Abstract
Human Rad51 (hRad51), the protein central to DNA pairing and strand exchange during homologous recombination, polymerizes on DNA to form nucleoprotein filaments. By making use of magnetic tweezers to manipulate individual DNA molecules, we measured the nucleation and growth of hRad51 nucleoprotein filaments, and their subsequent disassembly in real time. The dependence of the initial polymerization rate upon the concentration of hRad51 suggests that the rate-limiting step is the formation of a nucleus involving 5.5 ± 1.5 hRad51 monomers, corresponding to one helical turn of the hRad51 nucleoprotein filament. Polymerization is highly cooperative (i.e. a nucleation-limited reaction) at low concentrations and less cooperative (a growth-limited reaction) at high concentrations of the protein. We show that the observed preference of hRad51 to form nucleoprotein filaments on double-stranded DNA rather than on single-stranded DNA is due to the fact that it depolymerizes much faster from ssDNA than from dsDNA: indeed, hRad51 polymerizes faster on ssDNA than on dsDNA. Hydrolysis of ATP by hRad51 does not correlate with its dissociation from dsDNA. This suggests that hRad51 does not depolymerize rapidly from dsDNA after strand exchange but stays bound to the heteroduplex, highlighting the importance of partner proteins to facilitate hRad51 depolymerization from dsDNA.
INTRODUCTION
Among the various forms of DNA damage, double-strand breaks (DSB) are the most cytotoxic and genotoxic. Eukaryotic organisms use two major mechanisms to repair DSBs: non-homologous end-joining and homologous recombination. The non-homologous end-joining pathway brings any two broken DNA ends together and joins them; non-homologous end-joining can thus be mutagenic because the original DNA sequence is often altered before or during rejoining (1,2). By contrast, homologous recombination, which uses the intact, homologous sister chromatid DNA to direct the repair reaction, is predominantly error-free (2,3). The genetics and biochemistry of DSB repair by homologous recombination have been investigated extensively (4,5); it is promoted by enzymes of the Rad52 epistasis group, including, in mammalian cells, Rad51, Rad52, Rad54 and the five Rad51 paralogues (Rad51B, Rad51C, Rad51D, XRCC2 and XRCC3). In vitro, Rad51 is sufficient to promote pairing of homologous DNA molecules and to initiate strand exchange between them. By comparison with RecA, its homologue in Escherichia coli, however, Rad51 has low ATPase and strand-exchange activity in vitro (6–8). In E. coli bacteria, ATP hydrolysis is necessary to achieve strand exchange over more than a few kilobases, to realign mispaired repeats, to overcome heterologous sequences and to promote four-strand reactions. In mammals, the low ATPase activity of hRad51 may be compensated by the involvement of numerous partner proteins that are known to cooperate with Rad51 in an ATP-dependent manner during eukaryotic recombination in vivo (9–11).
When a DSB forms, the ends of the DNA break are resected by nucleases to yield 3′ ssDNA tails on which Rad51 polymerizes (4,5). Like RecA, Rad51 forms a right-handed helical nucleoprotein filament, in which the ssDNA is extended in length by a factor of 1.5 when compared to B-DNA (12–14). The pitch of the Rad51 nucleoprotein filament is 90–130 Å (13,14) and the Rad51 helix involves 6.4–6.5 monomers per helical turn (13,15,16). Each hRad51 monomer covers about three bases (on ssDNA) or base pairs (on dsDNA) (14–16). The Rad51–ATP–ssDNA complex then searches for homologous sequences among neighbouring dsDNA molecules and homologous DNA strands are exchanged when homology is found.
Although hRad51 shares many structural and functional similarities with RecA and the general process of recombination is similar for both, it differs significantly from RecA in certain biochemical characteristics. RecA has a very strong preference for nucleation on ssDNA (4,5,17) and ATP hydrolysis promotes its dissociation from ssDNA and dsDNA (17–20), whereas hRad51 shows a preferential affinity for dsDNA (21), it has a very low ATPase activity (6) and ATP hydrolysis does not seem to correlate with hRad51 dissociation from dsDNA (22,23). Since efficient strand exchange requires preferential binding to ssDNA rather than dsDNA (7,17), this might explain hRad51's weaker strand-exchange activity in vitro when compared to RecA, and suggests that one of the roles of the numerous other proteins involved in HR in vivo may be to favour formation of hRad51–ssDNA nucleoprotein filaments over dsDNA ones. Such a role has been shown for Rad54 (24,25), Rad52 (26), Rad55 and Rad57 (27) and BRCA2 (28–30).
