Abstract
For optimal compatibility with biopharmaceutical manufacturing and gene therapy, heterologous transgene control systems must be responsive to side-effect-free physiologic inducer molecules. The arginine-inducible interaction of the ArgR repressor and the ArgR-specific ARG box, which synchronize arginine import and synthesis in the intracellular human pathogen Chlamydia pneumoniae, was engineered for arginine-regulated transgene (ART) expression in mammalian cells. A synthetic arginine-responsive transactivator (ARG), consisting of ArgR fused to the Herpes simplex VP16 transactivation domain, reversibly adjusted transgene transcription of chimeric ARG box-containing mammalian minimal promoters (PART) in an arginine-inducible manner. Arginine-controlled transgene expression showed rapid induction kinetics in a variety of mammalian cell lines and was adjustable and reversible at concentrations which were compatible with host cell physiology. ART variants containing different transactivation domains, variable spacing between ARG box and minimal promoter and several tandem ARG boxes showed modified regulation performance tailored for specific expression scenarios and cell types. Mice implanted with microencapsulated cells engineered for ART-inducible expression of the human placental secreted alkaline phosphatase (SEAP) exhibited adjustable serum phosphatase levels after treatment with different arginine doses. Using a physiologic inducer, such as the amino acid l-arginine, to control heterologous transgenes in a seamless manner which is devoid of noticeable metabolic interference will foster novel opportunities for precise expression dosing in future gene therapy scenarios as well as the manufacturing of difficult-to-produce protein pharmaceuticals.
INTRODUCTION
Conditional transcription control systems which fine-tune heterologous transgene expression in mammalian cells are fundamental for gene-function analysis (1,2), drug discovery (3,4), expression dosing in gene therapy (5), design of synthetic gene networks and for manufacturing of difficult-to-produce protein pharmaceuticals (6,7). Currently available mammalian transgene control systems capitalize on a generic design consisting of a synthetic transactivator (transrepressor), typically derived from a prokaryotic response regulator fused to a mammalian transactivation (transrepression) domain and a chimeric promoter assembled by cloning transactivator-specific operator sites adjacent to a minimal (constitutive) eukaryotic promoter (8–10). Binding of an inducer molecule modulates the affinity of the transactivator (transrepressor) for the cognate promoter and either induces (ON-type systems, (11–14)) or represses (OFF-type systems, (14,15)) transcription of linked transgenes. Trigger molecules and parameters include antibiotics (10,14,16), hormones and hormone analogs (17–19), quorum sensing substances (12,20,21), temperature (22), the redox poise (20), immunosuppressive and antidiabetic drugs (23,24), gaseous acetaldehyde (11) and biotin (25). Although most of these established gene regulation systems show excellent regulation performance in vitro as well as in animals, the inducer molecules are often incompatible with gene therapy and biopharmaceutical manufacturing scenarios because of their side-effects, which are well documented for hormones and some of their analogs (26,27), antibiotics (28–30) and immunosuppressive drugs (31,32). While the biopharmaceutical manufacturing industry strives to develop gene regulation systems controlled by FDA-approved media components to prevent prohibitive downstream processing and licensing procedures (33), the gene therapy community dreams of capturing pathologic signals which can be converted into a well-tuned therapeutic intervention. The common interest is exploiting physiologic signal molecules for titration of heterologous product gene expression and prevent interference between endogenous and synthetic regulatory circuits.
As an obligate intracellular pathogen infecting the respiratory tracts and causing pneumonia and atherosclerotic heart disease (34–37), Chlamydia pneumoniae has evolved to capture physiologic signals and synchronize its metabolism with the host cell (38). For example, the coordination of arginine biosynthesis and catabolism with availability of this amino acid in host cells is managed by ArgR, a master regulatory molecule acting as an arginine-dependent apo-repressor that specifically binds to arginine-responsive transactivator (ARG) box operators anzd represses the glnPQ operon which encodes a putative arginine transport system in an arginine-responsive manner (39,40). ArgR, which is highly conserved in bacterial systems, can function both as transcriptional repressor or activator (41,42). In Escherichia coli, ArgR negatively regulates the expression of the arginine biosynthetic genes in response to intracellular l-arginine levels (43). When acting as a repressor, ArgR requires allosteric activation by l-arginine to bind to an 18-bp palindromic operator sequence known as the ARG box (39,43–45). While the N-terminal half of ArgR contains a winged helix-turn-helix family DNA binding domain, the C-terminal half is responsible for arginine binding.
We have engineered ArgR and the ARG box of C. pneumoniae for l-arginine-regulated transgene (ART) regulation system in mammalian cells. As an ON-type system by default, ART remained silent at minimal endogenous l-arginine concentrations and was adjustably induced by increasing l-arginine levels. Using a proteogenic amino acid to control heterologous transgene in mammalian cells grown in monolayer cultures or in mice should foster new opportunities in gene therapy and biopharmaceutical manufacturing.
MATERIALS AND METHODS
Plasmid construction
All plasmids and oligonucleotides constructed and used in this study are listed in Table 1. Details on vector design are also provided in Table 1.
Table 1.
Plasmids and oligonucleotides used and designed in this study
| Plasmid | Description and cloning strategy | Reference or source |
|---|---|---|
| pRevTRE | Oncoretroviral expression vector containing a tetracycline-responsive expression unit | Clontech, Palo Alto, CA, USA |
| pPur | Selection vector conferring puromycin resistance to eukaryotic cells | Clontech, Palo Alto, CA, USA |
| pSV2neo | Selection vector conferring neomycin resistance to eukaryotic cells | Clontech, Palo Alto, CA, USA |
| pBP10 | Vector encoding a PETR5-driven SEAP expression unit (PETR5-SEAP-pA; PETR5, ETR-2bp-PhCMVmin) | (9) |
| pBP11 | Vector encoding a PETR6-driven SEAP expression unit (PETR6-SEAP-pA; PETR6, ETR-4bp-PhCMVmin) | (9) |
| pBP12 | Vector encoding a PETR7-driven SEAP expression unit (PETR7-SEAP-pA; PETR7, ETR-6bp-PhCMVmin) | (9) |
| pBP13 | Vector encoding a PETR8-driven SEAP expression unit (PETR8-SEAP-pA; PETR8, ETR-8bp-PhCMVmin) | (9) |
| pBP14 | Vector encoding a PETR9-driven SEAP expression unit (PETR9-SEAP-pA; PETR9, ETR-10bp-PhCMVmin) | (9) |
| pMF111 | Vector encoding a PhCMVmin-driven SEAP expression unit (PhCMVmin-SEAP-pA) | (69) |
| pMT1227 | Vector encoding the C. pneumoniae CWL029 ArgR gene | (39) |
| pWW35 | Constitutive ET1 expression vector (PSV40-ET1-pA) | (10) |
| pWW42 | Constitutive ET2 expression vector (PSV40-ET2-pA) | (10) |
| pWW64 | Constitutive ET3 expression vector (PSV40-ET3-pA) | (9) |
| pSH91 | Constitutive ARG2 expression vector (PSV40-ARG2-pA) argR was amplified from pMT1227 using OSH49: 5′-gatcgaattcccaccATGA AAAAAAAAGTAAC TATAGATGAGG-3′ and OSH50: 5′-cttatggcgcgcggctgtacgc ggaATCCAAGAA AACTTGCAGTA AATTTG-3′ (upper case, annealing sequence; lower case italic, restriction sites), digested with EcoRI/BssHII and ligated into pWW35 (EcoRI/BssHII). | This work |
| pSH92 | Vector encoding OARG-0bp-PhCMVmin-ET1-pA OARG-0bp-PhCMVmin was amplified from pRevTRE using OSH51: 5′-gatcgacgtcAGTTTTCTTGGATTAATTGCATAAATATGATTTCATTATAAATAAATATGCATAAGAGGTCGGAGTGcctgcaggTCGAGCTCGGTACCCGGGTC-3′ and OWW22: 5′-gctagaattcCGCGGAGGCTGGATCGG-3′ (upper case, annealing sequence; lower case italic, restriction sites; upper case italic, OARG), digested with AatII/EcoRI and ligated into pWW35 (AatII/EcoRI). | This work |
| pSH93 | Vector encoding a PART1-driven expression unit (PART1-SEAP-pA; PART1, OARG-0bp-PhCMVmin) OARG-0bp-PhCMVmin was excised from pSH92 (SspI/EcoRI) and ligated into pMF111 (SspI/EcoRI). | This work |
| pSH105 | Vector encoding a PART2-driven expression unit (PART2-SEAP-pA; PART2, OARG-2bp-PhCMVmin) 2bp-PhCMVmin-SEAP was excised from pBP10 (SbfI/XhoI) and ligated into pSH93 (SbfI/XhoI). | This work |
