Abstract
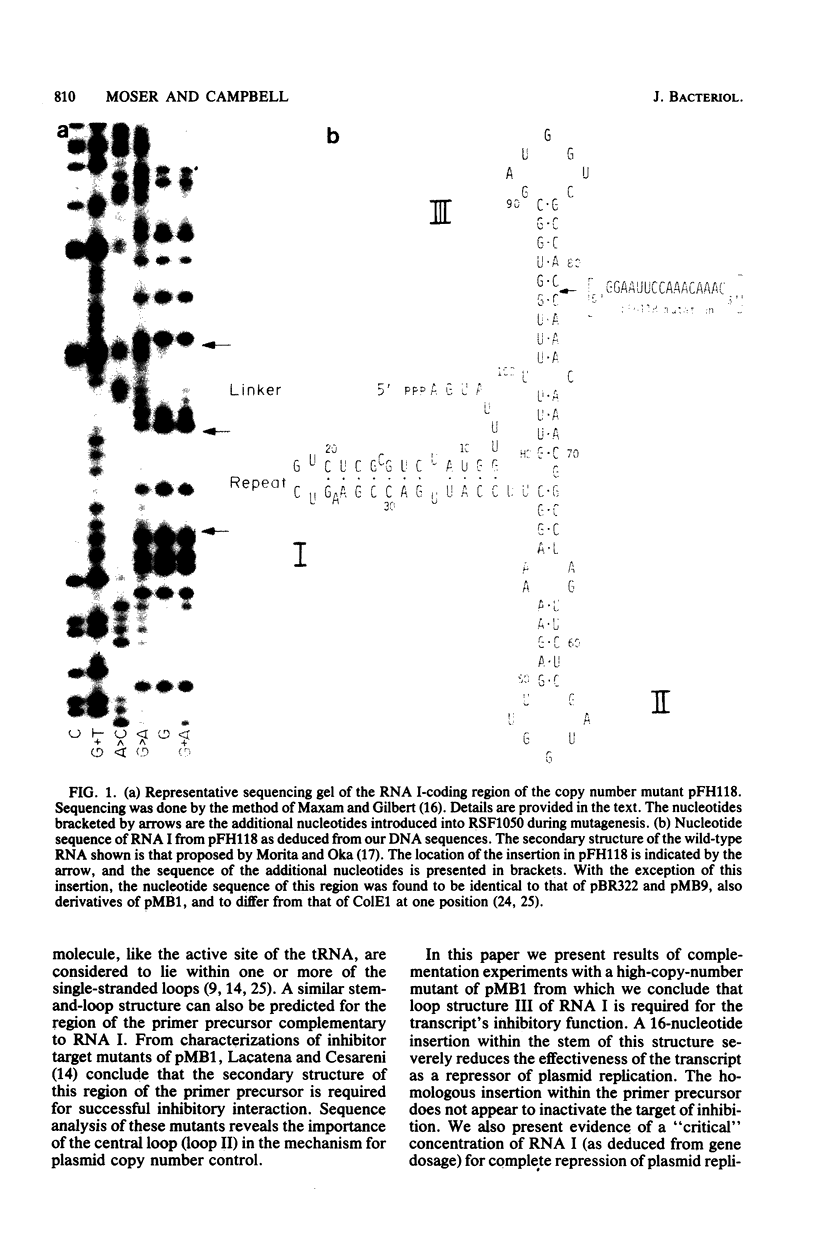
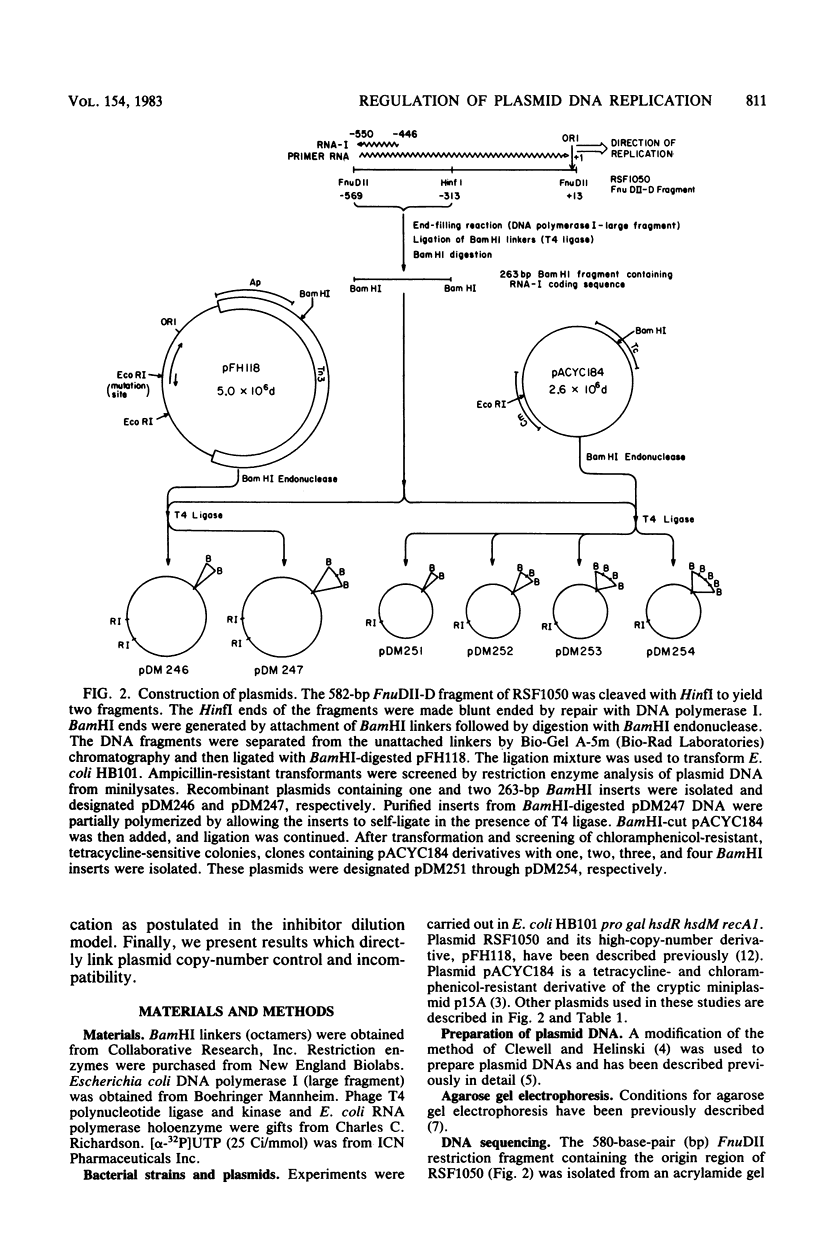
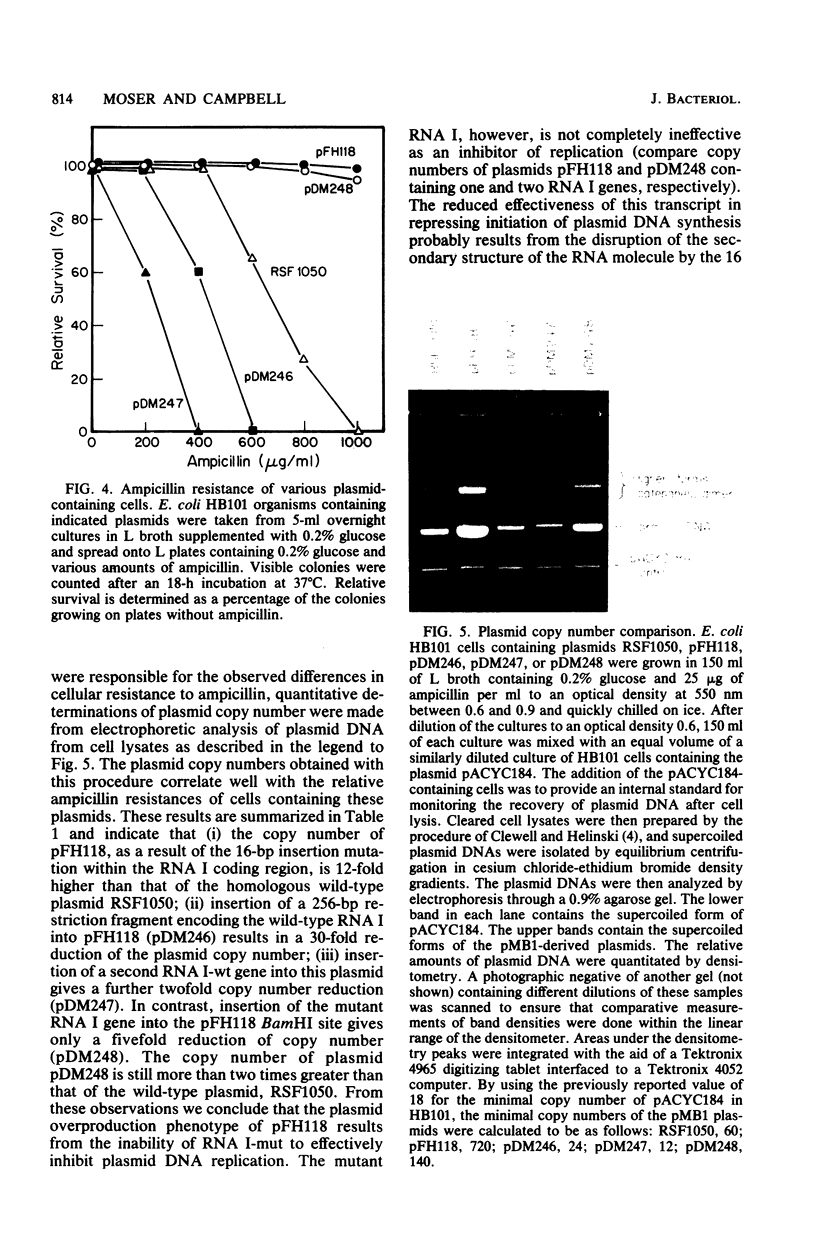
A 16-base-pair insertion has been identified as the mutation responsible for the high-copy-number phenotype of the plasmid copy number mutant pFH118. The mutation is located near the plasmid origin of replication in a region of the genome that encodes two overlapping RNA transcripts. One of these transcripts, RNA I, acts as a negative regulator of plasmid replication. The second transcript is the precursor to the primer for the initiation of DNA synthesis. We demonstrate through complementation that the pFH118 DNA overproduction phenotype is a consequence of the reduced effectiveness of the mutant RNA I at inhibiting plasmid replication and not a consequence of an altered target site on the primer precursor. In addition, a series of plasmids containing multiple RNA I-coding genes was constructed for investigating the effects of RNA I gene dosage on plasmid copy number and incompatibility. The results of this study strongly support the inhibitor dilution model of plasmid copy control with RNA I as the plasmid-specified inhibitor responsible for both copy number control and incompatibility.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Betlach M., Hershfield V., Chow L., Brown W., Goodman H., Boyer H. W. A restriction endonuclease analysis of the bacterial plasmid controlling the ecoRI restriction and modification of DNA. Fed Proc. 1976 Jul;35(9):2037–2043. [PubMed] [Google Scholar]