The activity of hRad51 in vitro depends on the biochemical conditions and, especially, on the ionic environment; the role of divalent cations such as Mg2+ and Ca2+ is a key to hRad51 activity (31–33). Mg2+ is required by RecA–Rad51 family members to form the active ATP-bound nucleoprotein filament, but RecA and hRad51 require distinct Mg2+ concentrations to stimulate their activities. At high Mg2+ concentrations, the hRad51–ATP–ssDNA filament is quickly converted to an inactive hRad51–ADP–ssDNA form, due to relatively rapid ATP hydrolysis and slow dissociation of ADP. Ca2+ exerts its stimulatory effect by reducing the ATPase activity of hRad51, so maintaining the hRad51–ATP–ssDNA filament in its active form (32,33). Furthermore, Ristic et al. (34) observed by using atomic force microscopy (AFM) that substituting Ca2+ for Mg2+ resulted in a reduced ATPase rate, and lead to more regular nucleoprotein filaments.
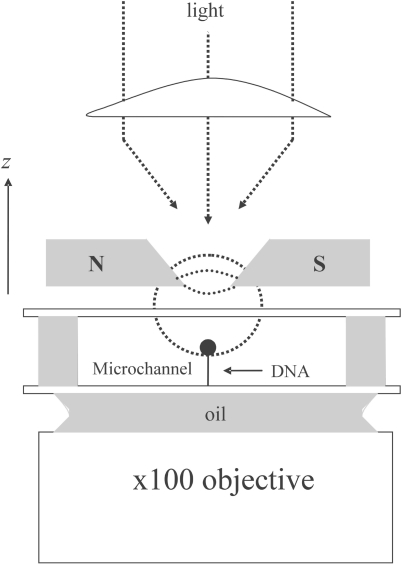
To fully understand homologous recombination, we must have knowledge of the kinetic, thermodynamic and mechanical aspects of hRad51 polymerization on both ssDNA and dsDNA. Here, we have followed this reaction in real time on single DNA molecules by monitoring the elongation of DNA manipulated with magnetic tweezers. The magnetic tweezers set-up we used to measure nucleoprotein filament length is illustrated in Figure 1 and was described in detail in previous articles (35,36). With this device, a single DNA molecule, immobilized on the surface of a microfluidic flow cell, is attached at its free end to a magnetic bead. This allows the DNA molecule to be held under tension and to be manipulated by magnets. The length of the DNA (end-to-end distance) can then be measured in real time using a three dimensional tracking of the bead's position (35).
Figure 1.
The magnetic tweezers set-up. A single DNA molecule is attached by one end to the bottom surface of a microfluidic flow cell and by the other end to a magnetic bead. A pair of magnets placed above the cell creates a magnetic field with a horizontal direction and a strong vertical gradient that pulls the bead upwards. By moving the magnets in the z dimension, one can tune the force stretching the molecule in the range of 0.001–15 pN with a precision of 10% (35, 36). The extension of the DNA molecule is given by the distance between the bead and the glass surface, measured by real-time analysis of the bead's image recorded at 120 Hz through a 100× objective, with accuracy typically better than 5 nm. By rotating the magnets, one can control the DNA topology. Here, molecules that can freely rotate around one of the backbone links due to a single-strand nick (unconstrained dsDNA) were selected by their flat length-versus-torsion profile (35).
We examined hRad51 polymerization on unconstrained dsDNA (molecules with free rotation around a single-strand nick). First, we observed hRad51 polymerization under various forces at a fixed hRad51 monomer concentration; second, we fixed the stretching force during polymerization and tested several hRad51 concentrations. We studied polymerization in the presence of ATP, ATPγS (a poorly hydrolysable analogue of ATP), and AMP-PNP (a non-hydrolysable analogue of ATP). We then examined hRad51 polymerization on ssDNA as a function of the force applied to the molecule, and in the presence of various divalent cations and cofactors. Our results highlight the differences between the binding properties of the human protein and its bacterial homologue RecA.
MATERIALS AND METHODS
Microchannel
A polydimethylsiloxane (PDMS) microchannel 2 cm long, 2 mm wide and 80 µm high was prepared according to Fulconis et al. (37), and placed on a glass coverslip of 24 mm × 40 mm (Erie Scientific Company, France) treated with anti-digoxigenin (Roche, France) for subsequent binding of digoxigenin-labelled DNA molecules. Prior to first use of the channel, bovine serum albumin (BSA; 10 mg ml−1) was injected into it and incubated overnight at 4°C to minimize adsorption of Rad51 onto the glass surface and onto the PDMS walls.
DNA construction
The DNA molecule held in the magnetic tweezers was a 14 400 base-pair fragment, ligated at one end to a multi-digoxigenin-labelled DNA fragment of 600 bp and at the other end to a multi-biotin-labelled fragment of about 820 bp. All the DNA fragments were obtained by polymerase chain reaction from phage λ DNA. The DNA molecules were then bound to a large excess of streptavidin-coated 2.8 µm magnetic beads (Dynal Biotech, Norway) in Binding Buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 50 mM NaCl) by interaction of the biotin label with the streptavidin. For experiments involving dsDNA, the biotinylated dsDNA molecules were incubated with the beads at room temperature. For experiments involving ssDNA, we heated the solution containing DNA molecules in Binding Buffer at 96°C for 90 s in order to denature the DNA duplexes, and then plunged the solution into ice to avoid rehybridization, prior to incubating the denatured DNA with the beads. In assays with dsDNA and ssDNA, the DNA-bound bead suspension was introduced at a controlled flow rate into the PDMS microchannel. After 30 min of incubation, most of the unbound beads were washed out of the channel with TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 7.5).