| pSH106 | Vector encoding a PART3-driven expression unit (PART3-SEAP-pA; PART3, OARG-4bp-PhCMVmin) 4bp-PhCMVmin-SEAP was excised from pBP11 (SbfI/XhoI) and ligated into pSH93 (SbfI/XhoI). | This work |
| pSH107 | Vector encoding a PART4-driven expression unit (PART4-SEAP-pA; PART4, OARG-6bp-PhCMVmin) 6bp-PhCMVmin-SEAP was excised from pBP12 (SbfI/XhoI) and ligated into pSH93 (SbfI/XhoI). | This work |
| pSH108 | Vector encoding a PART5-driven expression unit (PART5-SEAP-pA; PART5, OARG-8bp-PhCMVmin) 8bp-PhCMVmin-SEAP was excised from pBP13 (SbfI/XhoI) and ligated into pSH93 (SbfI/XhoI). | This work |
| pSH109 | Vector encoding a PART6-driven expression unit (PART6-SEAP-pA; PART6, OARG-10bp-PhCMVmin) 10bp-PhCMVmin-SEAP was excised from pBP14 (SbfI/XhoI) and ligated into pSH93 (SbfI/XhoI). | This work |
| pSH115 | Vector encoding AscI-OARG-MluI-0bp-PhCMVmin-ET1-pA AscI-OARG-MluI-0bp-PhCMVmin was amplified from pRevTRE using OSH69: 5′-gatcgacgtcggcgcgccAGTTTTCTTGGATTAATTGCATAAATATGATTTCATTATAAATAAATATGCATAAGAGGTCGGAGTGacgcgtcctgcaggTCGAGCTCGGTACCCGGGTC-3′ and OWW22: 5′-gctagaattcCGCGGAGGCTGGATCGG-3′ (upper case, annealing sequence; lower case italic, restriction sites; upper case italic, OARG), digested with AatII/EcoRI and ligated into pWW35 (AatII/EcoRI). | This work |
| pSH117 | Vector encoding a PARTm1-driven expression unit (PARTm1-SEAP-pA; PARTm1, AscI-OARG-MluI-0bp-PhCMVmin) AscI-OARG-MluI-0bp-PhCMVmin was excised from pSH115 (SspI/EcoRI) and ligated into pSH93 (SspI/EcoRI). | This work |
| pSH119 | Vector encoding a PARTm2-driven expression unit (PARTm2-SEAP-pA; PARTm2, AscI-OARG-7bp-OARG-MluI-0bp-PhCMVmin) AscI-OARG-MluI-0bp-PhCMVmin-SEAP was excised from pSH117 (AscI/XhoI) and ligated into pSH117 (MluI/XhoI). | This work |
| pSH120 | Constitutive ARG1 expression vector (PSV40-ARG1-pA) The NF-κB-derived transcription domain (p65) was excised from pWW42 (BssHII/BamHI) and ligated into pSH91 (BssHII/BamHI). | This work |
| pSH121 | Constitutive ARG3 expression vector (PSV40-ARG3-pA) The E2F4-derived transcription domain (E2F4) was excised from pWW64 (BssHII/BamHI) and ligated into pSH91 (BssHII/BamHI). | This work |
| pSH122 | Autoregulated vector encoding a PART1-driven expression unit (PART1-SEAP-IRESPV-ARG2-pA) ARG2 was excised from pSH91 (NotI/XbaI) and ligated into pSH93 (NotI/NheI). | This work |
| pSH126 | Vector encoding a PARTm3-driven expression unit (PARTm3-SEAP-pA; PARTm3, AscI-OARG-7bp-OARG-7bp-OARG–MluI-0bp-PhCMVmin) AscI-OARG-7bp-OARG–MluI-0bp-PhCMVmin-SEAP was excised from pSH119 (AscI/XhoI) and ligated into pSH117 (MluI/XhoI). | This work |
| pSH127 | Vector encoding a PARTm4-driven expression unit (PARTm4-SEAP-pA; PARTm4, AscI-OARG-7bp-OARG-7bp-OARG–7bp-OARG-MluI-0bp-PhCMVmin) AscI-OARG-7bp-OARG–MluI-0bp-PhCMVmin-SEAP was excised from pSH119 (AscI/XhoI) and ligated into pSH119 (MluI/XhoI). | This work |
ArgR, transactivator of the C. pneumoniae CWL029 of the glnPQ operon; ARG1, l-arginine-dependent transactivator (ArgR-p65); ARG2, l-arginine-dependent transactivator (ArgR-VP16); ARG3, l-arginine-dependent transactivator (ArgR-E2F4); E2F4, transactivation domain of the human E2F4; ET1, macrolide-dependent transactivator (E-VP16); ET2, macrolide dependent transactivator (E-p65); ET3, macrolide dependent transactivator (E-E2F4); ETR, operator module specific for MphR(A); IRESPV, internal ribosome entry site of polioviral origin; NF-κB, human transcription factor; OARG, ArgR-specific operator; p65, transactivation domain of NF-κB; pA, SV40-derived polyadenylation site; PART1-6, l-arginine-responsive promoters containing different spacers between OARG and PhCMVmin; PARTm1-4, l-arginine-responsive promoters containing one (PARTm1), two (PARTm2), three (PARTm3) or four (PARTm4) operator sequences upstream of PhCMVmin; PhCMV, human cytomegalovirus immediate early promoter; PETR5-9, macrolide-responsive promoters containing different spacers between ETR and PhCMVmin; PhCMVmin, minimal version of the human cytomegalovirus promoter; PSV40, constitutive simian virus 40 promoter; SEAP, human placental secreted alkaline phosphatase; VP16, H. simplex virus-derived transactivation domain.
Cell culture, transfection and construction of stable cell lines
Wild-type Chinese hamster ovary cells (CHO-K1, ATCC: CCL-61) and its derivatives [e.g. CHO-ET1-SEAP1; (10)] were cultivated in l-arginine-free ChoMaster® HTS (Cell Culture Technologies, Gravesano, Switzerland) supplemented with 10 mg/l l-arginine, 5% (v/v) fetal calf serum (FCS, PAN Biotech GmbH, Aidenbach, Germany, Cat. No. 3302, Lot No. P251110) and 1% (v/v) penicillin/streptomycin solution (Sigma, St Louis, MO, USA, Cat. No. P4458). Human fibrosarcoma cells (HT-1080, ATCC, CCL-121), African green monkey kidney cells (COS-7, ATCC CRL-1651) and human embryonic kidney cells, transgenic for the simian virus 40 (SV40) large T antigen [HEK293-T, (46)], were cultivated in l-arginine-free Hektor G (Cell Culture Technologies, Gravesano, Switzerland) supplemented with 10 mg/l l-arginine, 10% (v/v) FCS and 1% (v/v) penicillin/streptomycin solution. Mouse fibroblasts (NIH/3T3, ATCC: CRL-1658) were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen, Basel, Switzerland, Cat. No. 52100-39) supplemented with 10% FCS and 1% (v/v) penicillin/streptomycin solution. All cell types were cultivated at 37°C in a humidified atmosphere containing 5% CO2.
For DNA transfection, 1.2 μg DNA (for co-transfection an equal amount of each plasmid) was transfected into 55 000 cells pre-cultivated for 24 h in a well of a 24-well plate. For transfection of CHO-K1, the plasmid DNA was diluted into a total volume of 25 μl 0.5 M CaCl2 solution, mixed with 25 μl  solution (100 mM HEPES, 280 mM NaCl, 1.5 mM Na2HPO4, pH 7.1) and incubated for 15 min at room temperature. The DNA–calcium phosphate complex was added, and centrifuged onto the cells (5 min at 1200 g) to increase transfection efficiency. After incubation for 2 h, the cells were exposed to a glycerol shock for 30 s (l-arginine-free ChoMaster® HTS medium supplemented with 15% glycerol). After a single washing with phosphate-buffered saline (PBS, Dulbecco's phosphate-buffered saline, Invitrogen, Basel, Switzerland, Cat. No. 21600-0069), the cells were cultivated in ChoMaster® HTS supplemented with different l-arginine concentrations. For transfection of COS-7, HEK293-T, the standard calcium phosphate-based transfection protocol was used (21) and transfected populations were cultivated in Hektor G supplemented with different l-arginine concentrations. HT-1080 and NIH/3T3 were transfected with Fugene™ 6 (Roche Diagnostics AG, Basel, Switzerland, Cat. No. 11814443001) following the supplier's protocol and cultivated in Hektor G and DMEM, respectively. Production of the reporter protein was assessed in the culture supernatants 60 h after transfection, unless indicated otherwise.
solution (100 mM HEPES, 280 mM NaCl, 1.5 mM Na2HPO4, pH 7.1) and incubated for 15 min at room temperature. The DNA–calcium phosphate complex was added, and centrifuged onto the cells (5 min at 1200 g) to increase transfection efficiency. After incubation for 2 h, the cells were exposed to a glycerol shock for 30 s (l-arginine-free ChoMaster® HTS medium supplemented with 15% glycerol). After a single washing with phosphate-buffered saline (PBS, Dulbecco's phosphate-buffered saline, Invitrogen, Basel, Switzerland, Cat. No. 21600-0069), the cells were cultivated in ChoMaster® HTS supplemented with different l-arginine concentrations. For transfection of COS-7, HEK293-T, the standard calcium phosphate-based transfection protocol was used (21) and transfected populations were cultivated in Hektor G supplemented with different l-arginine concentrations. HT-1080 and NIH/3T3 were transfected with Fugene™ 6 (Roche Diagnostics AG, Basel, Switzerland, Cat. No. 11814443001) following the supplier's protocol and cultivated in Hektor G and DMEM, respectively. Production of the reporter protein was assessed in the culture supernatants 60 h after transfection, unless indicated otherwise.
CHO-ARG2-SEAP, transgenic for l-arginine-controlled SEAP expression, was constructed by sequential co-transfection and clonal selection of (i) pSH91 and pPur (Clontech, Cat. No. 6156-1) (CHO-ARG2) and (ii) pSH93 and pSV2neo (Clontech, Cat. No. 6172-1) (CHO-ARG2-SEAP). To assess the dose–response characteristics of ART-regulated gene expression, CHO-ARG2-SEAP was cultured at 110 000 cells/ml for 60 h in l-arginine-free ChoMaster® HTS supplemented with l-arginine ranging from 10 mg/l to 10 000 mg/l. Reversibility of l-arginine-mediated SEAP production was assessed by cultivating CHO-ARG2-SEAP (110 000 cells/ml) for 1 week while alternating l-arginine concentrations from 10 to 1000 mg/l every 48 h.
