Abstract
Eukaryotic DNA replication initiates at origins of replication by the assembly of the highly conserved pre-replicative complex (pre-RC). However, exact sequences for pre-RC binding still remain unknown. By chromatin immunoprecipitation we identified in vivo a pre-RC-binding site within the origin of bidirectional replication in the murine rDNA locus. At this sequence, ORC1, -2, -4 and -5 are bound in G1 phase and at the G1/S transition. During S phase, ORC1 is released. An ATP-dependent and site-specific assembly of the pre-RC at origin DNA was demonstrated in vitro using partially purified murine pre-RC proteins in electrophoretic mobility shift assays. By deletion experiments the sequence required for pre-RC binding was confined to 119 bp. Nucleotide substitutions revealed that two 9 bp sequence elements, CTCGGGAGA, are essential for the binding of pre-RC proteins to origin DNA within the murine rDNA locus. During myogenic differentiation of C2C12 cells, we demonstrated a reduction of ORC1 and ORC2 by immunoblot analyses. ChIP analyses revealed that ORC1 completely disappears from chromatin of terminally differentiated myotubes, whereas ORC2, -4 and -5 still remain associated.
INTRODUCTION
The initiation of DNA replication in eukaryotic cells is strictly coordinated within the cell cycle and starts from discrete sites defined as origins of replication. In the budding yeast Saccharomyces cerevisiae the hexameric origin recognition complex (ORC) binds to the origin in an ATP-dependent manner and serves as a recruitment factor for sequential loading of CDC6, CDT1 and minichromosome maintenance (MCM) proteins 2–7 to form the pre-replicative complex (pre-RC) in the early G1 phase of the cell cycle. Transition to S phase is initiated by activation of specific protein kinases and is marked by the conversion of pre-RCs to initiation complexes (1,2). Although most of what we know about initiation of DNA replication derives from studies using S. cerevisiae, the basic composition of core initiation proteins has been conserved among all eukaryotic organisms examined thus far (1,3,4).
In yeast, the ORC remains bound to origin DNA throughout the cell cycle. In higher eukaryotes, however, the association of the ORC with chromatin seems to be cell cycle regulated. In human cells, ORC1 dissociates from chromatin in S phase, whereas ORC2 remains bound (5). Likewise, hamster ORC1 is released from chromatin as cells enter S phase and is re-bound to chromatin during M to G1 transition (6,7). CDC6 is bound to chromatin in G1 phase in yeast and mammalian cells. As cells enter S phase, CDC6 is ubiquitinated and degraded in yeast (8–10) whereas mammalian CDC6 is phosphorylated, translocated to the cytoplasm and degraded in mitosis in an ubiquitin-dependent manner (11,12). The activity of CDT1 is essential for CDC6-dependent recruitment of the MCM proteins (13,14). Hence, CDT1 accumulates during G1 phase and is inactivated after onset of S phase to prevent re-replication (15–17). Mammalian MCM proteins associate with chromatin during G1 phase (11,18–20), after the G1/S transition human MCM proteins are progressively released (11).
During vertebrate ontogenesis, cells can exit the cell cycle early in G1 phase and enter a quiescent (G0 phase) or terminally differentiated state (21). It has been proposed that the reduction of proteins involved in DNA replication might contribute to this process (22). For instance, MCM2-7 are depleted both in G0 phase and permanently arrested cells. CDC6 also seems to be reduced in these cells. However, levels of ORC proteins appear to remain high during progression into quiescence (2).
In contrast to the conserved mechanism of initiation, the process that determines origin selection is highly divergent. In lower eukaryotes, origins can be defined by rather short consensus elements (23). Metazoan origins, however, are less specified and can probably extend over thousands of base pairs. So far about two-dozen metazoan origins have been described, but they do not share any consensus sequences (24–26). Even so, sequence-specific and ATP-dependent binding of Drosophila melanogaster ORC has been observed at a control element in the chorion amplification loci (27). Three described human origins, including the lamin B2 origin, the origin in the PRKDC/MCM4 intergenic region, and that in the region upstream of the human TOP1 gene have been shown to interact with the ORC in vivo (5,28,29). However, it has also been shown that ORC exhibits sequence-independent DNA binding. For instance, purified human ORC binds preferentially to synthetic AT-rich polynucleotides, but does not discriminate between natural DNA fragments with an origin and control fragments (30). Another work demonstrated that mammalian ORC binds to an extrachromosomally replicating plasmid in a sequence-independent manner (31). Even if it is commonly accepted that DNA replication initiates at specific sites, it is not clear whether specific consensus sequence elements are used to specify origins or whether higher order regulation, like chromatin organization, is more important.
As an experimental system to explore the initiation of DNA replication we used murine rRNA genes. Approximately 400 tandemly repeated rDNA units, each 44 kb in size, are clustered in the nucleolar organizing regions of mouse cells. In the murine rRNA genes, one origin of bidirectional replication (OBR) was mapped by nascent strand length determination within a 3 kb region centered ∼1.6 kb upstream of the rDNA transcription start site of RNA polymerase I (32).
In this report, we identified by chromatin immunoprecipitation (ChIP) assays in vivo a pre-RC-binding site in the OBR of the murine rDNA cistron. At this site, ORC subunits 2, 4 and 5 are bound throughout the cell cycle whereas ORC1 assembles in a cell cycle-dependent manner. By electrophoretic mobility shift assays (EMSA) with partially purified nuclear extracts this binding site was confirmed and confined to a 119 bp DNA sequence. A protein complex containing ORC2 showed site-specific and ATP-stimulated DNA binding. Nucleotide substitutions in two 9 bp sequence elements within the 119 bp pre-RC assembly site reduced the formation of this DNA/protein complex significantly. We further demonstrated by immunoblot analyses the loss of specific pre-RC proteins during myogenic differentiation of C2C12 cells. ChIP analyses revealed that ORC1 is lost from the pre-RC-binding site during terminal differentiation, but that a core complex of the pre-RC consisting of ORC2, -4, -5, still occupies the pre-RC-binding site in terminally differentiated myotubes.
MATERIALS AND METHODS
Cell culture and synchronization
Murine FM3A cells were maintained at a density of 2 × 105 cells/ml in 1 × RPMI-1640 medium (PAA, Austria), supplemented with 10% fetal calf serum at 37°C under 5% CO2. Cells were synchronized in G1 phase by adding mevastatin (10 μM, final), at the G1/S transition by adding mimosine (0.5 mM, final) and in S phase by adding hydroxyurea (1 mM, final) for at least 30 h. The synchronization in cell cycle was checked by propidium iodide staining of the cells followed by flow cytometry using FACScan analysis. Analysis was carried out on a BD LSR System (Becton-Dickinson) and cell cycle phases were estimated using the MultiPlus software (Phoenix Flow Systems). Data are presented as histograms showing relative DNA content (x-axis) and cell number (y-axis).
Mouse C2C12 myoblasts were maintained at subconfluent densities in Dulbecco's modified Eagle's medium (PAA, Austria) supplemented with 10% fetal calf serum (growth medium, GM) at 37°C under 10% CO2. To prepare differentiated multinucleated myotubes, myoblasts were shifted to DMEM containing 2% horse serum (differentiation medium, DM) when the cells reached 70–80% confluence. For the first 12 h, the differentiation medium included 10 µg/ml insulin (Sigma), afterwards 10 µM cytosine β-d-arabinofuranoside (Ara-C, Sigma) was added to eliminate still replicating non-differentiated cells. The media were changed every day.
Antibodies
Antibodies used in this work were generated against individual initiator proteins by immunization of rabbit (ORC2, ORC4, ORC5, ORC6) and sheep (ORC1) with recombinant proteins. Polyclonal antisera were affinity purified on antigen columns. The specificity of antibodies has been described earlier (33). As a negative control in ChIP experiments corresponding to rabbit and sheep preimmune sera, respectively, were used.