Preparation of hRad51 protein
The human Rad51 (hRad51) gene was inserted at the NdeI site of the pET15b expression vector (Novagen) and expressed in the E. coli JM109 (DE3) strain, which also carries an expression vector for the minor tRNAs (Codon (+) RIL, Novagen), to produce hexahistidine-N-terminus-tagged hRad51. The protein was purified on nickel–nitrilotriacetic acid (Ni–NTA)–agarose (Invitrogen, France). The hexahistidine tag was then removed from the hRad51 portion of the protein with thrombin protease (Amersham Biosciences, USA): SDS–PAGE (SDS–polyacrylamide gel electrophoresis) showed that more than 90% of the tag was eliminated. The solution was then dialysed against 100 mM Tris–acetate buffer pH 7.5 containing 5% glycerol and 7 mM spermidine to precipitate the protein. The precipitated protein was dissolved in 100 mM potassium phosphate buffer pH 7.0 containing 150 mM NaCl, 1 mM EDTA, 2 mM 2-mercaptoethanol and 10% glycerol and further purified by chromatography on a MonoQ column (Amersham Biosciences, USA). The purified hRad51 was dialysed against 20 mM HEPES-NaOH buffer pH 7.5 containing 150 mM NaCl, 0.1 mM EDTA, 2 mM 2-mercaptoethanol and 10% glycerol. The protein concentration was determined using the Bio-Rad protein assay kit using BSA as the standard protein.
Typical experimental conditions
Polymerization of hRad51 on dsDNA and ssDNA was tested in two different buffers: buffer A (2 mM MgCl2, 15 mM Tris–HCl, pH 7.5, 25 mM NaCl, 1 mM DTT, 0.05% Tween 20) and buffer B (5 mM CaCl2, 25 mM Tris–HCl, pH 7.5, 1 mM DTT, 0.05% Tween 20). The hRad51 protein was used at eight different concentrations: 3 µM, 200 nM, 150 nM, 125 nM, 100 nM, 50 nM, 25 nM and 20 nM. The hRad51 solution, prepared as described above, was injected at a constant flow (200 µl h−1) for 3 min into the PDMS microchannel to allow the protein solution to reach the DNA molecule held in the tweezer. During this buffer-exchange step, the length of the molecule could not be recorded accurately, because it was perturbed by lateral friction forces. After 3 min, when the flow was stopped, the length of the DNA was recorded by 3D tracking of the magnetic bead, as described in ref. (35).
Due to diffusion-convection processes occurring during injection of the protein into the microchannel, the protein concentration in the vicinity of the DNA is variable for a finite period. To estimate the duration of this period, we inferred the protein's diffusion coefficient from Einstein's equation:
where rRad51, the radius of one monomer (considered spherical), is ∼34 Å. This leads to DRad51 = 6 × 10−11 m2 s−1. Then, the period of concentration build-up is approximated as:
where τ is the duration of the injection step and ν is the average speed of the flow inside the channel. Taking into account the dimensions of the channel, the flow rate during the injection and the injection time (around 200 s), we calculate that the period of concentration build-up is about 1 s.
In all the experimental conditions used here, the amount of DNA bound to the coverslip in the microchannel was small enough to avoid any significant depletion of protein from its initial concentration. The volume of the cell is 3 µl; at the lowest hRad51 concentration used in our experiments (20 nM), the total number of hRad51 monomers inside the microchannel is therefore (3 × 10−6) ×(20 × 10−9) × (6 × 1023) = 3.6 × 1010. The number of DNA molecules attached to the surface of the microchannel, estimated by direct visual counting, is around 500. One DNA molecule, when completely covered by protein, captures around 5000 Rad51 monomers (with a stoichiometry of one monomer for 3 bp of DNA). Thus, the total number of proteins involved in nucleoprotein filaments cannot exceed 500 × 5000 =2.5 × 106, about 104 times less than the lowest bulk protein concentrations used in this study. The protein concentration in the bulk solution can then be considered constant throughout each experiment.
All the experiments reported in the ‘Results’ section were performed at 24 ± 2°C. Experiments performed at 20°C showed that the polymerization depends rather strongly on temperature: at 20°C, the initiation of polymerization was delayed compared to at 24°C and some DNA molecules were never covered by hRad51 within an experimental time of a few hours in the presence of ATP (data not shown).