Western blot analysis
Sixty hours after transfection of 350 000 HEK-293T cells with the vector encoding ARG2 (pSH91, PSV40-ARG2-pA), the cell lysate was heat denaturated in 5× SPS-PAGE reducing sample buffer (50% glycerol, 250 mM Tris, 10% sodium dodecylsulfate, 500 mM dithiothreitol, 0.01% bromophenolblue, pH 6.8) at 85°C for 3 min and size-fractionated on a 12% SDS-polyacrylamide gel. The gel was transferred to a polyvinylidene fluoride membrane (Millipore Corporation, Bedford, USA. Cat. No. IPVH20200) and blocked overnight with 5% low fat milk (Migros, Switzerland) in PBS. The target protein was detected with a primary monoclonal anti-VP16 (Santa Cruz Biotechnology Inc., California, Cat. No. sc-7545, 1 : 100 dilution in 5% low fat milk in PBS) and visualized using a secondary HRP-coupled anti-mouse IgG (Amersham Biosciences UK Limited, Buckinghamshire, UK, Cat. No. NA931V, 1 : 1000 dilution in 5% low fat milk in PBS) and a chemiluminescence-based assay (GE Healthcare Amersham UK Limited, Buckinghamshire, UK, Cat. No. RPN2132). Following the same protocol, tubulin-α was visualized as a loading control (Ab-2 (DM1A); Neomarkers, California, Cat. No. MS-581-P1).
Regulating l-arginine and l-arginine analogs
l-arginine stock solution was prepared in water by adjusting 50 g/l l-arginine base (0.287 M, AppliChem, Darmstadt, Germany, Cat. No. A3653,0100) to pH 7.2 with HCl and used at the final concentrations indicated. l-ornithine hydrochloride (Fluka, Buchs, Switzerland, Cat. No. 75470) and l-citrulline (Fluka, Buchs, Switzerland, Cat. No. 27510), l-homoarginine hydrochloride (Acros Organics, Basel, Switzerland, Cat. No. 169090050) were dissolved in water (0.287 M) with pH adjusted to 7.2 by NaOH. l-arginine methyl ester dihydrochloride (Fluka, Buchs, Switzerland, Cat. No. 11030), l-arginine ethyl ester dihydrochloride (Sigma-Aldrich, Steinheim, Germany, Cat. No. A2883), agmatine sulfate (Sigma-Aldrich, Cat. No. A7127) and l-canavanine sulfate (Sigma-Aldrich, Cat. No. C9758) stock solutions were prepared by adjusting the pH of a 0.144 M solution to pH 7.2.
Quantification of reporter gene expression
Production of the human placental secreted alkaline phosphatase (SEAP) was quantified using a p-nitrophenylphosphate-based light absorbance time course (47,48). Interferon-β quantification was determined using the human Interferon β-specific ELISA (PBL Laboratories, NJ, USA, Cat. No. 41400-1) according to the manufacturer's protocol.
Determination of osmolarity
The osmolarity of ChoMaster® HTS medium supplemented with different l-arginine concentrations was assessed with a Vapro® according to the manufacturer's instructions (Wescor Inc., Logan, UT, USA).
In vivo methods
CHO-K1 cells, engineered for l-arginine-controlled SEAP expression (CHO-ARG2-SEAP), were encapsulated into 400 μm alginate-poly-(l-lysine)-alginate beads (200 cells/capsule) using the Inotech Encapsulator Research IE-50R (Inotech Biotechnologies Ltd, Basel, Switzerland) according to the manufacturer's protocol and the following parameters: 0.2 mm nozzle, 20 ml syringe at a flow rate of 405 units, nozzle vibration frequency 1024 Hz, voltage for bead dispersion 900 V. Seven hundred microliters of l-arginine-free ChoMaster® HTS containing 2 × 106 encapsulated cells (104 capsules/mouse) were injected intraperitoneally into mice (oncins France souche 1, Charles River Laboratories, France). One hour after implantation, l-arginine was administered by injection at doses ranging from 0 to 100 mg/kg. l-arginine was prepared for in vivo administration by diluting the l-arginine stock solution (see above) with an iso-osmotic 0.9% NaCl solution to obtain the appropriate concentrations. Control mice were implanted with capsules containing wild-type CHO-K1 cells. Seventy-two hours after implantation, the mice were sacrificed and their blood collected. For SEAP quantification, serum was isolated using a microtainer SST tube (Beckton Dickinson, Plymouth, UK) according to the manufacturer's protocol. All experiments involving mice were performed according to the European Community Council directive (86/609/EEC), approved by the French Ministry of Agriculture and Fishery (Paris, France) and performed by M.D-E.-B. at the Institut Universitaire de Technologie, IUTA, F-69622 Villeurbanne Cedex, France.
RESULTS
Determination of the l-arginine concentration range compatible with host physiology and potential l-arginine-controlled gene expression
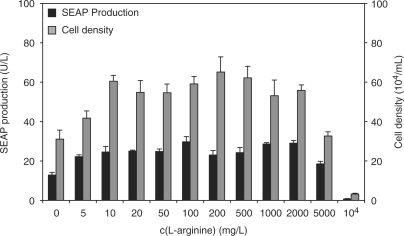
Since l-arginine plays a central role in the urea cycle, the synthesis of nitric oxide and for the synthesis of cytoplasmic and nuclear proteins (49), it is often the rate-limiting amino acid in fast-growing organisms (50,51). In order to prevent l-arginine becoming a physiologic bottleneck or endogenous l-arginine interfering with ART-controlled transgene expression in mammalian cells, we exposed a CHO-K1-derived SEAP-expressing cell line [CHO-ET1-SEAP1, (10)] to l-arginine concentrations ranging from 0 to 10 000 mg/l and scored SEAP levels and cell density after a 60 h cultivation period. A 10 mg/l l-arginine was sufficient to support robust wild-type-like cell growth with SEAP production levels equivalent to control cultivations in standard ChoMaster® HTS medium containing a default l-arginine concentration of 200 mg/l (Figure 1). This observation is quantitatively comparable with physiologic plasma concentrations of l-arginine found in mice and humans [∼17 mg/l, (52)]. Between 10 mg/l and 2 g/l of l-arginine, SEAP production and cell densities were comparable to populations cultivated in standard ChoMaster® HTS medium. At l-arginine concentrations above 2 g/l the culture media became hyperosmotic (>310 mOsm) which compromised cell viability and titer (Figure 1). Based on this data, we selected 10 mg/l and 1 g/l as low and high l-arginine concentrations in follow-up experiments.
Figure 1.
Impact of l-arginine on overall production capacity and cell density. CHO-ET1-SEAP1, constitutively expressing SEAP, were exposed to l-arginine concentrations ranging from 0 to 10 000 mg/l. SEAP production (black bars) and maximum cell densities (gray bars). Standard ChoMaster® HTS medium contains 200 mg/l l-arginine. For l-arginine concentration ranging from 0 to 2 g/l, the media is iso-osmotic (280 – 310 mOsm). Above 2 g/l l-arginine, the media shifts into hypersomotic condition (>310 mOsm).
Design of a l-arginine-responsive mammalian transcription control system
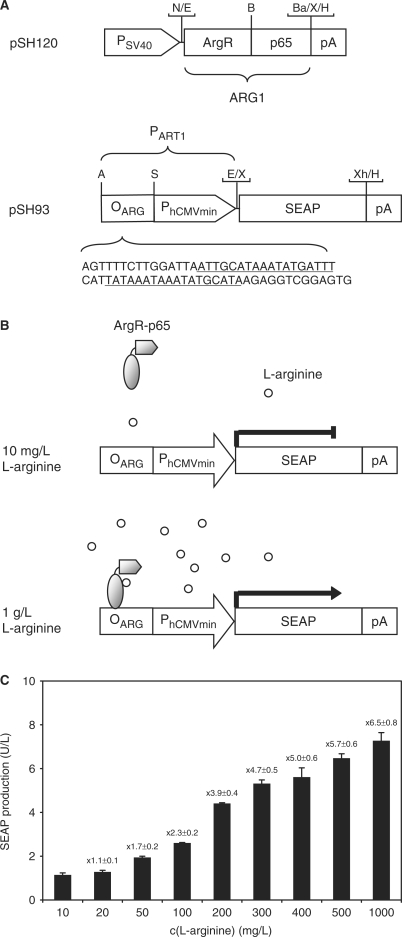
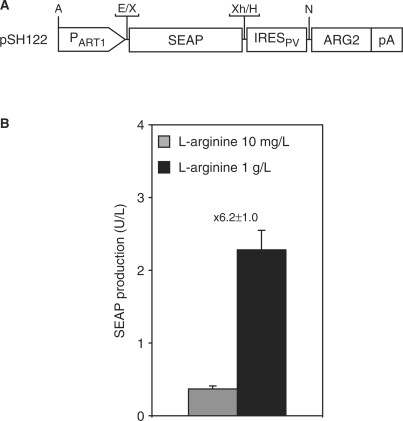
Capitalizing on the ArgR repressor of Chlamydia pneumoniae, which manages arginine metabolism by binding to specific operator sequences (OARG) upstream of the glnPQ operon in a l-arginine-inducible manner, we have designed a l-arginine-regulated transgene control system (ART) which fine-tunes transgene transcription in mammalian cells. ART consists of two components, a l-arginine-regulated transactivator (ARG1) and an ARG1-specific synthetic promoter (PART1): (i) ARG1 was designed by fusing ArgR C-terminally to the human NF-κB transactivation domain (ARG1, ArgR-p65; pSH120, PSV40-ARG1-pA) (8,39). (ii) PART1 was assembled by cloning an ArgR-specific operator module harboring two ARG boxes separated by a single base pair triplet (OARG, AGTTTTCTTGGATTAATTGCATAAATATGATTTCATTATAAATAAATATGCATAAGAGGTCGGAGTG; predicted ARG boxes underlined) 5′ of a minimal version of the human cytomegalovirus immediate early promoter (PhCMVmin; PART1, OARG–PhCMVmin; pSH93, PART1–SEAP-pA; Figure 2A) (15,39). Upon co-transfection of pSH120 (PSV40–ARG1-pA) and pSH93 (PART1–SEAP-pA) into CHO-K1, SEAP expression remained repressed in the presence of 10 mg/l and could be induced by addition of l-arginine up to 1 g/l, suggesting that ARG1 binds and transactivates PART1 in mammalian cells akin to l-arginine-triggered induction of the glnPQ operon by ArgR in Chlamydia pneumoniae (39) (Figure 2B and C).