PCR primer pairs
Fragment A: 5′-GAA AGC AAA TCA CTA TGA AGA G-3′ (2798f), 5′-GCA CAG TTA GGC ACA GTT AGG-3′ (3136r); fragment B: 5′-CCT AAC TGT GCC TAA CTG TGC-3′ (3116f), 5′-CGT TCC CGA AAC TTG TCA CC-3′ (3483r); fragment C: 5′-AAT GAG TGA GTG AAT GTG GCG-3′ (3380f), 5′-GTC CTC TCG GCC TCA GAT GTA-3′ (3815r); fragment D: 5′-CAC GTG TCT CGT TTC GTT CC-3′ (12361f), 5′-ACA ACC GCC CAC ACG TCT G-3′ (12790r); 5.Δ100: 5′-TAC TTT CGT TTT TGG GTG CC-3′ (3218f); 5.Δ200: 5′-GTA CGC TGC TCC GTC GTG-3′ (3313f); 3.Δ100: 5′-CTC ACT CAT TCT GTC TCT CCG-3′ (3389r); 3.Δ200: 5′-ACA AAC GGT CCC CAT CGT-3′ (3287r); 119 bp: 5′-ACG ATG GGG ACC GTT TGT-3′ (3270f), 5′-TCA CTC ATT CTG TCT CTC CGG-3′ (3388r). Numbers in parenthesis indicate the location of oligonucleotides within the Mus musculus 45S pre rRNA gene (accession number: X82564).
In vivo cross-linking and nuclear extract preparation
DNA/protein cross-linking was performed by incubating FM3A (2 × 107 per condition) with 1% (v/v) formaldehyde for 10 min at RT under gentle agitation. The reaction was quenched by adding glycine to a final concentration of 0.125 M. Cells were washed in PBS including protease inhibitors and lysed by incubation in cell lysis buffer (10 mM Tris, pH 8.0, 10 mM NaCl, 0.2% NP-40, 0.5 mM PMSF, 1× PI-Mix: 2 µg/ml pepstatin, 2 µg/ml leupeptin, 5 µg/ml aprotinin) for 10 min at 4°C, followed by centrifugation at 600g for 5 min. Nuclei were lysed in nuclei lysis buffer (50 mM Tris, pH 8.1, 10 mM EDTA, 1% SDS, 0.5 mM PMSF, 1× PI-Mix) and incubated with acid-washed glass beads for 40 min on a shaker. The extract was sonicated 7× for 30 s each time and centrifuged. The supernatant containing chromatin was used for chromatin immunoprecipitation.
Chromatin immunoprecipitation assay
DNA fragments of less than 500 bp were obtained by incubation with 45 U micrococcal nuclease S7 (Roche) per 200 μg nucleoprotein for 15 min at 37°C in the presence of 20 mM CaCl2. Reactions were stopped by adding EDTA (20 mM final). The extract was divided into three equal volumes. One aliquot was removed and used in the PCR analysis as input control. The remainder of chromatin was diluted with IP dilution buffer (20 mM Tris, pH 8.1, 150 mM NaCl, 3 mM EDTA, 0.01% SDS, 1% Triton X-100) and incubated overnight at 4°C with 10–15 μg antibodies or preimmune sera in a final volume of 800 μl. Thirty microliter of protein A-Sepharose were added and samples were incubated for 4 h at 4°C. Each batch of resin was washed 5× with wash buffer I (20 mM Tris, pH 8.1, 150 mM NaCl, 2 mM EDTA, 0.1% SDS 1% NP-40), 3× with wash buffer II (10 mM Tris pH 8.1, 250 mM LiCl, 1 mM EDTA, 1% NP-40, 1% Na-deoxycholic-acid) and 3× with TE (10 mM Tris, pH 8.0, 1 mM EDTA). DNA–protein complexes were eluted twice with 100 μl elution buffer (100 mM NaHCO3, 1% SDS) for 10 min at 37°C. After RNase A treatment (0.06 mg/ml) in the presence of NaCl (0.3 M) for 30 min at 37°C, cross-links were reverted for 30 min at 65°C. DNA was recovered by treating the samples overnight at 37°C with 0.2 mg/ml proteinase K, followed by two extractions with phenol/chloroform and ethanol precipitation. DNA was resuspended in 45 μl bdH2O and analyzed by PCR with primer pairs specific for fragment A, B, C and D, respectively. The amplification program consisted of 2 min at 94°C, followed by 32 cycles of 30 s at 94°C, 30 s at 52°C, 30 s at 72°C and finally 7 min at 72°C. PCR band intensities were analyzed by Aida v.3.5. A sequence-specific association of pre-RC proteins was assumed if the PCR signals obtained by specific antibodies were stronger than those by preimmune sera.
Preparation of nuclear extracts and protein purification by cation exchange and gel filtration chromatography
FM3A cells were washed in PBS and resuspended in hypotonic lysis buffer (10 mM HEPES, pH 7.8, 10 mM NaCl, 1.5 mM MgCl2, 5 mM β-mercaptoethanol, 1 × PI-Mix, 1 mM DTT). After the cell suspension was homogenized in a Dounce homogenizer, 1/10 volume of 10× salt buffer (300 mM HEPES, pH 7.8, 1400 mM KCl, 30 mM MgCl2, 5 mM β-mercaptoethanol, 1× PI-Mix, 1 mM DTT) was added. After centrifugation (24 000g, 30 min) the pellet containing the nuclei was resuspended in KEP buffer (20 mM HEPES, pH 7.9, 25% glycerol, 420 mM KCl, 0.2 mM EDTA, 1.5 mM MgCl2, 5 mM β-mercaptoethanol, 1× PI-Mix, 1 mM DTT) and rotated for 1 h at 4°C. After centrifugation (12 000g, 30 min) the nuclear extract was dialyzed overnight against buffer H (50 mM HEPES, pH 7.5, 1 mM EDTA, 1 mM EGTA, 5 mM Na-acetate, 10% glycerol, 5 mM β-mercaptoethanol, 1× PI-Mix, 1 mM DTT) containing 250 mM NaCl. The dialyzed sample was first purified on a 5 ml cation exchange column (HiTrap Heparin HP, Amersham Biosciences) and bound proteins were eluted with a linear gradient of NaCl (250–800 mM). Samples were analyzed on immunoblots for the presence of ORC2. Fractions containing ORC2 were combined and dialyzed overnight against buffer H containing 100 mM NaCl. The extract was further purified by chromatography over a 1 ml cation exchange column (HiTrap SP Sepharose Fast Flow, Amersham Biosciences) and bound proteins were eluted with a linear gradient of NaCl (100–500 mM). After immunoblot analysis using antibodies against ORC1 and -2 fractions containing ORC1 and ORC2 were concentrated on Centrifugal Filter Devices (Amicon® Ultra, Millipore) and subjected to gel filtration on a Superdex 200 PC 3.2/30 column (Amersham Biosciences).
Electrophoretic mobility shift assay
Oligonucleotides were labeled with γ32P-ATP using T4 polynucleotide kinase. PCR reactions were carried out as described above. EMSAs (10 µl) contained 10 mM HEPES, pH 7.5, 5 mM MgCl2, 1 mM CaCl2, 70 mM KCl, 250 ng poly dI+dC, 5 mM ATP (as indicated), 10 fmol γ32P-ATP 5′ end-labeled DNA fragments and 400 ng purified nuclear extract. Binding reactions were incubated for 45 min on ice, 10 min at 30°C, and again 45 min on ice. For supershift assays, 100 ng of either specific or control antibodies were added for a further 2 h on ice. Electrophoresis was performed on 4% native polyacrylamide gels at 100 V in 0.5× TBE and at 4°C. Following electrophoresis, gels were dried and autoradiographed.
