160 is localized to the cytoplasm in rat7-1 cells. As controls, we also visualized NLS-NES-GFP and ΔRGG-Nab2-GFP. As expected both control proteins accumulate in the nuclei of rat7-1 cells.
In vivo function of an Scp160 NLS mutant
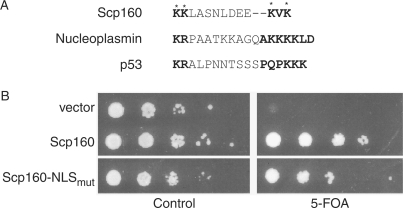
Based on its primary amino acid sequence, Scp160 has been predicted to contain a bipartite NLS (20). Like other previously characterized bipartite NLS sequences, Scp160 contains two stretches of basic amino acids separated by a short spacer region (Figure 5A). However, results of our localization studies suggest that the predicted NLS within Scp160 does not target the protein to the nucleus and, therefore, may not be functional. If this assessment is correct, amino acid substitutions within the key NLS residues in the sequence should not significantly impact the function of Scp160. To test this hypothesis, four basic residues within the predicted NLS of Scp160 were changed to alanines to create Scp160-NLSmut (residues changed are indicated by asterisks in Figure 5A). This Scp160 variant was then assayed for its ability to rescue the synthetic lethality of the Δscp160Δeap1 cells. Growth of Δscp160Δeap1 cells expressing Scp160-NLSmut was not significantly affected compared to growth of these cells expressing wild-type Scp160 (Figure 5B). In contrast, yeast-containing vector alone (pRS315) were unable to grow on 5-FOA medium. These results suggest that the predicted NLS does not affect the global in vivo function of Scp160.
Figure 5.
Functional analysis of Scp160-NLSmut. (A) An alignment of the predicted NLS sequence from Scp160 with functional NLS sequences from nucleoplasmin and p53 is shown. The lysine residues that were changed to alanines to create Scp160-NLSmut are indicated by asterisks. (B) To assess the functional importance of the predicted NLS within Scp160, we tested whether an Scp160 variant with a mutant NLS (Scp160-NLSmut) could replace wild-type Scp160 in Δscp160Δeap1 cells where Scp160 is required for viability (36). Cultures were grown to log phase and equal numbers of cells were serially diluted by orders of magnitude and spotted onto control plates lacking uracil and leucine (left) or test plates lacking leucine but containing 5-FOA (right). The top row shows cells carrying LEU2 vector (pRS315); the middle row shows cells containing a wild-type copy of Scp160; the bottom row shows cells that express Scp160-NLSmut as the sole copy of Scp160.
DISCUSSION
This study was designed to test whether the yeast RNA-binding protein, Scp160, is a nucleocytoplasmic shuttling protein and hence to examine the function of the predicted nuclear targeting signals within Scp160. Previous studies have revealed that Scp160 is localized to the cytoplasm at steady state (22–25) and also provided evidence that Scp160 associates with specific mRNAs (19). Given the presence of sequences reminiscent of both an NLS and an NES motif (21), one possible mechanism for Scp160 function, consistent with other shuttling RNA binding proteins (26,32), is that Scp160 could enter the nucleus, associate with RNA substrates, and then accompany these RNA transcripts to the cytoplasm. However, results of our analysis indicate that Scp160 is not likely to enter the nucleus. Furthermore, the predicted NLS is not required for Scp160 function. Taken together, these experiments argue for an alternative model for Scp160 function. More generally, this study underlines the importance of experimental analysis to support the identification of putative nuclear transport signals in a protein of interest.
In this study, we localized Scp160 in a number of conditional mutants to test the hypothesis that Scp160 is able to move into the nucleus and accompany transcripts to the cytoplasm by virtue of its predicted NLS and NES signals. We localized Scp160 in mutants blocked in well-characterized export pathways, including NES-protein export (crm1-3), mRNA biosynthesis (rbp1-1), mRNA export (mex67-5) and all known export pathways (rat7-1). Despite its putative targeting sequences, Scp160 did not accumulate in the cell nucleus when any of these pathways were blocked. Furthermore, we have shown that amino acid changes within the predicted NLS of Scp160 do not severely affect Scp160 function in vivo. Due to the slight growth defect we observed when four lysines within the NLS were changed to alanines, we cannot eliminate the possibility that these residues are important for some other function of Scp160, however, taken together our data suggest that Scp160 does not enter the nucleus. We cannot, however, eliminate the possibility that Scp160 enters the nucleus under specific growth conditions not tested here. There is also the possibility that Scp160 export could depend on a pathway not examined here, but the nuclear pore mutant, which blocks all known export pathways, makes this interpretation of our results unlikely.