Figure 2.
Diagram of the ART regulation system. (A) The bacterial repressor ArgR of Chlamydia pneumoniae, fused to the human NF-κB transactivation domain (ARG1, ArgR-p65), is expressed in a constitutive manner under the control of the simian virus 40 promoter (PSV40). The l-arginine-responsive promoter (PART1) harbors an ArgR-specific operator sequence (OARG, capital letters, ARG boxes underlined) upstream of the minimal version of the cytomegalovirus immediate early promoter (PhCMVmin) and drives expression of a reporter gene (e.g. human secreted alkaline phosphatase, SEAP) in a l-arginine-induced manner. All expression units are terminated by a polyadenylation site (pA). Selected restriction sites are indicated: A, AatII; B, BssHII; Ba, BamHI; E, EcoRI; H, HindIII; N, NotI; S, SbfI; X, XbaI, Xh, XhoI. (B) At a low l-arginine concentration (10 mg/l), the ARG1 is in a low-affinity DNA binding state and does not interact with its specific operator sequence (OARG); therefore, expression of the transgene remains silent. At a higher l-arginine concentration (1 g/l), the chimeric transactivator switches to a high-affinity conformation and activates transcription from PhCMVmin upon binding to PART1 through direct ARG1-OARG interaction, thus enabling the transcription of the reporter gene (e.g. SEAP). (C) CHO-K1 were transiently transfected with pSH120 (PSV40-ARG1-pA) and pSH93 (PART1-SEAP-pA) and cultivated for 60 h in medium adjusted to l-arginine concentration ranging from 10 to 1000 mg/l before SEAP production was profiled. The induction factor of SEAP expression is indicated on the top of each bar.
Optimizing the ART system I—different l-arginine-dependent transactivator variants
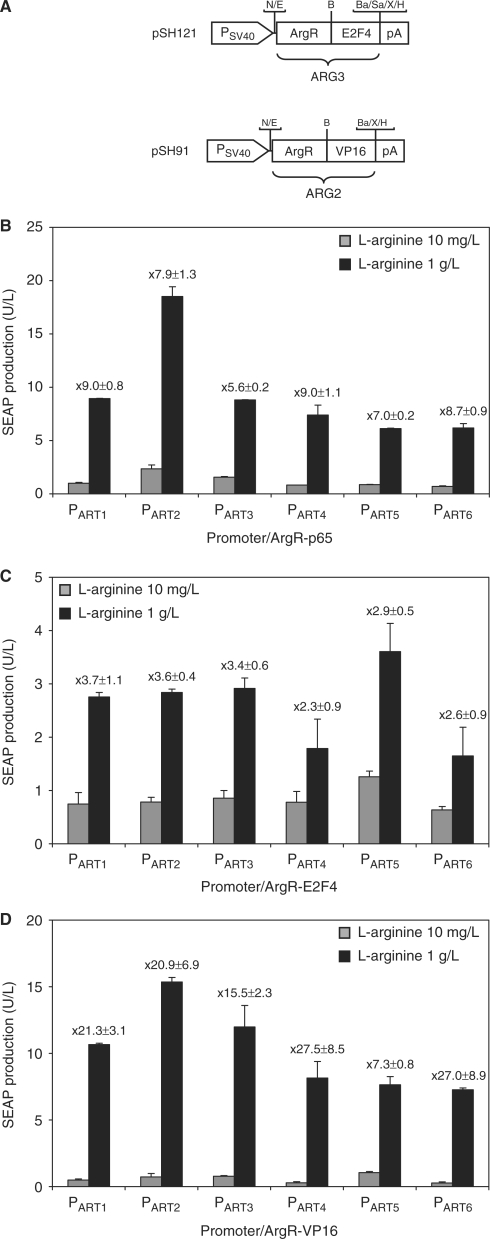
Weber and colleagues (9) have demonstrated that the type of transactivation domain fused to the response regulator may impact overall regulation performance. To characterize potential alternatives to the p65 transactivation domain, we fused the Chlamydia pneumoniae l-arginine-dependent transactivator to the VP16 domain from Herpes simplex (ARG2) (53) and the transactivation domain of the human E2F4 (ARG3) (54) (Figure 3A). For comparative analysis of the regulation performance, ARG1-, ARG2- and ARG3-encoding plasmids were co-transfected with pSH93 (PART1-SEAP-pA) into CHO-K1 and SEAP production was profiled after cultivation for 60 h in medium containing either 10 mg/l or 1 g/l l-arginine (pSH120 and pSH93: 1.12 ± 0.12 U/l (10 mg/l l-arginine), 7.25 ± 0.39 U/l (1 g/l l-arginine); pSH91 and pSH93: 0.50 ± 0.07 U/l (10 mg/l l-arginine), 10.66 ± 0.09 U/l (1 g/l l-arginine); pSH121 and pSH93: 0.75 ± 0.22 U/l (10 mg/l l-arginine), 2.76 ± 0.08 U/l (1 g/l l-arginine). SEAP expression values of the different transactivator–promoter combinations exhibited varied performance characteristics with the basal expression of ARG2 being slightly lower compared to ARG1 and ARG3 exhibiting the poorest performance characterized by lower maximum and higher basal expression levels. Taken together, co-transfection of PART1 with ARG2 enabled the highest induction performance for SEAP expression (Figure 3B–D, SEAP values with PART1).
Figure 3.
Regulation performance of PART1 variants with different l-arginine-dependent transactivators. (A) Schematic representation of the different l-arginine-dependent transactivators. The bacterial repressor ArgR of Chlamydia pneumoniae, fused to human E2F4 (ARG3, ArgR-E2F4) or H. simplex VP16 (ARG2, ArgR-VP16). The expression of both transactivators is driven by the simian virus 40 promoter (PSV40). Selected restriction sites are indicated: B, BssHII; Ba, BamHI; E, EcoRI; H, HindIII; N, NotI; Sa, SalI; X, XbaI. The l-arginine-responsive promoters harboring 0 (PART1), 2 (PART2), 4 (PART3), 6 (PART4), 8 (PART5) and 10 (PART6) bp linkers between the operator sequence and the minimal promoter were co-transfected with l-arginine-dependent transactivators containing the (B) human NF-κB (ARG1, ArgR-p65), (C) human E2F4 (ARG3, ArgR-E2F4) and (D) H. simplex VP16 (ARG2, ArgR-VP16) transactivation domains. Cells were grown at low (10 mg/l) or high (1 g/l) l-arginine concentrations and SEAP production was assessed 60 h after transfection. The regulation factor for each promoter/transactivator combination is specified.
Optimizing the ART system II—promoter variants which differ in the distance between ARG box operator and minimal promoter
The relative spacing of promoter components such as operator and minimal promoter influences the assembly of the transcription machinery and impacts basal as well as maximum transcription levels (55,56). We have, therefore, engineered PART1 variants containing an increasing number of 2 bp insertions between OARG and PhCMVmin which modifies the distance and the torsion angle between these promoter components. While PART1 harbors the default 21 bp between the putative ARG box (OARG) and PhCMVmin, PART2 (OARG-2 bp-PhCMVmin), PART3 (OARG-4 bp-PhCMVmin), PART4 (OARG-6 bp-PhCMVmin), PART5 (OARG-8 bp-PhCMVmin) and PART6 (OARG-10 bp-PhCMVmin) contain additional 2, 4, 6, 8 and 10 bp between OARG and PhCMVmin. All promoters were cloned 5’ of SEAP resulting in a genetic configuration which were isogenic to pSH93 (see Table 1). 60 h after co-transfection of pSH120 (PSV40-ARG1-pA), pSH121 (PSV40-ARG3-pA) or pSH91 (PSV40-ARG2-pA) and either pSH93 or one of its isogenic derivatives into CHO-K1 and cultivation in the presence of 10 mg/l or 1 g/l, SEAP production was quantified. When co-transfected with pSH120 or pSH91, PART2 drove maximum SEAP production while basal expression was minimal for PART6. Comparing PART2 through PART6, maximum SEAP expression levels seem to decrease with increasing distance of OARG and PhCMVmin (Figure 3B and D). For co-transfection of CHO-K1 with pSH121, E2F4 transactivation domain mediated the poorest regulation performance, characterized by higher leakiness (10 mg/l l-arginine) and lower maximum expression levels (1 g/l l-arginine) (Figure 3C). Overall, PART1 showed an optimal balance between basal and maximum expression.