Site-directed mutagenesis
The PCR mixture contained 20 ng template plasmid DNA (1545 bp rDNA fragment from −3476 to −1931 cloned into pUC18), 15 pmol of each oligonucleotide, 200 µM dNTPs, 2.5 U Pfu DNA polymerase (Stratagene) and buffer in a total volume of 50 µl. PCR was performed under the following conditions: denaturation at 95°C for 30 s, followed by 18 cycles of 95°C for 30 s, 55°C for 1 min and 72°C for 1.5 min/kb. The reaction was incubated with 10 U of DpnI for 1 h at 37°C to digest methylated, non-mutated parental DNA. An aliquot (1/5) of the PCR product was transformed in 100 µl of competent JM109 Escherichia coli cells (Stratagene). The presence of the mutation was confirmed by DNA sequencing of the constructs.
BrdU incorporation
BrdU staining was performed with proliferating or differentiated C2C12 cells grown on coverslips. In principle, BrdU was added to cell media at 10 µM for 3 h. After fixation with 70% ethanol (30 min), cells were incubated in 0.07 N NaOH for 2 min followed by a neutralization step with PBS, pH 8.5. Subsequently, cells were incubated with anti-BrdU (Becton Dickinson) in PBS containing 0.5% Tween 20 for 30 min.
Immunoblotting analysis
Extracts from proliferating and differentiating C2C12 cells were prepared by the following method. Cells were trypsinized and washed twice with PBS. Cells were lysed in cell lysis buffer (20 mM HEPES/NaOH, pH 7.9, 10 mM NaCl, 3 mM MgCl2, 0.1% NP40, 10% glycerol, 0.2 mM EDTA, 1× PI-Mix, 1 mM DTT) for 15 min on ice. After centrifugation (2000 r.p.m., 5 min) the soluble supernatant was transferred to another tube and the pellet containing the nuclei was washed once (20 mM HEPES/NaOH, pH 7.9, 20% glycerol, 0.2 mM EDTA, 1× PI-Mix, 1 mM DTT). Nuclear proteins were extracted with high salt buffer (20 mM HEPES/NaOH, pH 7.9, 400 mM NaCl, 0.2 mM EDTA, 40% glycerol, 1× PI-Mix, 1 mM DTT) for 45 min on ice. Protein concentrations were determined by the method of Bradford (1976). Equal amounts (20 µg) of soluble and nuclear extract were subjected to SDS-PAGE and transferred onto nitrocellulose membrane (Schleicher & Schuell). The membrane was probed with primary antibodies followed by a secondary antibody conjugated with horseradish peroxidase and detected by the ECL system (Amersham Bioscience).
RESULTS
A Pre-RC-binding site is located within the origin of bidirectional replication of the mouse ribosomal gene cluster
In a previous study, we have mapped an origin of bidirectional replication (OBR) in the murine rDNA locus by nascent strand determination analysis (Figure 1A) (32). Since replication is initiated within a region of 3 kb, we examined by ChIP assays whether ORC proteins are site-specifically assembled within this OBR. In asynchronously growing FM3A cells, proteins were cross-linked in vivo to DNA by formaldehyde and nucleoprotein complexes were immunoprecipitated with antibodies raised against ORC2 as well as with preimmune serum as a control. Precipitated DNA was amplified via PCR using primer pairs each flanking 1 kb of the OBR. A sequence-specific association of ORC2 proteins was assumed if the signals of PCR analyses obtained by specific antibodies exceeded those by preimmune sera.
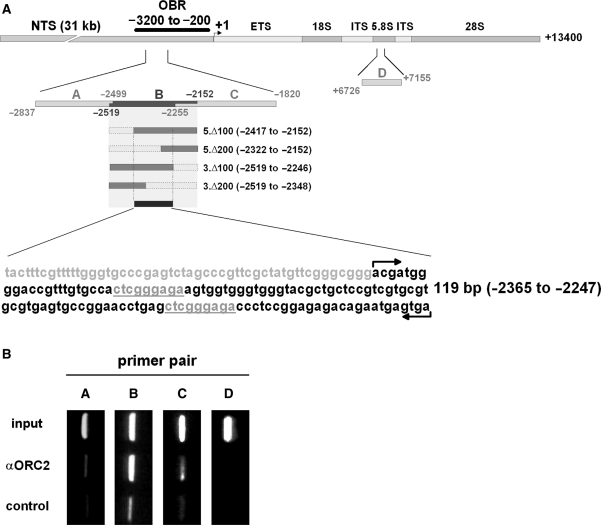
Figure 1.
Pre-RC proteins are associated with region B in the origin of bidirectional replication (OBR). (A) Scheme of the origin of replication in murine rDNA. The transcription start site is marked as +1. OBR designates the origin of bidirectional replication mapped by nascent strand determination analysis (32). DNA fragments used in ChIP and EMSA assays are denoted according to the nucleotide position at the 5′ and at the 3′ ends relative to the transcription start site. The 119 bp fragment used in EMSA experiments is marked by arrows. The conspicuous 9 bp repetitive sequence elements are underlined. (B) Cross-linked chromatin was immunoprecipitated with antibodies against ORC2 (αORC2) and preimmune serum (control). Isolated DNA was analyzed with primer pairs flanking regions A, B, C and D, respectively. PCR products of the corresponding chromatin samples before immunoprecipitation are shown (input).
In first experiments, ORC2 was found cross-linked with DNA sequences located −3 to −2 kb upstream of the transcription start site (data not shown). To further confine this binding site, three subfragments of this region were used in subsequent ChIP experiments (Figure 1A, fragments A: −2837 to −2499, B: −2519 to −2152 and C: −2255 to −1820). PCR analysis following immunoprecipitation with antibodies against ORC2 produced a strong signal with primer pairs for fragment B, whereas intensities of PCR products A or C were significantly weaker (Figure 1B). This suggests that a pre-RC binding site is located within fragment B, situated −2519 to −2152 upstream the transcription start site. As an additional negative control, PCR analyses were performed with primers specific for a region in the 5.8 S rRNA gene, fragment D. ORC2 was not to be found interacting with this DNA sequence in the coding region.
Cell cycle-dependent binding of pre-RC proteins in murine FM3A cells
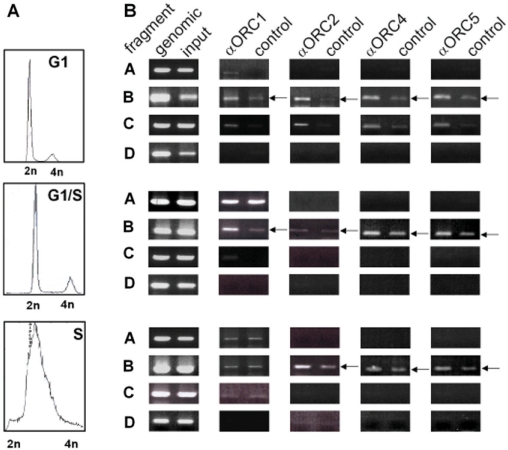
To study the binding of initiator proteins to fragment B in the murine rDNA locus during the cell cycle, ChIP assays with synchronized FM3A cells were performed. FM3A cells were arrested in G1 phase, at the G1/S transition and in S phase as described in Materials and Methods section. Cross-linked chromatin was immunoprecipitated with antibodies against ORC1, -2, -4 and -5, respectively, as well as with preimmune sera. During G1 phase and at G1/S transition ORC1, -2, -4 and -5 are bound to fragment B (Figure 2, upper and middle panel). During S phase, ORC1 dissociates from the pre-RC-binding site (Figure 2, lower panel). Densitometric values of PCR results are shown in Supplementary Figure 1. Apparently, fragment B has an intrinsic, unspecific protein-binding capacity resulting in a higher intensity of PCR bands with control antibodies and primer pair B compared to primer pairs A, C or D.