As a whole, these data suggest that Scp160 functions exclusively in the cytoplasm where it most likely associates with mRNA. A further extension of these findings is that the pleiotropic effects observed in Δscp160 cells (20,23,48), including increased DNA content per cell, missegregation of nutritional markers through meiosis and abnormal cell morphology, are likely due to indirect effects, perhaps caused by changes in expression of genes required for these pathways. Thus, deregulation of mRNA transcripts that bind to Scp160 may cause some of the phenotypes observed in Δscp160 cells. Alternatively, some phenotypes may be due to loss of direct interaction of Scp160 with other proteins within the cell resulting in incompletely formed mRNP complexes. Taken with the localization studies described here, we conclude that the predicted nuclear targeting sequences in Scp160 are non-functional.
Scp160 is most closely related to the vigilin class of proteins, which, like Scp160, contain a large array of 14 to 15 repeated KH domains (21). Vigilins control gene expression through multiple mechanisms including regulating mRNA stability in the cytoplasm (49,50) and modulating heterochromatin formation in the nucleus (51,52). One piece of data that links Scp160 function with related proteins from higher eukaryotes is the finding that the Drosophila vigilin protein, Ddp1, can complement one phenotype associated with deletion of the SCP160 gene in S. cerevisiae (48). Specifically, this study demonstrated that expression of Ddp1 in yeast cells lacking Scp160 could complement the ploidy defect observed when Scp160 is absent. However, this study did not test whether the Ddp1 entered the yeast nucleus and thus the mechanism underlying the phenotypic suppression remains unclear. Furthermore, given the vast differences in centromere sequence and organization in S. cerevisiae and Drosophila (53,54), it seems extremely unlikely that Ddp1 could complement the Δscp160 yeast mutant by interacting with yeast centromeres. Thus, Ddp1 may share other evolutionarily conserved functions with Scp160 that have yet to be defined. Taken together, these findings suggest that the vigilin family of proteins controls gene expression through multiple mechanisms and that those functions that are located within the nucleus may be present only in higher eukaryotes. It is interesting that the role of vigilins in modulating heterochromatin is related to the RNAi pathway and that this pathway is absent from S. cerevisiae (55). Perhaps the nuclear function of vigilins arose in concert with the RNAi pathways.
Beyond the implications for Scp160 and vigilin function, this study highlights the importance of experimentally testing predicted targeting signals. Many proteins contain putative classical nuclear transport signals presumed to contribute to protein function in the absence of corroborating experimental evidence. Thus, caution must be used when incorporating in silico predictions into models for protein function.
ACKNOWLEDGEMENTS
We gratefully acknowledge Allison Lange for her helpful discussions about site-directed mutagenesis and nuclear transport; Sara Leung for microscopy reagents, protocols and advice, and all other members of the Corbett and Fridovich-Keil laboratories for their many helpful discussions and contributions. This work was supported in part by funds from the NSF (J.F.K.), funds from the Emory University Research Committee (J.F.K.), and a grant from the NIH (A.H.C.). Funding to pay the Open Access publication charges for this article was waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 2.Suntharalingam M, Wente SR. Peering through the pore. Nuclear pore complex structure, assembly, and function. Dev. Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- 3.Lim RY, Fahrenkrog B. The nuclear pore complex up close. Curr. Opin. Cell Biol. 2006;18:342–347. doi: 10.1016/j.ceb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalderon D, Richardson WD, Markham AF, Smith AE. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 6.Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 7.Dingwall C, Laskey RA. Nuclear targeting sequences – a consensus? Trends Biochem. Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 8.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 9.Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 10.Bayliss R, Littlewood T, Strawn LA, Wente SR, Stewart M. GLFG and FxFG nucleoporins bind to overlapping sites on importin-beta. J. Biol. Chem. 2002;277:50597–50606. doi: 10.1074/jbc.M209037200. [DOI] [PubMed] [Google Scholar]