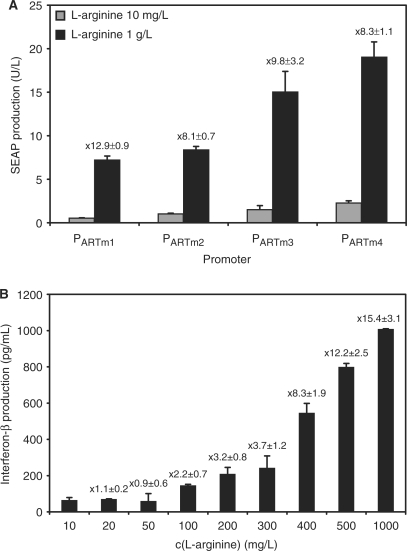
Optimizing the ART system III—promoter variants which differ in the number of ARG box operator modules
Increasing the number of tandem operator modules within a trigger-inducible promoter may increase maximum expression levels as more transactivators can be recruited to the promoter (57). The transcriptional activity of the promoters harboring the operator in a monomeric (pSH117, PARTm1-SEAP-pA; PARTm1, AscI-OARG-MluI-0bp-PhCMVmin), dimeric (pSH119, PARTm2-SEAP-pA; PARTm2, AscI-OARG-7bp-OARG–MluI-0bp-PhCMVmin), trimeric (pSH126, PARTm3-SEAP-pA; PARTm3, AscI-OARG-7bp-OARG-7bp-OARG–MluI-0bp-PhCMVmin) or tetrameric (pSH127, PARTm4-SEAP-pA; PARTm4, AscI-OARG-7bp-OARG-7bp-OARG–7bp-OARG-MluI-0bp-PhCMVmin) configuration were assessed in CHO-K1 cells co-transfected with pSH91 (pSH91, PSV40-ARG2-pA). In general, increasing the number of operator modules resulted in higher maximum expression levels but also higher basal expression (Figure 4A). Having defined the parameters for optimal transgene regulation, the ART system was further validated for expression of the multiple sclerosis therapeutic interferon-β. Interferon-β was cloned downstream of PART1 (pSH113, PART1-INF-β-pA), which was transactivated by ARG2 (pSH91, PSV40-ARG2-pA). CHO-K1 transiently co-transfected with pSH113 and pSH91 enabled adjustable INF-β expression, when exposed to l-arginine concentrations ranging from 10 mg/l to 1g/l (Figure 4B).
Figure 4.
(A) Validation of PART1 variants containing a different number of ARG-specific operator modules. SEAP expression vector encoding l-arginine-responsive promoters harboring monomeric (pSH117, PARTm1-SEAP-pA; PARTm1, AscI-OARG-MluI-0bp-PhCMVmin), dimeric (pSH119, PARTm2-SEAP-pA; PARTm2, AscI-OARG-7bp-OARG–MluI-0bp-PhCMVmin), trimeric (pSH126, PARTm3-SEAP-pA; PARTm3, AscI-OARG-7bp-OARG-7bp-OARG–MluI-0bp-PhCMVmin) or tetrameric (pSH127, PARTm4-SEAP-pA; PARTm4, AscI-OARG-7bp-OARG-7bp-OARG–7bp-OARG-MluI-0bp-PhCMVmin) operator modules were co-transfected with pSH91 (PSV40-ARG2-pA) into CHO-K1 and SEAP production was profiled after 60 h. The induction factor is shown on the top of each bar. (B) Dose–response profile of interferon-β expression in CHO-K1. Cells were transiently co-transfected with pSH113 (pSH113, PART1-INF-β-pA) and pSH91 (PSV40-ARG2-pA) and grown for 48 h at different l-arginine concentrations before quantification of the interferon-β production in the supernatant. Fold induction is shown on the top of each bar.
Validation of the ARG2—PART1 configuration in different cell lines
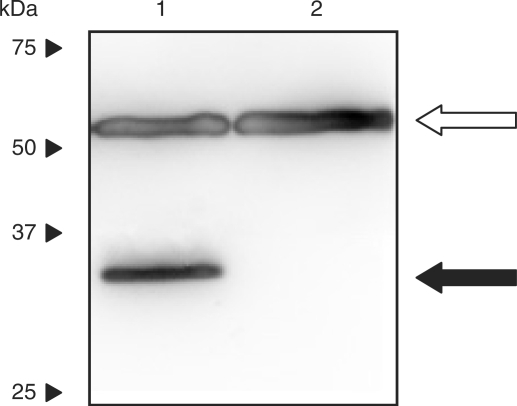
Since the combination of ART2 and PART1 provided the best combination of low leaky and maximum expression levels, we sought to further validate this system. Immortalized cell lines of human, rodent and monkey origin were co-transfected with pSH91 and pSH93. Sixty hours post transfection, cells exposed to 10 mg/l l-arginine displayed low SEAP expression, whereas 1 g/l l-arginine induced high-level production of the reporter gene (Table 2). Moreover, expression integrity of ARG2 in mammalian cells was confirmed by western blot analysis using HEK293-T transiently transfected with pSH91 (PSV40-ARG2-pA) (Figure 5).
Table 2.
Quantitative SEAP expression profiles under the control of PART1 in immortalized cell lines with pSH91, an ARG2 encoding vector and pSH93, a PART1-driven SEAP expression cassette, SEAP expression levels were determined in cell culture media (U/l), 60 h after co-transfection
| Cell line | SEAP activity (U/l) | |
|---|---|---|
| 10 mg/l l-arginine | 1 g/l l-arginine | |
| CHO-K1 | 0.69 ± 0.45 | 8.08 ± 0.80 |
| HEK-293T | 1.67 ± 0.25 | 21.83 ± 3.19 |
| NIH/3T3 | (0.72 ± 0.39) × 10−3 | (25.05 ± 2.00) × 10−3 |
| COS-7 | 0.15 ± 0.07 | 2.47 ± 0.20 |
| HT-1080 | 0.04 ± 0.00 | 0.72 ± 0.06 |
Figure 5.
Western blot analysis of ARG2 expression in HEK-293T cells transfected with pSH91 (pSH91, PSV40-ARG2-pA) and cultivated for 60 h. Lane 1, lysate from HEK-293T transfected with pSH91 (pSH91, PSV40-ARG2-pA); lane 2, lysate from untransfected control cells. The 35 kDa band, indicative for the fusion protein ARG2 is shown with a black arrow. The loading control (tubulin-α, 57kDa) is indicated with an open arrow. Migration of molecular mass markers (kDa) is indicated on the left.
Autoregulated l-arginine-inducible transgene expression in CHO-K1
The ART system was also validated in an autoregulated positive feedback configuration enabling one-step installation of regulated transgene expression in mammalian cells using a single-vector format. This configuration mediates simultaneous expression of transactivator and transgene, both driven by the transactivator-dependent promoter and was found to be instrumental for the design of noise resistant gene networks (58). Leaky transcripts mediated by the PART1 promoter initiate production of relatively few transactivator molecules which trigger full expression of PART1-driven transgenes in the presence of l-arginine. SEAP expression was assessed in CHO-K1 transiently transfected with pSH122 (pSH122, PART1-SEAP-IRESPV-ARG2-pA, Figure 6A). PART1 transcription mediates basal SEAP and ARG2 production which results in ARG2-triggered auto-induction of the dicistronic expression unit in the presence of high l-arginine concentrations (Figure 6B).
Figure 6.
(A) Diagram of the autoregulated l-arginine-inducible SEAP expression vector (pSH122). pSH122 harbors the l-arginine-responsive promoter (PART1) which drives transcription of the dicistronic expression unit encoding the the human placental alkaline phosphatase (SEAP) in the first and the l-arginine-dependent transactivator (ARG2) in the second cistron. Whereas translation of SEAP occurs via a classic cap-dependent mechanism, translation-initation of ARG2 is mediated by an internal ribosome entry site of polioviral origin (IRESPV). pA is the polyadenylation signal. Selected restriction sites are indicated: A, AatII; E, EcoRI; N, NotI; X, XbaI, Xh, XhoI. (B) pSH122 was transiently transfected into CHO-K1, cultivated in media containing 10 mg/l or 1 g/l l-arginine prior to SEAP quantification. Fold induction is indicated on the top of the bars.
Regulation performance of l-arginine derivatives and secondary products
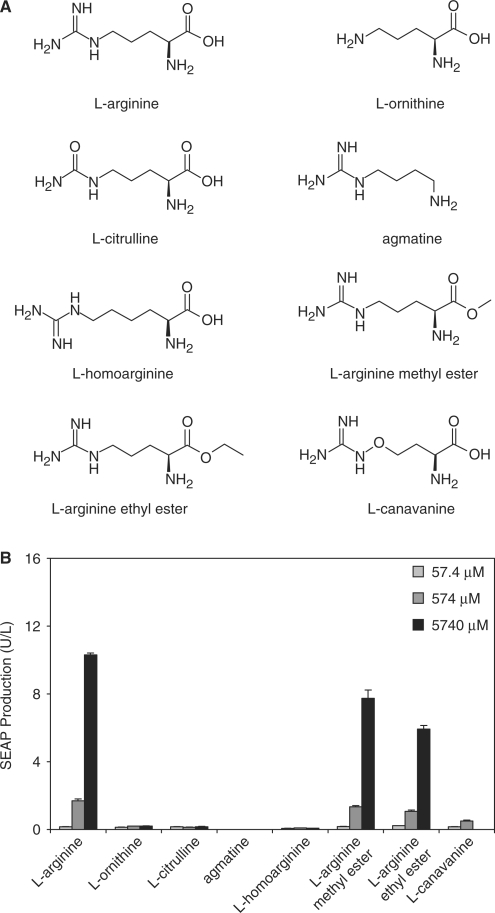
In order to assess the specificity of ART-controlled transgene expression, we cultivated CHO-K1, co-transfected with pSH91 (PSV40-ARG2-pA) and pSH93 (PART1-SEAP-pA), in the presence of increasing concentrations of l-arginine and several of its derivatives (l-canavanine, l-homoarginine, l-arginine methyl ester and l-arginine ethyl ester) and secondary metabolic products (l-ornithine, l-citrulline, agmatine) and profiled SEAP production after 60 h (Figure 7A). Of all these compounds only l-arginine and its methyl and ethyl esters induced SEAP to significant levels while retaining typical dose–response characteristics (Figure 7B).