Conversion of the polymerization velocity from nm·s−1 into monomers·s−1
To convert velocities of elongation from nm s−1 into monomers s−1, we needed to determine how many monomers are involved in elongating the nucleoprotein fibre by 1 nm. Considering that each hRad51 monomer covers 3 bp and stretches these base pairs by a factor of 1.5, the protein contribution to the end-to-end distance of the filament is calculated as 3 × 1.5 × d0, where d0 is 0.34 nm, the rise per base pair in B-DNA. In other words, we neglect the thermal fluctuations of the nucleoprotein complex and assume that, under the stretching forces used for studying hRad51 polymerization, between 2.8 and 7 pN, the nucleoprotein filament is essentially fully extended. This assumption is justified by the previously reported values of the persistence length of DNA–hRad51 nucleoprotein filaments (32), and its self-consistency with our own results will be discussed in the Results section. We also assume that the mean length of 3 bp of naked DNA along the vertical axis is 3 × d0 × L(F)/Lc, where L(F) is the distance between the ends of a the DNA molecule pulled at a force F, and Lc is its crystallographic length. L(F) is given by the force–extension response curve of the DNA molecule, and is always smaller than Lc, since deformations of the molecule by Brownian motion tends to reduce its end-to-end distance (35). Then, one monomer induces a change in the distance between the ends of the DNA molecule of 3 × d0 × [1.5−L(F)/Lc].
Calculation of the portion of DNA covered by hRad51
To calculate the portion of DNA covered by hRad51 at the end of polymerization, p, we assumed that the measured length is a linear combination of the naked DNA part and of the DNA part covered by hRad51, the covered regions being extended by a factor of 1.5. Thus,
| 1 |
where linitial is the DNA length measured before hRad51 injection, lfinal is the length of the nucleoprotein filament measured at the end of the experiment and Lc is the DNA crystallographic length (see Supplementary Data for details).
Determination of the nucleoprotein filament's persistence length
The persistence length quantifies the bending stiffness of a polymer. The persistence length of nucleoprotein filaments was measured by fitting their experimental force versus length behaviour to the worm-like chain (WLC) model (35,36) in a large range of forces (typically from 10−3 to 10 pN) after full completion of the polymerization. The nucleoprotein filament being not totally homogeneous, the persistence length extracted from the WLC model is only an average value, and it will be called effective persistence length in the following.
RESULTS
Polymerization on and depolymerization from unconstrained dsDNA
The effect of applied force on the formation of a nucleoprotein filament
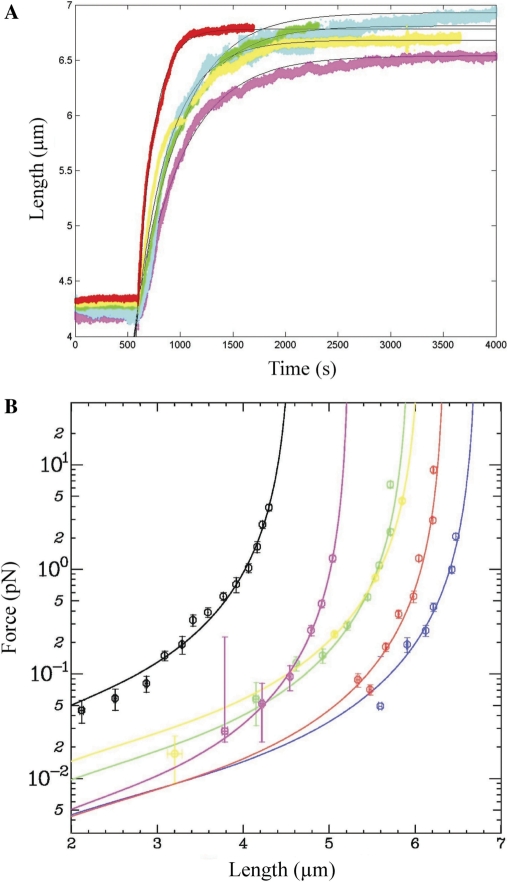
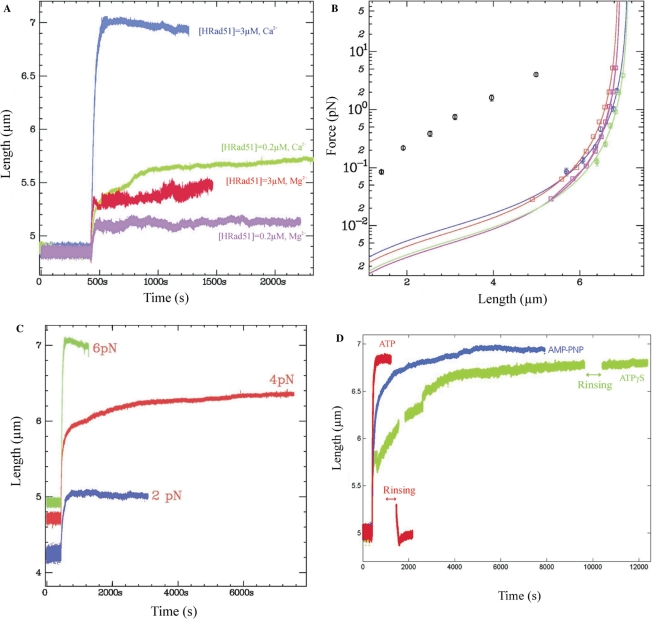
We first analysed the polymerization kinetics of hRad51 on an unconstrained dsDNA molecule held in the tweezers at various applied forces ranging from 0.5–7 pN (Figure 2). After recording the length of a single DNA molecule for 600 s, we injected 100 µM ATP and 200 nM hRad51 protein in buffer A (see ‘Materials and Methods’ section) into the microchannel. Assembly of hRad51 started immediately as indicated by elongation of the DNA molecule (Figure 2A). Each experiment was repeated at least twice for each force (total of 11 experiments). The difference between experiments performed at the same force never exceeded 5%.