- Cesareni G., Muesing M. A., Polisky B. Control of ColE1 DNA replication: the rop gene product negatively affects transcription from the replication primer promoter. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6313–6317. doi: 10.1073/pnas.79.20.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad S. E., Campbell J. L. Characterization of an improved in vitro DNA replication system for Escherichia coli plasmids. Nucleic Acids Res. 1979 Jul 25;6(10):3289–3304. doi: 10.1093/nar/6.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad S. E., Campbell J. L. Role of plasmid-coded RNA and ribonuclease III in plasmid DNA replication. Cell. 1979 Sep;18(1):61–71. doi: 10.1016/0092-8674(79)90354-4. [DOI] [PubMed] [Google Scholar]
- Conrad S. E., Wold M., Campbell J. L. Origin and direction of DNA replication of plasmid RSF1030. Proc Natl Acad Sci U S A. 1979 Feb;76(2):736–740. doi: 10.1073/pnas.76.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Gotoh T., Timmis K. N. Incompatibility properties of Col E1 and pMB1 derivative plasmids: random replication of multicopy replicons. Cell. 1981 Jan;23(1):229–238. doi: 10.1016/0092-8674(81)90287-7. [DOI] [PubMed] [Google Scholar]
- Heffron F., Bedinger P., Champoux J. J., Falkow S. Deletions affecting the transposition of an antibiotic resistance gene. Proc Natl Acad Sci U S A. 1977 Feb;74(2):702–706. doi: 10.1073/pnas.74.