Figure 7.
Impact of l-arginine and its derivatives on ART-controlled gene expression. (A) The structure of the l-arginine derivatives used in this study. (B) Sixty hours after co-transfection with pSH91 (PSV40-ARG2-pA) and pSH93 (PART1-SEAP-pA), SEAP expression of CHO-K1 was measured. Cells were incubated in cell culture media containing 10 mg/l l-arginine and supplemented with 57.4, 574 or 5740 μM of l-arginine, l-ornithine, l-citrulline, agmatine, l-homoarginine, l-arginine methyl ester, l-arginine ethyl ester or l-canavanine (5740 μM correspond to 1 g/l l-arginine).
Adjustability and reversibility of the ART-controlled transgene expression
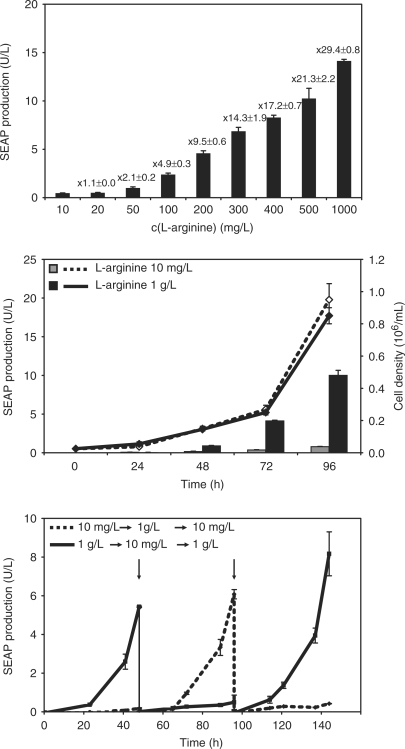
In order to determine the adjustability and reversibility of l-arginine-controlled transgene expression, we generated a stable CHO-K1-derived cell line transgenic for constitutive ARG2 expression and PART1-controlled SEAP production (CHO-ARG2-SEAP). CHO-ARG2-SEAP was cultivated for 60 h in medium containing increasing concentrations of l-argininine before SEAP was profiled in the culture supernatant. As shown in Figure 8A, SEAP expression could be precisely adjusted to specific levels which correlate with particular l-arginine doses. Besides adjustability, rapid response kinetics and reversibility are key assets for a mammalian transgene control system. In order to assess the expression kinetics of the ART system, we profiled both SEAP expression and cell growth for over 96 h. CHO-ARG2-SEAP cells cultivated in medium supplemented with either 10 mg/l or 1 g/l l-arginine displayed similar growth rates, whereby significant SEAP expression was detected only in CHO-ARG2-SEAP cultivated in the presence of 1 g/l l-arginine (Figure 8B). The reversibility was determined by cultivating CHO-ARG2-SEAP for up to 1 week while alternating the inducing (1 g/l) or repressing (10 mg/l) culture condition every other day. The SEAP production profiles revealed reproducible sequential expression kinetics, precise reversibility and tight repression after ON-to-OFF switching (Figure 8C).
Figure 8.
Dose-dependent control of gene expression and reversibility. (A) CHO-ARG2-SEAP cells, transgenic for l-arginine-controlled SEAP expression, were cultured for 60 h at different l-arginine concentrations before SEAP expression profiling. (B) Growth (line) and production (bars) kinetics of ART-controlled SEAP production were scored for CHO-ARG2-SEAP for 96 h. (C) Reversibility of l-arginine-mediated transgene expression following periodic addition and removal of l-arginine. CHO-ARG2-SEAP cells (110 000 cells/ml) were cultivated in 10 mg/l (dotted line) or in 1 g/l (solid line) l-arginine at time 0. Every 48 h (vertical arrows), cells were readjusted to 110 000 cells/ml and grown in fresh media with reversed l-arginine concentrations.
l-arginine-inducible transgene expression in mice
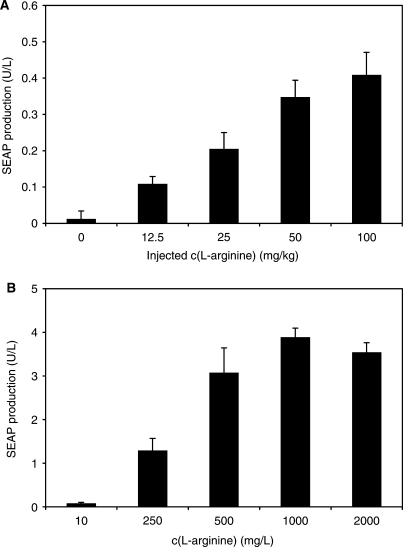
For validation of ART-controlled transgene expression in vivo, we have encapsulated CHO-ARG2-SEAP into coherent alginate-poly-(l-lysine)-alginate microcapsules (200 cells per capsule) and implanted them (104 capsules per mouse) intraperitoneally into mice. Implanted mice were given a daily dose of l-arginine ranging from 0 to 100 mg/kg for 72 h before quantifying serum SEAP levels. The SEAP levels reached in the serum of treated mice was adjustable and dependent on the injected l-arginine dose. The serum SEAP levels of control mice implanted with microencapsulated CHO-K1 (data not shown) or mice implanted with microencapsulated CHO-ARG2-SEAP but not receiving exogenous l-arginine showed insignificant SEAP expression level (Figure 9A). In parallel, the experiment was also performed in vitro with the same batch of encapsulated cells, which confirmed the dose–response characteristics of microencapsulated CHO-ARG2-SEAP (Figure 9B).
Figure 9.
ART-inducible SEAP expression in mice. (A) CHO-ARG2-SEAP cells were microencapsulated in alginate-poly-(l-lysine)-alginate beads and implanted intraperitoneally into female OF1 mice (2 × 106 cells per mouse). Implanted mice were exposed daily to different arginine concentrations. 72 h post-implantation, the level of SEAP in the serum of mice serum was determined. (B) SEAP expression of microencapsulated CHO-ARG2-SEAP cells cultured in vitro. CHO-ARG2-SEAP microencapsulated cells, originating from the same batch as for the transplantation in mice, were cultured for 72 h at the indicated l-arginine concentration before quantifying SEAP expression.
DISCUSSION
Currently available transgene control systems have been designed for optimal regulation performance in vitro or in a mouse model (59). To enable interference-reduced operation within a complex biochemical reaction network of the host cell, heterologous or modified endogenous transcription factors have been engineered to modulate transcription of specific target promoters (60,61). Initially, inducer molecules only required reasonable pharmacokinetics and needed to be non-toxic at regulation-effective concentrations, criteria which was best met by clinically licensed small-molecule drugs (16,18,62). However, the use of clinically licensed substances to control biopharmaceutical manufacturing of difficult-to-produce protein pharmaceuticals fails to comply with administrative regulations or requires prohibitive downstream processing standards (33,63,64). Also, long-term administration of drugs at (sub-)clinical doses has been associated in many cases with side-effects which limits their use in future gene therapy trials (26,28–30). In contrast, physiologic molecules such as amino acids are FDA-licensed media components and an integral part of host metabolism. Due to possible interference effects, such physiologic trigger molecules have so far not been considered for transgene control.
As an obligate intracellular pathogen, Chlamydia pneumoniae has evolved to optimally plug in its metabolism into the biochemical networks of host cells (38). As a major part of this metabolic crosstalk, the biosensor ArgR quantifies l-arginine levels and manages import of this amino acid (40). It is, therefore, reasonable to assume that the sensitivity of ArgR is attuned to physiologic l-arginine levels of mammalian cells. Indeed, ART-controlled expression of reporter and product genes shows excellent regulation performance (rapid induction kinetics, full adjustability, reproducible reversibility) in different cell lines as well as in mice with an arginine concentration window between 10 mg/l (required to maintain metabolism, transgene repressed) and 1 g/l (full induction). The l-arginine level in the plasma of mice is around 17 mg/l, which is well below the threshold concentration required to fully activate the ART system (52). By default, ART is an ON-type system, which means that it is only induced in the presence of increased l-arginine levels and remains repressed at physiologic concentrations of this amino acid.
ART is exclusively regulated by l-arginine and its ester forms which are rapidly converted to l-arginine by endogenous esterases (65). Secondary products of the l-arginine metabolism or the synthetic l-homoarginine failed to regulate the ART system. Even l-canavanine, which is stereochemically similar to l-arginine and was shown to fit in the amino acid binding pocket of ArgR was unable to modulate ART-controlled transgene expression (66).
Since l-arginine is a licensed component of cell culture media, the ART system will be a extremely valuable tool for the biomanufacturing of therapeutic proteins. Moreover, l-arginine is considered to be compatible with prolonged therapeutic use. In fact, l-arginine is currently therapeutically administered for pulmonary hypertension of newborns (67). It also improves endothelial function in animal models of hypercholesterolaemia and atherosclerosis with no life-threatening side-effects reported so far (68).