Figure 2.
Sequence-dependent association of pre-RC proteins with the OBR throughout the cell cycle in vivo. (A) FACS analyses showing cell cycle arrest of FM3A cells in G1 phase (82.6%), at the G1/S boundary (82.8%) and in S phase (66.8%). (B) Synchronized FM3A cells were treated with formaldehyde, cell lysates were prepared and subjected to immunoprecipitation using antibodies against ORC1, -2, -4 and -5 or using preimmune sera (control). Representative results of three experiments are shown.
Isolated pre-RC proteins associate with origin DNA in vitro
In order to further verify the specificity of the pre-RC-binding site at fragment B, we performed EMSAs with partially purified nuclear extract and the DNA fragments described above. Proteins from FM3A nuclear extracts were fractionated by three successive chromatographic steps as described in Materials and Methods section.
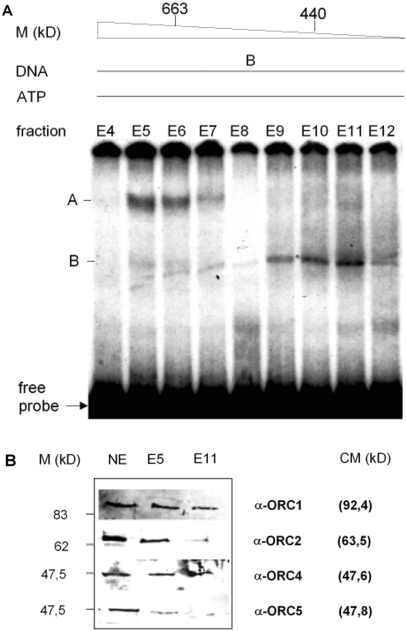
To analyze the DNA-binding activity, purified proteins of >300 kDa mass after gel filtration were incubated with radiolabeled fragment B and assayed by EMSA (Figure 3A). Proteins of several fractions bind to DNA fragment B, forming two major protein/DNA complexes of distinct electrophoretic mobility, designated complexes A and B, respectively. The fractions exerting the most prominent shifts, i.e. E5 (molecular mass of >663 kDa) and E11 (<440 kDa), were examined by immunoblotting using various antibodies specific for components of the pre-RC (Figure 3B). ORC1, -2, -4 and -5 were confirmed in fraction E5. Fraction E11 contained the same proteins but at a lower level than in fraction E5. By this three-step chromatography, ORC containing protein complexes could be enriched from nuclear extracts. Fractions E5 and E11 comprised highest DNA-binding activity resulting in the formation of complex A and B, respectively. Although the presence of pre-RC components was demonstrated in these protein fractions, the data could not provide conclusive evidence whether the complexes are caused primarily by binding of pre-RC components to fragment B or whether they are due to the interaction with unknown proteins.
Figure 3.
Purification and analysis of proteins from murine nuclear extracts. (A) Nuclear extracts from FM3A cells were purified twice on cationic exchange columns and once by gel filtration on a Superdex™200 10/300 GL column, calibrated with thyroglobulin and ferritin. To test for DNA-binding activity aliquots of fractions E4 to E15 were subjected to EMSA with 10 fmol γ32P-ATP 5′ end-labeled fragment B in the presence of 5 mM ATP and 250 ng poly dI+dC. Protein/DNA complexes A and B are indicated. (B) Immunoblot analysis of fractions E5 and E11 with antibodies raised against ORC1, -2, -4 and -5, respectively.
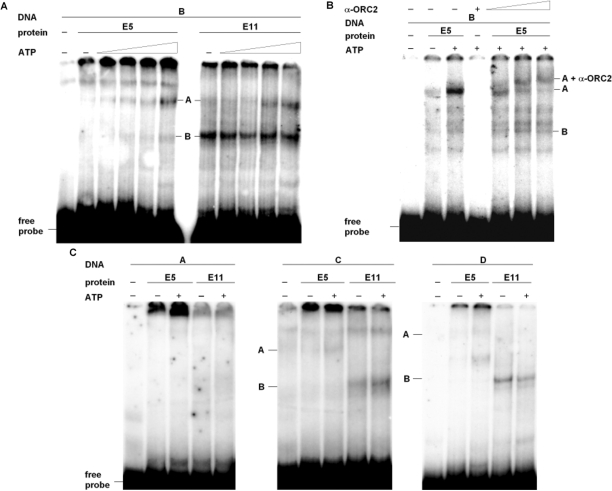
Next, the effect of ATP on the formation of complexes A and B was examined. The presence of two ATP-binding subunits in eukaryotic ORCs (27,34–36) lead us to expect an ATP-dependent increase in DNA binding if complex A or B contain these pre-RC components. The addition of increasing amounts of ATP to fraction E5 results in a dose-dependent increase of the formation of complex A, not however, on the formation of complex B (Figure 4A). This indicates that complex A, but not complex B, is probably caused by the binding of pre-RC proteins to origin DNA.
Figure 4.
DNA/protein complex A binds ATP dependently and sequence selectively to fragment B and is supershifted with antibodies against ORC2. (A) EMSAs were performed with proteins of fractions E5 or E11 (300 ng) and γ32P-ATP 5′ end-labeled fragment B with increasing concentrations of ATP (0.5, 1, 2.5 or 5 mM). (B) For supershifting, binding reactions were performed by incubating γ32P-ATP 5′ end-labeled fragment B and proteins of fraction E5 in the presence of 5 mM ATP with increasing amounts of antibodies against ORC2 as indicated. (C) EMSAs were performed with proteins of fractions E5 or E11 and fragments A, C and D in the absence or presence of 5 mM ATP.
To prove an involvement of pre-RC proteins in the formation of complex A more directly, we performed supershift experiments using increasing amounts of antibodies against ORC2, as a representative for pre-RC components. Including ORC2 antibodies into the DNA/protein-binding reaction led to a further retardation of the mobility of complex A (Figure 4B), indicating that ORC2 is part of complex A. In contrast, antibodies against ORC2 had no effect on the mobility of complex B (data not shown), again hinting to an ORC-independent complex formation in the latter case.
Sequence specificity of pre-RC binding
To investigate whether proteins in fraction E5 and E11 bind to fragment B in a sequence-specific manner, EMSA studies were carried out comparing binding to DNA fragments A, C and D (Figure 1A), both in the presence and absence of ATP. Neither proteins in fraction E5 nor E11 bound fragment A (Figure 4C, left panel). Proteins of fraction E5 showed no significant binding activity to fragments C or D as well (Figure 4C, middle and right panel). Thus, proteins of fraction E5 bind preferentially to fragment B, suggesting that complex A is formed sequence selectively. In contrast, complex B is formed on fragments B, C and D indicating sequence-unspecific DNA binding of proteins in fraction E11.
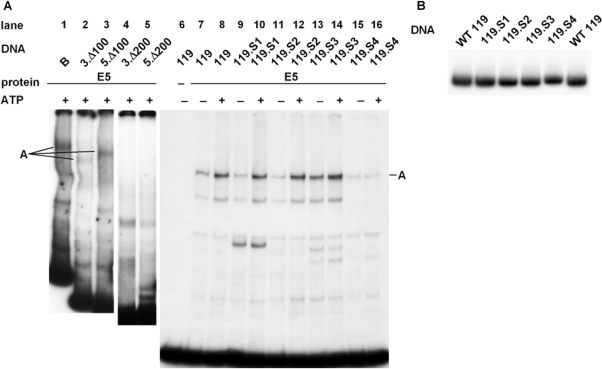
To further confine the pre-RC-binding site within fragment B, this fragment was shortened by 100 and 200 bp, respectively, either at the 3′ or at the 5′ end, (Figure 1; 3.Δ100; 3.Δ200; 5.Δ100; 5.Δ200). As shown in Figure 5A, lane 2 and 3, complex A is formed with fragments truncated 100 bp at their 3′ or 5′ ends. However, if 200 bp are deleted, formation of the protein/DNA complex is abrogated in both cases (Figure 5A, lanes 4 and 5). This indicates that a DNA sequence in the middle of fragment B is essential for the formation of complex A. EMSA experiments with a 119 bp fragment located in the middle of fragment B (−2365 to −2247, see Figure 1) showed that complex A is still formed in an ATP-dependent manner (Figure 5A, lanes 7 and 8). Thus, this 119 bp DNA fragment is sufficient for pre-RC/DNA complex formation.