Figure 2.
The effect of applied force on the formation of a hRad51–dsDNA nucleoprotein filament. (A) Polymerization of 200 nM hRad51 in buffer A containing 100 µM ATP on a single unconstrained dsDNA molecule held at 7 pN (red); 6 pN (yellow); 4 pN (green); 3 pN (cyan) or 2.8 pN (magenta). Black lines represent the fits obtained from Equation (2c) (see ‘Results’ section). (B) Extension versus force curves after polymerization at different forces. Magenta, blue, green, yellow and red curves correspond to hRad51 nucleofilaments formed respectively at 2.4, 2.6, 4.0, 5.0 and 7.0 pN. The black curve corresponds to a naked dsDNA.
When the length of the molecule reached equilibrium, the fractional DNA coverage by hRad51 proteins was calculated using Equation (1). In the range of 3–7 pN, we obtained a coverage of 80 ± 4% (mean ± SD, n = 11) of the DNA with no apparent systematic dependence upon the force applied during incubation (Table 1). This suggests that the coverage of dsDNA by hRad51 is strongly favoured thermodynamically, consistent with earlier studies (16,21).
Table 1.
The influence of the force applied on an unconstrained dsDNA on hRad51 nucleoprotein filament polymerization
| Force during incubation (pN) | Percentage of hRad51 filament (%) | Initial polymerization velocity (nm s−1) |
|---|---|---|
| 0.5 ± 0.1 | 65 ± 5 | 2.1 ± 0.1 (69 ± 1 monomers min−1) |
| 2.8 ± 0.2 | 74 ± 5 | 5.0 ± 0.1 (520 ± 10 monomers min−1) |
| 3.0 ± 0.3 | 87 ± 5 | 6.2 ± 0.1 (640 ± 10 monomers min−1) |
| 4.0 ± 0.4 | 82 ± 5 | 6.3 ± 0.1 (660 ± 10 monomers min−1) |
| 6.0 ± 0.6 | 78 ± 5 | 7.8 ± 0.2 (840 ± 20 monomers min−1) |
| 7.0 ± 0.7 | 79 ± 5 | 14.5 ± 0.4 (1550 ± 40 monomers min−1) |
We measured the initial polymerization velocity and the final filament length for each force, and calculated the relative extension (percentage of coverage by active filament) as described in the Results section.
We measured the initial polymerization rate (initial slope of the length versus time curves; see Table 1). In contrast to the extent of coverage, the initial polymerization velocity increases with the force applied, from 2.1 ± 0.1 nm s−1 (69 ± 1 monomers·min−1) at 0.5 ± 0.1 pN to 14.5 ± 0.4 nm s−1 (1550 ± 40 monomers·min−1) at 7 ± 0.7 pN.
Finally, the force versus extension response of the hRad51–ATP–dsDNA nucleoprotein filament was measured after completion of polymerization (Figure 2B), and quantitatively fitted by the extended series expansion expression for the WLC model (38). The effective persistence length presents a large variation, but no simple-to-interpret systematic trend (see Supplementary Data, Table ST1, Figure SF1 and the associated text). Averaging over all experiments involving filaments polymerized at different forces, we found an average effective persistence length of 390 ± 150 nm.