As a prototype transgene control system responsive to a physiologic trigger molecule, the ART system has shown that metabolic and synthetic control circuits can functionally co-exist while sharing the same inducer. Although such systems may be of immediate use in gene-function analysis, gene network design as well as for biopharmaceutical manufacturing, the ART may develop its highest impact in future therapies: synchronization of therapeutic gene expression with fluctuations of endogenous molecules or the self-sufficient treatment of a pathological situation by exploiting signal molecules for a self-controlling genetic intervention are highly appealing strategies for future gene therapy applications.
ACKNOWLEDGEMENTS
We thank David Greber for critical comments on the manuscript. This work was supported by the Swiss National Science foundation (grant no. 3100A0-112549), The EC Framework 7 (COBIOS) and Cistronics Cell Technology GmbH, Einsteinstrasse 1-5, P.O.B 145, CH-8093 Zurich, Switzerland. Funding to pay the Open Access publication charges for this article was provided by ETH Zurich.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kawaguchi N, Xu X, Tajima R, Kronqvist P, Sundberg C, Loechel F, Albrechtsen R, Wewer UM. ADAM 12 protease induces adipogenesis in transgenic mice. Am. J. Pathol. 2002;160:1895–1903. doi: 10.1016/S0002-9440(10)61136-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paszty C, Brion CM, Manci E, Witkowska HE, Stevens ME, Mohandas N, Rubin EM. Transgenic knockout mice with exclusively human sickle hemoglobin and sickle cell disease. Science. 1997;278:876–878. doi: 10.1126/science.278.5339.876. [DOI] [PubMed] [Google Scholar]
- 3.Aubel D, Morris R, Lennon B, Rimann M, Kaufmann H, Folcher M, Bailey JE, Thompson CJ, Fussenegger M. Design of a novel mammalian screening system for the detection of bioavailable, non-cytotoxic streptogramin antibiotics. J. Antibiot. (Tokyo) 2001;54:44–55. doi: 10.7164/antibiotics.54.44. [DOI] [PubMed] [Google Scholar]
- 4.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat. Rev. Drug Discov. 2006;5:741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 5.Gersbach CA, Le Doux JM, Guldberg RE, Garcia AJ. Inducible regulation of Runx2-stimulated osteogenesis. Gene Ther. 2006;13:873–882. doi: 10.1038/sj.gt.3302725. [DOI] [PubMed] [Google Scholar]
- 6.Ulmer JB, Valley U, Rappuoli R. Vaccine manufacturing: challenges and solutions. Nat. Biotechnol. 2006;24:1377–1383. doi: 10.1038/nbt1261. [DOI] [PubMed] [Google Scholar]
- 7.Umana P, Jean-Mairet J, Bailey JE. Tetracycline-regulated overexpression of glycosyltransferases in Chinese hamster ovary cells. Biotechnol. Bioeng. 1999;65:542–549. [PubMed] [Google Scholar]
- 8.Urlinger S, Helbl V, Guthmann J, Pook E, Grimm S, Hillen W. The p65 domain from NF-kappaB is an efficient human activator in the tetracycline-regulatable gene expression system. Gene. 2000;247:103–110. doi: 10.1016/s0378-1119(00)00112-8. [DOI] [PubMed] [Google Scholar]
- 9.Weber W, Kramer BP, Fux C, Keller B, Fussenegger M. Novel promoter/transactivator configurations for macrolide- and streptogramin-responsive transgene expression in mammalian cells. J. Gene Med. 2002;4:676–686. doi: 10.1002/jgm.314. [DOI] [PubMed] [Google Scholar]
- 10.Weber W, Fux C, Daoud-el Baba M, Keller B, Weber CC, Kramer BP, Heinzen C, Aubel D, Bailey JE, Fussenegger M. Macrolide-based transgene control in mammalian cells and mice. Nat. Biotechnol. 2002;20:901–907. doi: 10.1038/nbt731. [DOI] [PubMed] [Google Scholar]
- 11.Weber W, Rimann M, Spielmann M, Keller B, Daoud-El Baba M, Aubel D, Weber CC, Fussenegger M. Gas-inducible transgene expression in mammalian cells and mice. Nat. Biotechnol. 2004;22:1440–1444. doi: 10.1038/nbt1021. [DOI] [PubMed] [Google Scholar]
- 12.Neddermann P, Gargioli C, Muraglia E, Sambucini S, Bonelli F, De Francesco R, Cortese R. A novel, inducible, eukaryotic gene expression system based on the quorum-sensing transcription factor TraR. EMBO Rep. 2003;4:159–165. doi: 10.1038/sj.embor.embor734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gossen M, Bujard H. Efficacy of tetracycline-controlled gene expression is influenced by cell type: commentary. Biotechniques. 1995;19:213–216. discussion 216–217. [PubMed] [Google Scholar]
- 14.Fussenegger M, Morris RP, Fux C, Rimann M, von Stockar B, Thompson CJ, Bailey JE. Streptogramin-based gene regulation systems for mammalian cells. Nat. Biotechnol. 2000;18:1203–1208. doi: 10.1038/81208. [DOI] [PubMed] [Google Scholar]
- 15.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gossen M, Bonin AL, Freundlieb S, Bujard H. Inducible gene expression systems for higher eukaryotic cells. Curr. Opin. Biotechnol. 1994;5:516–520. doi: 10.1016/0958-1669(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 17.Roscilli G, Rinaudo CD, Cimino M, Sporeno E, Lamartina S, Ciliberto G, Toniatti C. Long-term and tight control of gene expression in mouse skeletal muscle by a new hybrid human transcription factor. Mol. Ther. 2002;6:653–663. [PubMed] [Google Scholar]
- 18.Nordstrom JL. Antiprogestin-controllable transgene regulation in vivo. Curr. Opin. Biotechnol. 2002;13:453–458. doi: 10.1016/s0958-1669(02)00356-7. [DOI] [PubMed] [Google Scholar]
- 19.Palli SR, Kapitskaya MZ, Potter DW. The influence of heterodimer partner ultraspiracle/retinoid X receptor on the function of ecdysone receptor. FEBS J. 2005;272:5979–5990. doi: 10.1111/j.1742-4658.2005.05003.x. [DOI] [PubMed] [Google Scholar]
- 20.Weber W, Link N, Fussenegger M. A genetic redox sensor for mammalian cells. Metab. Eng. 2006;8:273–280. doi: 10.1016/j.ymben.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Weber W, Schoenmakers R, Spielmann M, El-Baba MD, Folcher M, Keller B, Weber CC, Link N, van de Wetering P, et al. Streptomyces-derived quorum-sensing systems engineered for adjustable transgene expression in mammalian cells and mice. Nucleic Acids Res. 2003;31:e71. doi: 10.1093/nar/gng071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber W, Marty RR, Link N, Ehrbar M, Keller B, Weber CC, Zisch AH, Heinzen C, Djonov V, et al. Conditional human VEGF-mediated vascularization in chicken embryos using a novel temperature-inducible gene regulation (TIGR) system. Nucleic Acids Res. 2003;31:e69. doi: 10.1093/nar/gng069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chong H, Ruchatz A, Clackson T, Rivera VM, Vile RG. A system for small-molecule control of conditionally replication-competent adenoviral vectors. Mol. Ther. 2002;5:195–203. doi: 10.1006/mthe.2002.0531. [DOI] [PubMed] [Google Scholar]
- 24.Pollock R, Clackson T. Dimerizer-regulated gene expression. Curr. Opin. Biotechnol. 2002;13:459–467. doi: 10.1016/s0958-1669(02)00373-7. [DOI] [PubMed] [Google Scholar]
- 25.Weber W, Stelling J, Rimann M, Keller B, Daoud-El Baba M, Weber CC, Aubel D, Fussenegger M. A synthetic time-delay circuit in mammalian cells and mice. Proc. Natl Acad. Sci. USA. 2007;104:2643–2648. doi: 10.1073/pnas.0606398104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkar NN. Mifepristone: bioavailability, pharmacokinetics and use-effectiveness. Eur. J. Obstet. Gynecol. Reprod. Biol. 2002;101:113–120. doi: 10.1016/s0301-2115(01)00522-x. [DOI] [PubMed] [Google Scholar]
- 27.Russo IH, Russo J. Role of hormones in mammary cancer initiation and progression. J. Mammary Gland Biol. Neoplasia. 1998;3:49–61. doi: 10.1023/a:1018770218022. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez AR, Rogers R.S., 3rd, Sheridan PJ. Tetracycline and other tetracycline-derivative staining of the teeth and oral cavity. Int. J. Dermatol. 2004;43:709–715. doi: 10.1111/j.1365-4632.2004.02108.x. [DOI] [PubMed] [Google Scholar]
- 29.Lautermann J, Dehne N, Schacht J, Jahnke K. [Aminoglycoside- and cisplatin-ototoxicity: from basic science to clinics] Laryngorhinootologie. 2004;83:317–323. doi: 10.1055/s-2004-814280. [DOI] [PubMed] [Google Scholar]
- 30.Alanis AJ. Resistance to antibiotics: are we in the post-antibiotic era? Arch. Med. Res. 2005;36:697–705. doi: 10.1016/j.arcmed.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 31.Kahan BD. Sirolimus: a comprehensive review. Expert Opin. Pharmacother. 2001;2:1903–1917. doi: 10.1517/14656566.2.11.1903. [DOI] [PubMed] [Google Scholar]
- 32.Kuypers DR. Benefit-risk assessment of sirolimus in renal transplantation. Drug Saf. 2005;28:153–181. doi: 10.2165/00002018-200528020-00006. [DOI] [PubMed] [Google Scholar]