- 11.Dasso M. The Ran GTPase: theme and variations. Curr. Biol. 12:R502–R508. doi: 10.1016/s0960-9822(02)00970-3. [DOI] [PubMed] [Google Scholar]
- 12.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell. Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 13.Stewart M. Insights into the molecular mechanism of nuclear trafficking using nuclear transport factor 2 (NTF2) Cell Struct. Funct. 2006;25:217–225. doi: 10.1247/csf.25.217. [DOI] [PubMed] [Google Scholar]
- 14.Matsuura Y, Stewart M. Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 2005;24:3681–3689. doi: 10.1038/sj.emboj.7600843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson AW, Lund E, Dahlberg J. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 2002;27:580–585. doi: 10.1016/s0968-0004(02)02208-9. [DOI] [PubMed] [Google Scholar]
- 16.Strom AC, Weis K. Importin-beta-like nuclear transport receptors. Genome Biol. 2001;2:3008.1–3008.9. doi: 10.1186/gb-2001-2-6-reviews3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerace L. Nuclear export signals and the fast track to the cytoplasm. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- 18.Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 19.Li AM, Watson A, Fridovich-Keil JL. Scp160p associates with specific mRNAs in yeast. Nucleic Acids Res. 2003;31:1830–1837. doi: 10.1093/nar/gkg284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber V, Wernitznig A, Hager G, Harata M, Frank P, Wintersberger U. Purification and nucleic-acid-binding properties of a Saccharomyces cerevisiae protein involved in the control of ploidy. Eur. J. Biochem. 1997;249:309–317. doi: 10.1111/j.1432-1033.1997.00309.x. [DOI] [PubMed] [Google Scholar]
- 21.Currie JR, Brown WT. KH domain-containing proteins of yeast: absence of a fragile X gene homologue. Am. J. Med. Genet. 1999;84:272–276. [PubMed] [Google Scholar]
- 22.Li AM, Vargas CA, Brykailo MA, Openo KK, Corbett AH, Fridovich-Keil JL. Both KH and non-KH domain sequences are required for polyribosome association of Scp160p in yeast. Nucleic Acids Res. 2004;32:4768–4775. doi: 10.1093/nar/gkh812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wintersberger U, Kuhne C, Karwan A. Scp160p, a new yeast protein associated with the nuclear membrane and the endoplasmic reticulum, is necessary for maintenance of exact ploidy. Yeast. 1995;11:929–944. doi: 10.1002/yea.320111004. [DOI] [PubMed] [Google Scholar]
- 24.Frey S, Pool M, Seedorf M. Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem. 2001;276:15905–15912. doi: 10.1074/jbc.M009430200. [DOI] [PubMed] [Google Scholar]
- 25.Lang BD, Li A, Black-Brewster HD, Fridovich-Keil JL. The brefeldin A resistance protein Bfr1p is a component of polyribosome-associated mRNP complexes in yeast. Nucleic Acids Res. 2001;29:2567–2574. doi: 10.1093/nar/29.12.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA. 2005;11:517–531. doi: 10.1261/rna.7291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunn EF, Hammell CM, Hodge CA, Cole CN. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev. 2005;19:90–103. doi: 10.1101/gad.1267005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Meth. Enzymol. 1991;194:281–302. [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Seedorf M, Damelin M, Kahana J, Taura T, Silver PA. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol. Cell. Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marfatia KA, Crafton EB, Green DM, Corbett AH. Domain analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. Dissecting the requirements for Nab2p-facilitated poly(A) RNA export. J. Biol. Chem. 2003;278:6731–6740. doi: 10.1074/jbc.M207571200. [DOI] [PubMed] [Google Scholar]
- 33.Matsuura Y, Lange A, Harreman MT, Corbett AH, Stewart M. Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J. 2003;22:5358–5369. doi: 10.1093/emboj/cdg538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Meth. Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 35.Cosentino GP, Schmelzle T, Hagnighat A, Helliwell SB, Hall MN, Sonenberg N. Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein. Mol. Cell. Biol. 2000;20:4604–4613. doi: 10.1128/mcb.20.13.4604-4613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendelsohn BA, Li AM, Vargas CA, Riehman K, Watson A, Fridovich-Keil JL. Genetic and biochemical interactions between SCP160 and EAP1 in yeast. Nucleic Acids Res. 2003;31:5838–5847. doi: 10.1093/nar/gkg810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neville M, Rosbash M. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan C, Lee LH, Davis LI. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 1998;17:7416–7429. doi: 10.1093/emboj/17.24.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taura T, Krebber H, Silver P. A member of the Ran-binding protein family, Yrb2p, is invovled in nuclear protein export. Proc. Natl Acad. Sci., USA. 1998;95:7427–7432. doi: 10.1073/pnas.95.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 41.Nonet M, Scafe C, Sexton J, Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrman R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hurt E, Strasser K, Segref A, Bailer S, Schlaich N, Presutti C, Tollervey D, Jansen R. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J. Biol. Chem. 2000;275:8361–8363. doi: 10.1074/jbc.275.12.8361. [DOI] [PubMed] [Google Scholar]
- 44.Green DM, Marfatia KA, Crafton EB, Zhang X, Cheng X, Corbett AH. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is Regulated by arginine methylation via Hmt1p. J. Biol. Chem. 2002;277:7752–7760. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- 45.Krebber H, Taura T, Lee MS, Silver PA. Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export. Genes Dev. 1999;13:1994–2004. doi: 10.1101/gad.13.15.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gorsch LC, Dockendorff TC, Cole CN. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell. Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Del Priore V, Snay CA, Bahr A, Cole CN. The product of the Saccharomyces cerevisiae RSS1 gene, identified as a high-copy suppressor of the rat7-1 temperature-sensitive allele of the RAT7/NUP159 nucleoporin, is required for efficient mRNA export. Mol. Biol. Cell. 1996;7:1601–1621. doi: 10.1091/mbc.7.10.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cortes A, Huertas D, Fanti L, Pimpinelli S, Marsellach FX, Pina B, Azorin F. DDP1, a single-stranded nucleic acid-binding protein of Drosophila, associates with pericentric heterochromatin and is functionally homologous to the yeast Scp160p, which is involved in the control of cell ploidy. EMBO J. 1999;18:3820–3833. doi: 10.1093/emboj/18.13.3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dodson RE, Shapiro DJ. Vigilin, a ubiquitous protein with 14 K homology domains, is the estrogen-inducible vitellogenin mRNA 3'-untranslated region-binding protein. J. Biol. Chem. 1997;272:12249–12252. doi: 10.1074/jbc.272.19.12249. [DOI] [PubMed] [Google Scholar]
- 50.Dodson RE, Shapiro DJ. Regulation of pathways of mRNA destabilization and stabilization. Prof. Nucleic Acid Res. Mol. Biol. 2002;72:129–164. doi: 10.1016/s0079-6603(02)72069-2. [DOI] [PubMed] [Google Scholar]
- 51.Huertas D, Cortes A, Casanova J, Azorin F. Drosophila DDP1, a multi-KH-domain protein, contributes to centromeric silencing and chromosome segregation. Curr. Biol. 2004;14:1611–1620. doi: 10.1016/j.cub.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 52.Wang Q, Zhang Z, Blackwell K, Carmichael GG. Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr. Biol. 2005;15:384–391. doi: 10.1016/j.cub.2005.01.046. [DOI] [PubMed] [Google Scholar]
- 53.Lamb JC, Birchler JA. The role of DNA sequence in centromere formation. Genome Biol. 2003;4:214. doi: 10.1186/gb-2003-4-5-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sunkel CE, Coelho PA. The elusive centromere: sequence divergence and functional conservation. Curr. Opin. Genet. Dev. 1995;5:756–767. doi: 10.1016/0959-437x(95)80008-s. [DOI] [PubMed] [Google Scholar]
- 55.Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double stranded RNA. Nat. Rev. Genet. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- 56.Palancade B, Zuccolo M, Loeillet S, Nicolas A, Doye V. Pml39, a novel protein of the nuclear periphery required for nuclear retention of improper messenger ribonucleoparticles. Mol. Biol. Cell. 2005;16:5258–5268. doi: 10.1091/mbc.E05-06-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brykailo MA, Corbett AH, Fridovich-Keil JL. Functional overlap between conserved and diverged KH domains in Saccharomyces cerevisiae SCP160. Nucleic Acids Res. 2007;35:1108–1118. doi: 10.1093/nar/gkl1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multi-functional yeast high-copy-number shuttle vectors. Gene. 110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]