Figure 5.
The pre-RC-binding site is confined to a 119 bp region of fragment B. (A) EMSAs analyses with deleted fragment B (3.Δ100, 5.Δ100, 3.Δ200, 5.Δ200; see Figure 1A) and wild-type or mutant 119 bp fragments (119, 119.S1–S4). Sequences of the mutant constructs are given in Table 1. (B) Migration of protein-free 119 bp DNA fragments, WT and S1 to S4, in a 4% PAA gel.
Inspecting this central 119 bp region closely, two conspicuous 9 bp sequence elements, CTCGGGAGA, were observed which are repeated at intervals of 63 bp (−2343 to −2335; −2280 to −2272) and which are completely or partially disrupted by the 200 bp deletions. To examine whether these sequence elements play a role in formation of complex A, we altered the sequence by introducing C to A, T to G and vice versa substitutions using site-specific mutagenesis. Four successive substitutions of 4 bp each were constructed, whereby the obtained constructs were used as template DNA for the subsequent mutagenesis (119.S1, 119.S2, 119.S3 and 119.S4; for sequences, see Table 1). EMSA experiments carried out with the mutated 119 bp DNA fragments showed that substitutions 1 (119.S1; lanes 9 and 10), 2 (119.S2, lanes 11 and 12) and 3 (119.S3, lanes 13 and 14) did not affect formation of the DNA/protein complexes. However, if 8 out of 9 bp in both sequence elements (119.S4) are substituted, formation of complex A is significantly reduced (lanes 15 and 16) indicating that the CTCGGGGAGA sequences might influence binding of pre-RC proteins to DNA in the murine rDNA locus. Interestingly, a naked DNA fragment containing all four substitutions (Figure 5B, 119.S4) migrates distinctly slower than protein-free WT 119 and 119.S1–119.S3 (lanes 1–4 and 6) suggesting that the secondary structure of the DNA is altered as a result of the S4 nucleotide substitutions.
Table 1.
Sequences of the mutated 119 bp fragments
| 119 (original sequence) | 5′-ACGATGGGGACCGTTTGTGCCACTCGGGAGAAGTGGTGGGTGG GTACGCTGCTCCGTCGTGCGTGCGTGA GTGCCGGAACCTGAGCTCGGGAGACCCTCCGGAGAGACAGAATGAGTGA_3′ |
| 119.S1 | 5′-ACGATGGGGACCGTTTGTGCCAAGATGGAGAAGTGGTGGGTGG GTACGCTGCTCCGTCGTGCGTGCGTGA GTGCCGGAACCTGAGCTC GGGAGACCCTCCGGAGAGACAGAATGAGTGA-3′ |
| 119.S2 | 5′-ACGATGGGGACCGTTTGTGCCAAGATTTCTAAGTGGTGGGTGG GTACGCTGCTCCGTCGTGCTGCGTGAG TGCCGGAACCTGAGCTC GGGAGACCCTCCGGAGAGACAGAATGAGTGA-3′ |
| 119.S3 | 5′-ACGATGGGGACCGTTTGTGCCAAGATTTCTAAGTGGTGGGTGG GTACGCTGCTCCGTCGTGCGTGCGTGA GTGCCGGAACCTGAGAGA TGGAGACCCTCCGGAGAGACAGAATGAGTGA-3′ |
| 119.S4 | 5′-ACGATGGGGACCGTTTGTGCCAAGATTTCTAAGTGGTGGGTGG GTACGCTGCTCCGTCGTGCGTGCGTGAGTGCCGGAACCTGAGAGAT TTCTACCCTCCGGAGAGACAGAATGA GTGA-3′ |
Conspicuous sequence elements are depicted in bold letters, introduced mutations are underlined.
Taken together, we were able to reproduce in vitro by EMSA the sequence-specific binding of pre-RC proteins to DNA shown in vivo by ChIP experiments. Formation of complex A, which contains ORC2 as confirmed by supershifting, is stimulated by ATP and occurs in a sequence-specific manner, preferring fragment B over A, C and D. Moreover, mutational analyses of fragment B revealed that (i) the pre-RC-binding site is situated within 119 bp, from −2365 to −2247 upstream the transcription start site and that (ii) two sequence elements, CTCGGGAGA, are crucial for the binding of pre-RC proteins to DNA.
Myogenic differentiation of C2C12 cells
Recent studies on various organisms have shown that in terminally differentiated cells or tissues some components of the pre-RC are absent (2). The loss of pre-RC components upon cell cycle exit could be a regulatory mechanism to suppress cell proliferation.
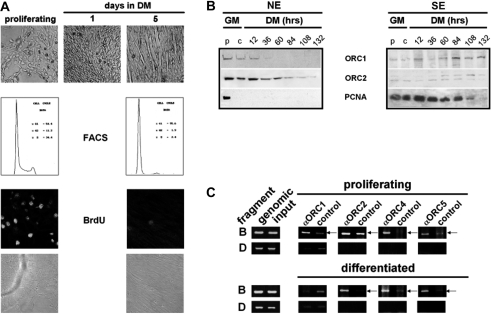
To address this issue experimentally, we analyzed protein levels of pre-RC components during myogenic differentiation and determined whether components of the pre-RC are attached to the OBR in terminally differentiated myotubes. For that we used murine C2C12 cells, which can be induced to terminal differentiation into myotubes by exposure to low mitogen medium for several days. Differentiation was assessed by morphological examination. After 5 days in differentiation medium (DM), predominantly elongated myotubes were present in the culture (Figure 6A, upper panel). The percentage of S phase cells considerably decreased during myogenic differentiation (34.4% in proliferating versus 2.4% in differentiated cells) concomitantly with a significant increase in the percentage of myotubes in the G0/G1 phase of the cell cycle (54.4% in proliferating versus 95.6% in differentiated cells, middle panel). The cessation of DNA replication in myotubes could further be verified by BrdU incorporation. After five days in differentiation medium only two persisting myoblasts were replicating in the examined area (lower panel).
Figure 6.
Distinct pre-RC proteins are absent in terminally differentiated myotubes. (A) Myogenic differentiation of C2C12 cells. Phase contrast microscopy showing cultures containing terminally differentiated myotubes by day 5 in differentiation medium (DM) (upper panel). Cell cycle profiles of proliferating (54.4% G1 phase, 34.4% S phase, 11.2% G2 phase) and differentiated (95.6% G1 phase, 2.4% S phase, 1.9% G2 phase) C2C12 cells (middle panel). BrdU incorporation: representive areas examined by indirect immunofluorescence microscopy demonstrate the cessation of DNA synthesis in differentiated C2C12 cells (lower panel). (B) Immunoblot analyses of pre-RC proteins during differentiation of C2C12 cells. Nuclear (NE) and soluble (SE) extracts were prepared from proliferating (p), confluent (c) and differentiating C2C12 cells at the times indicated in differentiation medium (DM). Same amounts (20 µg) of extracts were dissolved by SDS-PAGE and immunoblot analyses were performed using antibodies raised against pre-RC proteins. GM: growth medium. (C) ChIP analyses of pre-RC components interacting with fragment B in the OBR region in proliferating C2C12 myoblasts or terminally differentiated C2C12 myotubes after five days in DM. ChIP analysis was performed as described above with the antibodies indicated. Representative results of three experiments are shown.