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Heffron F., So M., McCarthy B. J. In vitro mutagenesis of a circular DNA molecule by using synthetic restriction sites. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6012–6016. doi: 10.1073/pnas.75.12.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A. 1980 May;77(5):2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacatena R. M., Cesareni G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature. 1981 Dec 17;294(5842):623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Oka A. The structure of a transcriptional unit on colicin E1 plasmid. Eur J Biochem. 1979 Jul;97(2):435–443. doi: 10.1111/j.1432-1033.1979.tb13131.x. [DOI] [PubMed] [Google Scholar]
- Richardson C. C. Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc Natl Acad Sci U S A. 1965 Jul;54(1):158–165. doi: 10.1073/pnas.54.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard H. M., Gelfand D. H., Polisky B. Analysis of a recessive plasmid copy number mutant: evidence for negative control of Col E1 replication. Cell. 1979 Oct;18(2):267–275. doi: 10.1016/0092-8674(79)90046-1. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6096–6100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T., Selzer G., Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Uhlin B. E., Nordström K. R plasmid gene dosage effects in Escherichia coli K-12: copy mutants of the R plasmic R1drd-19. Plasmid. 1977 Nov;1(1):1–7. doi: 10.1016/0147-619x(77)90003-8. [DOI] [PubMed] [Google Scholar]