Effect of protein concentration on the formation of the nucleoprotein filament
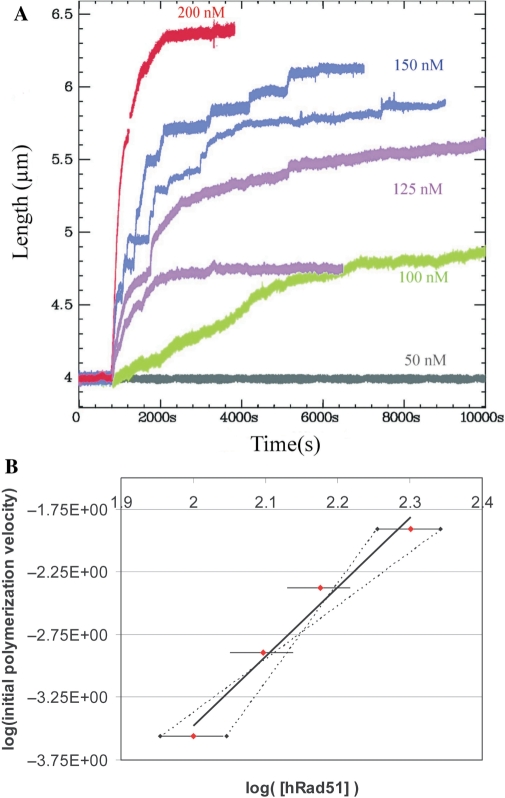
We next analysed the polymerization kinetics as a function of hRad51 concentration (Figure 3). An unconstrained dsDNA molecule was maintained in the magnetic tweezers under a force of 6 pN and the length of the DNA was measured as a function of time. After 600 s, a solution containing hRad51 and 100 µM ATP in buffer A was injected into the microchannel. The kinetics of nucleoprotein filament growth and the maximal length were measured on independent DNA molecules at seven different hRad51 concentrations: 20, 25, 50, 100, 125, 150 and 200 nM. All experiments were reproduced at least twice (Figure 3A; data obtained for two independent measurements under the same experimental conditions are represented with the same colour). For all experiments at hRad51 concentrations below 50 nM (three measurements in total), we observed no significant change in the filament length up to 3 h (Figure 3A, grey curve). At concentrations of 100–200 nM (Figure 3A, green, magenta, blue and red curves), the filament length increased significantly. The initial polymerization rate and the final length of the nucleoprotein filament depended directly on the protein concentration. Table 2 summarizes the initial polymerization rate and the percent coverage at the end of the polymerization for each hRad51 concentration. The shape of the length versus time curves also varied depending upon the protein concentration.
At 100 nM hRad51, we observed a linear increase in nucleoprotein filament length with two distinct slopes (Figure 3A, green curve), the initial polymerization rate being 50 times smaller than the initial polymerization rate at 200 nM hRad51. The final percent coverage at 100 nM hRad51 reached 28 ± 5% of the DNA molecule (Table 2).
At 125 nM hRad51, the plot of length versus time showed abrupt jumps and variability in the final length from one experiment to the other (Figure 3A, magenta curves). The final percent coverage reached 36 ± 13% of the DNA molecule (Table 2, average of the two experiments represented in magenta, Figure 3A). Note that the final length did not vary even when the duration of the incubation was extended to 5 h.
At 150 nM hRad51, variations in the final length were smaller than at 125 nM, but we observed again a stepwise increase in the length (Figure 3A, blue curves). We always observed the jumps in length at this concentration, but the position and the slope of these jumps varied (Supplementary Data). The final coverage at 150 nM reached 60 ± 3% of the DNA molecule (Table 2, average of the two experiments represented in blue, Figure 3A).
At 200 nM hRad51, we no longer observed stepwise increases in the length of the filament; the plot had a regular and very reproducible shape (Figure 3A, red curve). The coverage reached 78 ± 5% of the DNA molecule (Table 2). Moreover, once the nucleoprotein filament length reached equilibrium, it was stable for several hours in the presence of hRad51 monomers and ATP.
Figure 3.
The effect of hRad1 protein concentration on the formation of the nucleoprotein filament. (A) Polymerization on unconstrained dsDNA at 6 pN applied force in buffer A containing100 µM ATP and different concentrations of hRad51 : 50 nM hRad51 (grey); 100 nM hRad51 (green); 125 nM hRad51 (magenta); 150 nM hRad51 (blue) and 200 nM hRad51 (red). Data obtained under the same experimental conditions are represented with the same colour. (B) hRad51 concentration versus the initial slope of the curves shown in (A). The black straight line is a linear fit of the experimental data. The dotted lines represent the minimum and maximum slopes of the linear fit.
Table 2.
The influence of hRad51 concentration on the polymerization rate and the percentage of coverage. The dsDNA is maintained at 6 pN in all experiments
| hRad51 | Initial polymerization velocity (nm s−1 and monomers min−1) | Percentage of hRad51 coverage (%) |
|---|---|---|
| 20 | 0 | 0 |
| 25 | 0 | 0 |
| 50 | 0 | 0 |
| 100 | 0.3 ± 0.1 (30 ± 10 monomers min−1) | 28 ± 5 |
| 125 | 1.3 ± 0.1 (140 ± 10 monomers min−1) | 36 ± 13 |
| 150 | 4.2 ± 0.1 (450 ± 10 monomers min−1) | 60 ± 3 |
| 200 | 7.8 ± 0.2 (840 ± 20 monomers min−1) | 78 ± 5 |
The initial polymerization velocity also depended on the protein concentration. A log–log plot of hRad51 initial polymerization rate versus hRad51 concentration (Figure 3B) revealed a strongly non-linear dependence of the initial slope; a power-law fit yielded an exponent of 5.5 ± 1.5.