Thus, a system of terminally differentiated cells was available to investigate the expression of pre-RC components during differentiation and their interaction with the OBR region in terminally differentiated myotubes.
Expression of pre-RC proteins during myogenic differentiation of C2C12 cells
Immunoblot analyses were performed with nuclear and soluble extracts, respectively, prepared from proliferating, confluent or differentiating C2C12 cells. Subsequently immunoblot analyses of pre-RC components were carried out using specific antibodies raised against ORC1 and ORC2 (Figure 6B). Sixty hours post-differentiation, the amount of ORC1 in the nucleus was at undetectable levels. In contrast, soluble ORC1 levels exhibited a peak at 84 h and were then reduced. ORC2 expression remained unaffected until 60 h, and even after 132 h ORC2 was detectable at low levels in the nucleus. A soluble variant of ORC2 migrating slower than nuclear ORC2 was observed during differentiation, probably due to phosphorylation. Immunoblot analysis of proliferating cell nuclear antigen (PCNA) demonstrated nuclear localization exclusively in proliferating myoblasts, not however, in differentiated myotubes. A slow reduction of soluble PCNA with complete disappearance after 132 h in DM was observed.
In summary, protein levels of ORC1 and ORC2 significantly decreased in murine C2C12 myotubes during differentiation. ORC1 could hardly be detected in terminally differentiated myotubes at all, whereas ORC2 was detectable, albeit at lower levels than in proliferating myoblasts. These data demonstrate that the loss of proliferation capacity of mammalian cells during differentiation is associated with down-regulation of constituents of the pre-RC.
In vivo occupancy of the OBR region with pre-RC proteins in terminally differentiated myotubes
We performed ChIP analyses comparing chromatin of proliferating cells to that of differentiated cells to elucidate if members of the pre-RC still occupy the OBR region in non-replicating cells. Formaldehyde cross-linked nuclear proteins of proliferating and differentiated C2C12 cells after 5 days in DM were prepared. Chromatin was immunoprecipitated with antibodies raised against ORC1, -2, -4 and -5 and was analyzed with primer pairs specific to the pre-RC assembly region (fragment B) and to the control region in the 5.8 S rRNA gene (fragment D). Preimmune serum served as control. Densitometric values of PCR results are shown in Supplementary Figure 2. All investigated proteins are bound to OBR sequences in asynchronously growing cells (Figure 6C, upper panel). When cells exit the cell cycle during differentiation, ORC1 is released from chromatin (Figure 6C, lower panel). ORC2, -4 and -5 remain bound to the OBR region in terminally differentiated cells. The control region in the 5.8 S rRNA gene showed no interaction with any of these proteins.
These results probably indicate that chromatin association of ORC1 is regulated during transition from proliferation to terminal differentiation.
DISCUSSION
In the current work, we demonstrated that a pre-RC-binding site is located in a region positioned from −2519 to −2152 upstream the transcription start site of RNA polymerase I within the OBR of the murine rDNA gene cluster. This pre-RC-binding site has been verified by three independent approaches. First, we showed in vivo by ChIP experiments a cell cycle-dependent association of pre-RC proteins with this sequence in murine FM3A cells. Second, we demonstrated in vitro by EMSA a sequence-specific and ATP-stimulated binding of protein complexes containing ORC proteins to this DNA region. Deletion mutagenesis revealed that this binding site is located within 119 bp at −2365 to −2247 and that a repeated element, CTCGGGAGA, is crucial for the assembly of pre-RC proteins. Third, we showed by ChIP experiments in terminally differentiated murine C2C12 cells that this binding site still exists, but that only a subset of pre-RC proteins is bound at this region in differentiated myotubes.
Cell cycle-dependent binding of initiation proteins to fragment B
Using synchronized FM3A cells, we demonstrated that an ORC core complex consisting of ORC 2, -4, -5 was stably bound to fragment B throughout the cell cycle, whereas ORC1 exhibited a cell cycle-dependent association. During G1 phase and at the G1/S transition, ORC1, -2, -4 and -5, were bound to fragment B. In S phase, ORC1 dissociated from the initiation complex.
Compared to yeast and Drosophila, where ORC remains associated with chromatin throughout the cell cycle (37–39), ORC binding in mammalia is cell cycle-dependent. The association patterns we observed in murine cells are similar to those described for other species. In hamster cells, ORC1 is released from chromatin as cells enter S phase and is re-bound to chromatin during M to G1 transition (6,7). Similarly, we found that murine ORC1 is bound to chromatin in G1 and at the G1/S transition, but not in S phase. Furthermore, we demonstrated that ORC2 remains bound to chromatin throughout the cell cycle. In a recent study, it was shown also that human ORC1 and -2 bind at an origin of DNA replication in G1 phase but in S phase only ORC2 remains bound (5).
The pre-RC has an intrinsic sequence-selective DNA-binding activity
We have demonstrated that purified nuclear extracts containing ORC proteins bind ATP dependently and sequence specifically to fragment 119. The question arises why this sequence is preferred compared to adjacent sequences.
A possibility for ORC to find its binding site is the presence of ancillary factors promoting chromatin binding. It has been proposed that CDC6 might reduce the affinity of ORC to non-specific DNA and therefore acts as an essential determinant for origin specificity in metazoans (40,41). In the case of Epstein-Barr virus, the viral transcription factor EBNA-1 binds ORC and may be involved in the recruitment of ORC to oriP (42). It is highly probable that we copurified CDC6 or a yet unknown protein, which directs ORC to its binding site and that ORC itself is not responsible for sequence-specific binding to fragment B. The identification of all proteins involved in the formation of the DNA/protein complexes by mass spectrometry would be very interesting and remains to be done.
We confined the pre-RC-binding site to 119 bp within fragment B and found that a repetitive 9 bp sequence element, CTCGGGAGA, plays a crucial role in ensuring efficient loading of pre-RC proteins to DNA. In contrast to yeast where the ORC binds to defined sequences called ARS, origins in higher eukaryotes do not appear to contain consensus sequences, although some origins include AT-rich sequences (43). Fragment 119, however, is not particularly AT-rich, since its AT content is only 37% and no AT-rich nucleotide clusters are to be found. By substitutions S1–S4, the AT content is stepwise increased. On S4, the fragment with the highest AT content (44%), nearly no binding of pre-RC proteins was observed. We therefore propose that a higher AT content is not essential for ORC–DNA interaction within fragment 119.
If the 9 bp elements represent a consensus binding site for pre-RC proteins or for ancilliary factors recruiting pre-RC proteins to origin DNA, these elements are to be found in other origin regions. However, these elements could neither be detected with 100% identity in the origins of the human MCM4/PRKD (U90415), TOP1 (AL035652) and Lamin B2 (M94363) genes nor in the Sciara puff II/9A origin (AF332610). In the TOP1 gene two sequences with 88.8% (CTGGGGAGA) and 77.7% (CTCCGGAGT) and in the MCM4/PRKD gene with 77.7% (CCCGGGAAA) and 66.6% (CCTCCCGGA) identity, respectively, were found. However, these elements are 588 and 620 bp, respectively, apart from each other, and hence are located at a greater distance compared to those in the murine rRNA genes. Interestingly, the distance of 63 bp between both sequence elements corresponds to six helical turns of B DNA (10.5 bp/helical turn). Intrinsically bent or curved DNA molecules result when distinct base sequences or structural motifs are repeated in phase with the DNA helical repeat (44). Significantly, substitution 4 within the 9 bp sequence element leads to a slight, but measurable decrease of the relative electrophoretic mobility of the naked DNA, indicating an altered secondary structure as a result of substitution S4. Therefore, the 9 bp sequence element probably does not constitute a consensus binding sequence per se, but rather has effects on the formation of specific secondary DNA structures, which could allow binding of the pre-RC or its recruitment factor to this region of the murine rDNA.