- 33.Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat. Biotechnol. 2004;22:1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- 34.Heiskanen-Kosma T, Korppi M, Laurila A, Jokinen C, Kleemola M, Saikku P. Chlamydia pneumoniae is an important cause of community-acquired pneumonia in school-aged children: serological results of a prospective, population-based study. Scand. J. Infect. Dis. 1999;31:255–259. doi: 10.1080/00365549950163536. [DOI] [PubMed] [Google Scholar]
- 35.Heath PT. Epidemiology and bacteriology of bacterial pneumonias. Paediatr. Respir. Rev. 2000;1:4–7. doi: 10.1053/prrv.2000.0001. [DOI] [PubMed] [Google Scholar]
- 36.Mussa FF, Chai H, Wang X, Yao Q, Lumsden AB, Chen C. Chlamydia pneumoniae and vascular disease: an update. J. Vasc. Surg. 2006;43:1301–1307. doi: 10.1016/j.jvs.2006.02.050. [DOI] [PubMed] [Google Scholar]
- 37.Krull M, Maass M, Suttorp N, Rupp J. Chlamydophila pneumoniae. Mechanisms of target cell infection and activation. Thromb. Haemost. 2005;94:319–326. doi: 10.1160/TH05-04-0261. [DOI] [PubMed] [Google Scholar]
- 38.Bonanomi A, Dohm C, Rickenbach Z, Altwegg M, Fischer J, Gygi D, Nadal D. Monitoring intracellular replication of Chlamydophila (Chlamydia) pneumoniae in cell cultures and comparing clinical samples by real-time PCR. Diagn Microbiol. Infect. Dis. 2003;46:39–47. doi: 10.1016/s0732-8893(02)00572-2. [DOI] [PubMed] [Google Scholar]
- 39.Schaumburg CS, Tan M. Arginine-dependent gene regulation via the ArgR repressor is species specific in chlamydia. J. Bacteriol. 2006;188:919–927. doi: 10.1128/JB.188.3.919-927.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makarova KS, Mironov AA, Gelfand MS. Conservation of the binding site for the arginine repressor in all bacterial lineages. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-4-research0013. research0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu CD, Abdelal AT. Role of ArgR in activation of the ast operon, encoding enzymes of the arginine succinyltransferase pathway in Salmonella typhimurium. J. Bacteriol. 1999;181:1934–1938. doi: 10.1128/jb.181.6.1934-1938.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu CD, Yang Z, Li W. Transcriptome analysis of the ArgR regulon in Pseudomonas aeruginosa. J Bacteriol. 2004;186:3855–3861. doi: 10.1128/JB.186.12.3855-3861.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maas WK. The arginine repressor of Escherichia coli. Microbiol. Rev. 1994;58:631–640. doi: 10.1128/mr.58.4.631-640.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin L, Xue WF, Fukayama JW, Yetter J, Pickering M, Carey J. Asymmetric allosteric activation of the symmetric ArgR hexamer. J. Mol. Biol. 2005;346:43–56. doi: 10.1016/j.jmb.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 45.Ni J, Sakanyan V, Charlier D, Glansdorff N, Van Duyne GD. Structure of the arginine repressor from Bacillus stearothermophilus. Nat. Struct. Biol. 1999;6:427–432. doi: 10.1038/8229. [DOI] [PubMed] [Google Scholar]
- 46.Mitta B, Rimann M, Ehrengruber MU, Ehrbar M, Djonov V, Kelm J, Fussenegger M. Advanced modular self-inactivating lentiviral expression vectors for multigene interventions in mammalian cells and in vivo transduction. Nucleic Acids Res. 2002;30:e113. doi: 10.1093/nar/gnf112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berger J, Hauber J, Hauber R, Geiger R, Cullen BR. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- 48.Schlatter S, Rimann M, Kelm J, Fussenegger M. SAMY, a novel mammalian reporter gene derived from Bacillus stearothermophilus alpha-amylase. Gene. 2002;282:19–31. doi: 10.1016/s0378-1119(01)00824-1. [DOI] [PubMed] [Google Scholar]
- 49.Scott L, Lamb J, Smith S, Wheatley DN. Single amino acid (arginine) deprivation: rapid and selective death of cultured transformed and malignant cells. Br J. Cancer. 2000;83:800–810. doi: 10.1054/bjoc.2000.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wheatley DN. Arginine deprivation and metabolomics: important aspects of intermediary metabolism in relation to the differential sensitivity of normal and tumour cells. Semin. Cancer Biol. 2005;15:247–253. doi: 10.1016/j.semcancer.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 51.Wheatley DN, Scott L, Lamb J, Smith S. Single amino acid (arginine) restriction: growth and death of cultured HeLa and human diploid fibroblasts. Cell. Physiol. Biochem. 2000;10:37–55. doi: 10.1159/000016333. [DOI] [PubMed] [Google Scholar]
- 52.Zimmermann N, Rothenberg ME. The arginine-arginase balance in asthma and lung inflammation. Eur. J. Pharmacol. 2006;533:253–262. doi: 10.1016/j.ejphar.2005.12.047. [DOI] [PubMed] [Google Scholar]
- 53.Triezenberg SJ, Kingsbury RC, McKnight SL. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 54.Akagi K, Kanai M, Saya H, Kozu T, Berns A. A novel tetracycline-dependent transactivator with E2F4 transcriptional activation domain. Nucleic Acids Res. 2001;29:E23. doi: 10.1093/nar/29.4.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muller J, Oehler S, Muller-Hill B. Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J. Mol. Biol. 1996;257:21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- 56.Sathya G, Li W, Klinge CM, Anolik JH, Hilf R, Bambara RA. Effects of multiple estrogen responsive elements, their spacing, and location on estrogen response of reporter genes. Mol. Endocrinol. 1997;11:1994–2003. doi: 10.1210/mend.11.13.0039. [DOI] [PubMed] [Google Scholar]
- 57.Malphettes L, Weber CC, El-Baba MD, Schoenmakers RG, Aubel D, Weber W, Fussenegger M. A novel mammalian expression system derived from components coordinating nicotine degradation in arthrobacter nicotinovorans pAO1. Nucleic Acids Res. 2005;33:e107. doi: 10.1093/nar/gni107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Becskei A, Kaufmann BB, van Oudenaarden A. Contributions of low molecule number and chromosomal positioning to stochastic gene expression. Nat. Genet. 2005;37:937–944. doi: 10.1038/ng1616. [DOI] [PubMed] [Google Scholar]
- 59.Weber W, Fussenegger M. Pharmacologic transgene control systems for gene therapy. J. Gene Med. 2006;8:535–556. doi: 10.1002/jgm.903. [DOI] [PubMed] [Google Scholar]
- 60.Weber W, Fussenegger M. Novel gene switches. Handb. Exp. Pharmacol. 2007;178:73–105. doi: 10.1007/978-3-540-35109-2_4. [DOI] [PubMed] [Google Scholar]
- 61.Kramer BP, Fischer M, Fussenegger M. Semi-synthetic mammalian gene regulatory networks. Metab. Eng. 2005;7:241–250. doi: 10.1016/j.ymben.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 62.Tascou S, Sorensen TK, Glenat V, Wang M, Lakich MM, Darteil R, Vigne E, Thuillier V. Stringent rosiglitazone-dependent gene switch in muscle cells without effect on myogenic differentiation. Mol. Ther. 2004;9:637–649. doi: 10.1016/j.ymthe.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 63.Boehm R. Bioproduction of therapeutic proteins in the 21st century and the role of plants and plant cells as production platforms. Ann. N. Y. Acad. Sci. 2007;1102:121–134. doi: 10.1196/annals.1408.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daniell H, Streatfield SJ, Wycoff K. Medical molecular farming: production of antibodies, biopharmaceuticals and edible vaccines in plants. Trends Plant Sci. 2001;6:219–226. doi: 10.1016/S1360-1385(01)01922-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Markert CL, Hunter RL. The distribution of esterases in mouse tissues. J. Histochem. Cytochem. 1959;7:42–49. doi: 10.1177/7.1.42. [DOI] [PubMed] [Google Scholar]
- 66.Van Duyne GD, Ghosh G, Maas WK, Sigler PB. Structure of the oligomerization and L-arginine binding domain of the arginine repressor of Escherichia coli. J. Mol. Biol. 1996;256:377–391. doi: 10.1006/jmbi.1996.0093. [DOI] [PubMed] [Google Scholar]
- 67.Morris CR, Morris S.M., Jr, Hagar W, Van Warmerdam J, Claster S, Kepka-Lenhart D, Machado L, Kuypers FA, Vichinsky EP. Arginine therapy: a new treatment for pulmonary hypertension in sickle cell disease? Am. J. Respir. Crit. Care Med. 2003;168:63–69. doi: 10.1164/rccm.200208-967OC. [DOI] [PubMed] [Google Scholar]
- 68.Boger RH, Bode-Boger SM. The clinical pharmacology of L-arginine. Annu. Rev. Pharmacol. Toxicol. 2001;41:79–99. doi: 10.1146/annurev.pharmtox.41.1.79. [DOI] [PubMed] [Google Scholar]
- 69.Fussenegger M, Moser S, Mazur X, Bailey JE. Autoregulated multicistronic expression vectors provide one-step cloning of regulated product gene expression in mammalian cells. Biotechnol. Prog. 1997;13:733–740. doi: 10.1021/bp970108r. [DOI] [PubMed] [Google Scholar]