Depolymerization of hRad51 from a dsDNA nucleoprotein filament in the presence of ATP
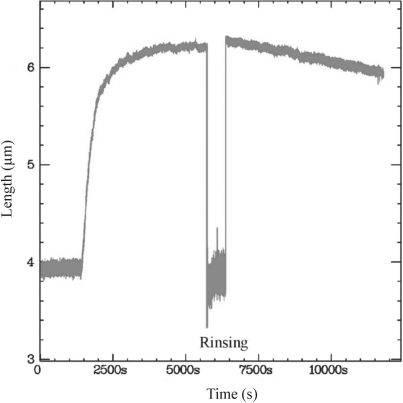
To analyse the contribution of hRad51 depolymerization to our assay, we monitored the length of a preassembled hRad51 nucleoprotein filament in the absence of hRad51 monomers (Figure 4). After polymerization at 200 nM hRad51 on dsDNA at 3 ± 0.3 pN, we removed the hRad51 monomers by rinsing the microchannel with buffer A containing 100 µM ATP for 10 min. We then measured the variation of the filament's length as a function of time, keeping the applied force at 3 pN. Immediately after the rinsing step, the nucleoprotein filament's length was unchanged: hRad51 monomers bound to the dsDNA molecule apparently did not dissociate from the DNA during rinsing. Subsequently, the filament's length decreased slowly at a constant velocity of 6.22 × 10−2 ± 1 × 10−4 nm s−1. If this length decrease was a consequence of depolymerization (see ‘Discussion’ section), it would correspond to a depolymerization rate of 6 ± 1 monomers min−1. This rate is two orders of magnitude slower than the initial polymerization rate (6.2 ± 0.1 nm s−1 or 640 ± 10 monomers min−1 when the DNA is pulled at 3.0 ± 0.3 pN; see Table 1).
Figure 4.
Depolymerization of hRad51 from dsDNA in the presence of buffer A. The dsDNA was maintained at 3 pN applied force throughout the experiment. After polymerization of 200 nM hRad51, we rinsed out the free hRad51 monomers with buffer A containing 100 µM ATP. During the rinsing step, the nucleofilament was bent due to the flow, so the apparent length decreased. After rinsing, the length of the nucleofilament was monitored for a further 100 min.
To measure the dependence of the depolymerization rate on the force exerted on the DNA, we performed similar rinsing experiments to that illustrated in Figure 4, exerting a force of 6, 3 or 1.5 pN on the filament during depolymerization (Table 3 and Supplementary Data Figure SF2). Note that in all cases polymerization was carried out under a force of 6 pN to ensure similar initial conditions. We observed that the depolymerization rate decreased exponentially with the force.
Table 3.
The effects of nucleotide cofactors on the structure of the hRad51–DNA complex
| ATP | AMP-PNP | ||
|---|---|---|---|
| dsDNA | Persistence length | 390 ± 150 nm | 190 ± 60 nm |
| Initial polymerization velocity at 6 pN | 7.8 ± 0.2 nm s−1 | 1.91 ± 0.02 nm s−1 | |
| 840 ± 20 monomers min−1 | 205 ± 2 monomers s−1 | ||
| Percentage of extension | 82 ± 2% | 45 ± 4% | |
| Depolymerization velocity at 6 pN | 2.5 × 10−2 ± 2 × 10−3 nm s−1 | 2.6 × 10−2 ± 1 × 10−3 nm s−1 | |
| 3 ± 1 monomers min−1 | 3 ± 1 monomers min−1 | ||
| Depolymerization velocity at 3 pN | 6.22 × 10−2 ± 1 × 10−4 nm s−1 | 0.40 ± 0.01 nm s−1 | |
| 6 ± 1 monomers min−1 | 41 ± 1 monomers min−1 | ||
| Depolymerization velocity at 1.5 pN | 9.1 × 10−2 ± 3 × 10−3 nm s−1 | 0.46 ± 0.01 nm s−1 | |
| 9 ± 1 monomers min−1 | 45 ± 1 monomers min−1 | ||
| SsDNA | Persistence length | 360 ± 30 nm | 460 ± 40 nm |
| Initial polymerization velocity at 6 pN (nm s−1) | 80 ± 10 nm s−1 | 29.3 ± 0.4 nm s−1 | |
| 8000 ± 1000 monomers min−1 | 2800 ± 40 monomers min−1 | ||
| Percentage of extension | 88 ± 2% | 81 ± 5% | |
| Initial depolymerization velocity at 6 pN | 4.7 ± 0.5 nm s−1 | <0.075 nm s−1 | |
| 450 ± 50 monomers min−1 |
In all these experiments, the nucleoprotein filament was held in the magnetic tweezers at an applied force of 6 pN during polymerization and the depolymerization in the presence of 0.1 mM ATP or the non-hydrolysable ATP analogue AMP-PNP. The hRad51 concentration was 200 nM in the experiments on dsDNA and 3 µM on ssDNA. For dsDNA, the filament was also maintained successively at 6, 3 and 1.5 pN to measure the force's influence on the depolymerization rate. The error bars are given by the fit for each individual experiment.