Remarkably, the 9 bp sequence element is to be found a third time within the 119 bp fragment (−2269 to −2261: CTCCGGAGA) with 88.8% identity and inverted CTCGGGAGA elements are found three times in the murine rRNA genes on the coding (+1794 to +1802: AGAGGGCTC) and non-coding strand (+11238 to +11246: TCTCCCGAG; +12980 to +12988: TCTCCCGAG). It has been described that more active initiation sites exists in the murine rDNA located either between + 2.0 and 8.0 kb or approximately at + 13 kb downstream of the transcription start site (32). Interestingly, an ORC2-binding site was localized by ChIP in the region from +1642 to +2082 including one of the 9 bp sequence elements described above (unpublished data). Also, in the region ∼+13 kb an inverted CTCGGGAGA element was found on the non-coding strand. However, so far this region has not been studied in respect of pre-RC-binding sites. Since pre-RC proteins exhibited relatively weak binding to fragment B in vitro, attempts to detect a specific DNA sequence by DNase I footprint experiments were unsuccessful (data not shown). Despite the sequence-specific binding of pre-RC proteins to fragment 119 and the evidence that the 9 bp sequence elements play a crucial role in this process, it is possible that the in vivo binding of pre-RCs to origins might be additionally affected by surrounding DNA sequences, by chromatin structure or by epigenetic events.
Initiation proteins in terminally differentiated cells
As cells undergo terminal differentiation, they enter an irreversible cell cycle arrest and generally lose the capacity to go through DNA replication and cell division. In previous studies, it has been proposed that the absence of proteins involved in initiation of DNA replication in quiescent or differentiated cells seems to be crucial for the maintenance of the ‘out-of-cycle’ state (22). To investigate the fate of replication proteins upon cell cycle exit, we took advantage of the in vitro differentiation of murine C2C12 myoblast cells to myotubes. These cells undergo terminal differentiation when exposed to a low mitogen medium. Using immunoblot analysis we demonstrated that ORC1 and -2 were abundant in proliferating cells, but the levels of these proteins were reduced after cells underwent differentiation. The reduction of ORC1 and -2 levels during differentiation is in agreement with a previous report showing that ORC1 and -2 are lacking in terminally differentiated Xenopus erythrocytes (45). ORC1 was reduced notably faster than ORC2, which suggests that ORC1 might be the limiting component of the active murine pre-RC. This is affirmed by the fact that during normal cell cycle transit the activity of ORC1, as the only ORC subunit, is regulated by ubiquitination. This modification results either in degradation of human ORC1 (11,46–48) or in dissociation of ORC1 from chromatin in hamster cells (6). In a similar manner, the degradation of ORC1 in terminally differentiated cells might contribute to the maintenance of the postmitotic state. Contrary to studies showing that ORC2 levels remain unaffected in human differentiated or quiescent cells (22,49,50) we observed that levels of ORC2 were reduced, but did not completely disappear, during terminal differentiation of murine C2C12 cells. A slower migrating soluble variant of ORC2 was observed in the course of differentiation, probably a result of phosphorylation. In S. cerevisiae (51), S. pombe (52) and Xenopus laevis (53) ORC2 is phosphorylated by CDK/cyclin complexes. The phosphorylation might result in conformational changes within the ORC and thereby probably inhibits binding of additional factors and prevents re-replication (54–56). ChIP experiments revealed that ORC1 dissociated from the pre-RC-binding site during differentiation, whereas ORC2, -4 and -5 were still bound to the OBR. The loss of ORC1 from chromatin in terminally differentiated cells is in agreement with the fast reduction of ORC1 shown by immunoblotting and could explain the function of this protein as regulator of the initiation of DNA replication in proliferating cells. Since ORC2 was present in the nucleus of myotubes at a very low level, but was detected by ChIP experiments at the OBR, this suggests that a persisting fraction of ORC2 remains bound at origins previously used in proliferating cells. This observation could also imply that ORC2 is not yet fully degraded upon differentiation and that soluble ORC2 is reduced first and chromatin-bound ORC2 later. ORC4 and -5 remain bound to chromatin in differentiated cells. Formerly, it was also demonstrated that Orc2, -3 and -5 can be detected in non-proliferating human cells (50). Alternatively, the persistence of ORC subunits in non-cycling cells could be due to the fact that despite their function in initiation of DNA replication these proteins play additional roles, for example in transcriptional silencing (39,57,58).
Taken together, the reduction and the chromatin dissociation of several proteins involved in the events of initiation of DNA replication could be demonstrated during myogenic differentiation of C2C12 cells. The loss of the important regulatory initiation protein ORC1 in terminally differentiated cells might contribute to the establishment and maintenance of the ‘out-of-cycle’ state, as a part of a highly sophisticated and regulated network. Nevertheless, the function of pre-RC proteins remaining at the OBR in terminally differentiated C2C12 cells remains unknown.
ACKNOWLEDGEMENTS
We are grateful to E. Dinkl for technical help and to R. Friedel for the performance of cell cycle analyses. We thank A. Chari, B. Laggerbauer and S. Meister for comments and discussions. This work was supported by the Deutsche Forschungsgemeinschaft. Funding to pay the Open Access publication charges for this article was provided by by the University of Wuerzburg.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Blow JJ, Hodgson B. Replication licensing – defining the proliferative state? Trends Cell Biol. 2002;12:72–78. doi: 10.1016/s0962-8924(01)02203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bielinsky AK, Blitzblau H, Beall EL, Ezrokhi M, Smith HS, Botchan MR, Gerbi SA. Origin recognition complex binding to a metazoan replication origin. Curr. Biol. 2001;11:1427–1431. doi: 10.1016/s0960-9822(01)00444-4. [DOI] [PubMed] [Google Scholar]
- 4.Stillman B. Origin recognition and the chromosome cycle. FEBS Lett. 2005;579:877–884. doi: 10.1016/j.febslet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Ladenburger EM, Keller C, Knippers R. Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol. Cell. Biol. 2002;22:1036–1048. doi: 10.1128/MCB.22.4.1036-1048.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li CJ, DePamphilis ML. Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol. 2002;22:105–116. doi: 10.1128/MCB.22.1.105-116.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Natale DA, Li CJ, Sun WH, DePamphilis ML. Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G(1) transition in mammals. EMBO J. 2000;19:2728–2738. doi: 10.1093/emboj/19.11.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drury LS, Perkins G, Diffley JF. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elsasser S, Chi Y, Yang P, Campbell JL. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol. Biol. Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perkins G, Drury LS, Diffley JF. Separate SCF(CDC4) recognition elements target Cdc6 for proteolysis in S phase and mitosis. EMBO J. 2001;20:4836–4845. doi: 10.1093/emboj/20.17.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen BO, Wagener C, Marinoni F, Kramer ER, Melixetian M, Denchi EL, Gieffers C, Matteucci C, Peters JM, et al. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 2000;14:2330–2343. doi: 10.1101/gad.832500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maiorano D, Moreau J, Mechali M. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature. 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- 14.Cook JG, Chasse DA, Nevins JR. The regulated association of Cdt1 with minichromosome maintenance proteins and Cdc6 in mammalian cells. J. Biol. Chem. 2004;279:9625–9633. doi: 10.1074/jbc.M311933200. [DOI] [PubMed] [Google Scholar]
- 15.Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 16.Nishitani H, Taraviras S, Lygerou Z, Nishimoto T. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J. Biol. Chem. 2001;276:44905–44911. doi: 10.1074/jbc.M105406200. [DOI] [PubMed] [Google Scholar]
- 17.Li A, Blow JJ. Cdt1 downregulation by proteolysis and geminin inhibition prevents DNA re-replication in Xenopus. EMBO J. 2005;24:395–404. doi: 10.1038/sj.emboj.7600520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimitrova DS, Todorov IT, Melendy T, Gilbert DM. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimitrova DS, Prokhorova TA, Blow JJ, Todorov IT, Gilbert DM. Mammalian nuclei become licensed for DNA replication during late telophase. J. Cell Sci. 2002;115:51–59. doi: 10.1242/jcs.115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaarschmidt D, Ladenburger EM, Keller C, Knippers R. Human Mcm proteins at a replication origin during the G1 to S phase transition. Nucleic Acids Res. 2002;30:4176–4185. doi: 10.1093/nar/gkf532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pardee AB. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 22.Stoeber K, Tlsty TD, Happerfield L, Thomas GA, Romanov S, Bobrow L, Williams ED, Williams GH. DNA replication licensing and human cell proliferation. J. Cell Sci. 2001;114:2027–2041. doi: 10.1242/jcs.114.11.2027. [DOI] [PubMed] [Google Scholar]
- 23.Bell SP. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- 24.Gilbert DM. Making sense of eukaryotic DNA replication origins. Science. 2001;294:96–100. doi: 10.1126/science.1061724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aladjem MI, Fanning E. The replicon revisited: an old model learns new tricks in metazoan chromosomes. EMBO Rep. 2004;5:686–691. doi: 10.1038/sj.embor.7400185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cvetic C, Walter JC. Eukaryotic origins of DNA replication: could you please be more specific?Semin. Cell Dev. Biol. 2005;16:343–353. doi: 10.1016/j.semcdb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Austin RJ, Orr-Weaver TL, Bell SP. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 1999;13:2639–2649. doi: 10.1101/gad.13.20.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdurashidova G, Danailov MB, Ochem A, Triolo G, Djeliova V, Radulescu S, Vindigni A, Riva S, Falaschi A. Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J. 2003;22:4294–4303. doi: 10.1093/emboj/cdg404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller C, Ladenburger EM, Kremer M, Knippers R. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J. Biol. Chem. 2002;277:31430–31440. doi: 10.1074/jbc.M202165200. [DOI] [PubMed] [Google Scholar]
- 30.Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaarschmidt D, Baltin J, Stehle IM, Lipps HJ, Knippers R. An episomal mammalian replicon: sequence-independent binding of the origin recognition complex. EMBO J. 2004;23:191–201. doi: 10.1038/sj.emboj.7600029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gogel E, Langst G, Grummt I, Kunkel E, Grummt F. Mapping of replication initiation sites in the mouse ribosomal gene cluster. Chromosoma. 1996;104:511–518. doi: 10.1007/BF00352115. [DOI] [PubMed] [Google Scholar]
- 33.Kneissl M, Putter V, Szalay AA, Grummt F. Interaction and assembly of murine pre-replicative complex proteins in yeast and mouse cells. J. Mol. Biol. 2003;327:111–128. doi: 10.1016/s0022-2836(03)00079-2. [DOI] [PubMed] [Google Scholar]
- 34.Rao H, Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc. Natl Acad. Sci. USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 36.Chesnokov I, Remus D, Botchan M. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc. Natl Acad. Sci. USA. 2001;98:11997–12002. doi: 10.1073/pnas.211342798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 38.Ogawa Y, Takahashi T, Masukata H. Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol. Cell Biol. 1999;19:7228–7236. doi: 10.1128/mcb.19.10.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 40.Mizushima T, Takahashi N, Stillman B. Cdc6p modulates the structure and DNA binding activity of the origin recognition complex in vitro. Genes Dev. 2000;14:1631–1641. [PMC free article] [PubMed] [Google Scholar]
- 41.Harvey KJ, Newport J. Metazoan origin selection: origin recognition complex chromatin binding is regulated by CDC6 recruitment and ATP hydrolysis. J. Biol. Chem. 2003;278:48524–48528. doi: 10.1074/jbc.M307661200. [DOI] [PubMed] [Google Scholar]
- 42.Schepers A, Ritzi M, Bousset K, Kremmer E, Yates JL, Harwood J, Diffley JF, Hammerschmidt W. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 2001;20:4588–4602. doi: 10.1093/emboj/20.16.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aladjem MI, Rodewald LW, Kolman JL, Wahl GM. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- 44.Crothers DM, Haran TE, Nadeau JG. Intrinsically bent DNA. J. Biol. Chem. 1990;265:7093–7096. [PubMed] [Google Scholar]
- 45.Lu ZH, Xu H, Leno GH. DNA replication in quiescent cell nuclei: regulation by the nuclear envelope and chromatin structure. Mol. Biol. Cell. 1999;10:4091–4106. doi: 10.1091/mbc.10.12.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tatsumi Y, Ohta S, Kimura H, Tsurimoto T, Obuse C. The ORC1 cycle in human cells: I. cell cycle-regulated oscillation of human ORC1. J. Biol. Chem. 2003;278:41528–41534. doi: 10.1074/jbc.M307534200. [DOI] [PubMed] [Google Scholar]
- 47.Fujita M, Ishimi Y, Nakamura H, Kiyono T, Tsurumi T. Nuclear organization of DNA replication initiation proteins in mammalian cells. J. Biol. Chem. 2002;277:10354–10361. doi: 10.1074/jbc.M111398200. [DOI] [PubMed] [Google Scholar]
- 48.Ritzi M, Tillack K, Gerhardt J, Ott E, Humme S, Kremmer E, Hammerschmidt W, Schepers A. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell Sci. 2003;116:3971–3984. doi: 10.1242/jcs.00708. [DOI] [PubMed] [Google Scholar]
- 49.Musahl C, Holthoff HP, Lesch R, Knippers R. Stability of the replicative Mcm3 protein in proliferating and differentiating human cells. Exp. Cell Res. 1998;241:260–264. doi: 10.1006/excr.1998.4041. [DOI] [PubMed] [Google Scholar]
- 50.Thome KC, Dhar SK, Quintana DG, Delmolino L, Shahsafaei A, Dutta A. Subsets of human origin recognition complex (ORC) subunits are expressed in non-proliferating cells and associate with non-ORC proteins. J. Biol. Chem. 2000;275:35233–35241. doi: 10.1074/jbc.M005765200. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- 52.Lygerou Z, Nurse P. The fission yeast origin recognition complex is constitutively associated with chromatin and is differentially modified through the cell cycle. J. Cell Sci. 1999;112(Pt 21):3703–3712. doi: 10.1242/jcs.112.21.3703. [DOI] [PubMed] [Google Scholar]
- 53.Romanowski P, Marr J, Madine MA, Rowles A, Blow JJ, Gautier J, Laskey RA. Interaction of Xenopus Cdc2 x cyclin A1 with the origin recognition complex. J. Biol. Chem. 2000;275:4239–4243. doi: 10.1074/jbc.275.6.4239. [DOI] [PubMed] [Google Scholar]
- 54.Leatherwood J. Emerging mechanisms of eukaryotic DNA replication initiation. Curr. Opin. Cell Biol. 1998;10:742–748. doi: 10.1016/s0955-0674(98)80117-8. [DOI] [PubMed] [Google Scholar]
- 55.Vas A, Mok W, Leatherwood J. Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex. Mol. Cell. Biol. 2001;21:5767–5777. doi: 10.1128/MCB.21.17.5767-5777.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carpenter PB, Mueller PR, Dunphy WG. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature. 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- 57.Bell SP, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- 58.Fox CA, Loo S, Dillin A, Rine J. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev. 1995;9:911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]