Effect of nucleotide cofactors on nucleoprotein filament polymerization and depolymerization
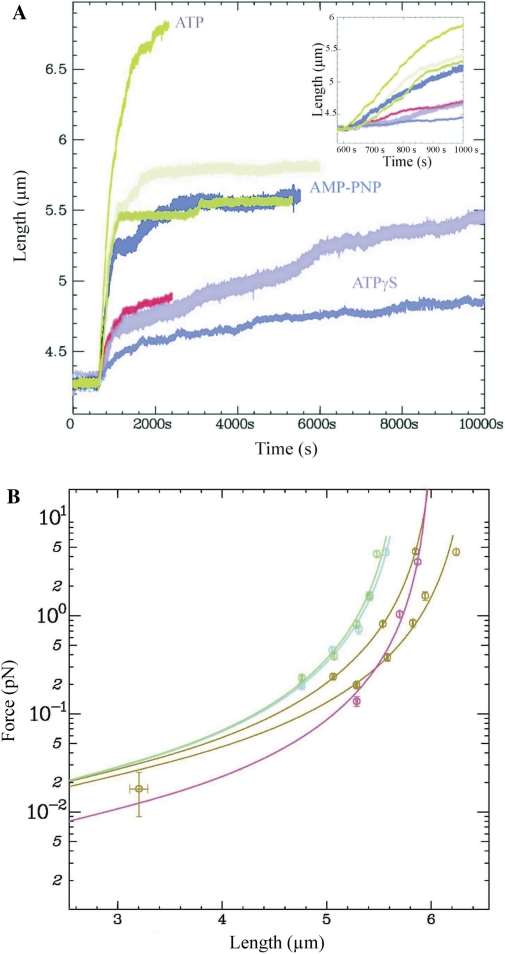
Finally, we analysed the effects of various nucleotide cofactors on the kinetics of hRad51–dsDNA filament formation. Holding a single dsDNA molecule at an applied force of 4 pN in the microchannel, we injected a solution containing 200 nM hRad51 in buffer A and 100 µM ATP, ATPγS or AMP-PNP and measured nucleoprotein filament growth with time (Figure 5). Three independent measurements were made for each nucleotide. Figure 5A presents typical length versus time curves obtained for each cofactor. In the presence of ATP, we observed a variation of less than 5% between measurements of the initial polymerization rate and of the equilibrium filament length (data not shown), whereas in the presence of ATPγS or AMP-PNP, the measurements were less reproducible (three curves obtained under the same conditions on independent molecules are plotted for both ATPγS and AMP-PNP). In each case, we evaluated the initial polymerization velocity and the percentage of extension (see supplementary discussion on the calculation of hRad51 coverage in the presence of AMP-PNP and ATPγS). The percentage of extension (Table 3) shows that, in the presence of AMP-PNP, hRad51 does not bind dsDNA as efficiently as in the presence of ATP: indeed, the final percentage of extension reached only 45 ± 4% of the DNA molecule (average of three experiments).
Figure 5.
The effect of nucleotide cofactor on the formation of the nucleoprotein filament. (A) Polymerization of 200 nM hRad51 on unconstrained dsDNA held at 4 pN in buffer A containing: 100 µM ATP (brown); 100 µM AMP-PNP (cyan, green and blue), or 100 µM ATPγS (pink, red and magenta). The insert shows the polymerization curves during the first 500s. The experiment was reproduced three times on independent molecules under the same conditions. (B) Extension versus force curves after polymerization in the presence of different cofactors as in (A): ATP (brown); AMP-PNP (blue); ATPγS (pink).
Figure 5B presents force versus extension curves of the nucleoprotein filaments formed with each cofactor; effective persistence lengths were extracted in each case by applying the WLC model (Table 3). We measured a persistence length of 190 ± 60 nm (average of seven experiments on independent molecules) in the case of nucleoprotein formed in the presence of 100 µM AMP-PNP and stretching the DNA at 6 pN. Thus, hRad51-AMP-PNP-dsDNA nucleofilaments are less rigid than hRad51-ATP-dsDNA ones: this is consistent with their lower hRad51 coverage. In the presence of ATPγS, the final extension of the hRad51-ATPγS-ssDNA was even shorter (Figure 5A) and we measured an average persistence length of 145 ± 20 nm (average of three experiments on independent molecules).
We performed, on hRad51–dsDNA filaments preassembled with AMP-PNP as the nucleotide cofactor, depolymerization experiments similar to those performed on filaments preassembled with ATP (Figure 4). After assembly, we rinsed the free hRad51 monomers out of the microchannel and followed the length of the filament over time when a force of 6, 3 or 1.5 pN was applied (Table 3). At 6 pN, depolymerization was comparable to that observed for hRad51-ATP-dsDNA. At 3 and 1.5 pN, depolymerization was six times faster in the case of hRad51-AMP-PNP-dsDNA than hRad51-ATP-dsDNA. HRad51-AMP-PNP-dsDNA nucleoprotein filaments seem to depolymerize quickly at low force whereas hRad51-ATP-dsDNA stay stable at the different forces tested.
Polymerization on and depolymerization from ssDNA
Polymerization kinetics on ssDNA
In a first series of experiments, ssDNA molecules were stretched at a force of 6 pN to unfold their secondary structures and the polymerization of various concentrations of hRad51 was studied in the presence of different divalent cations (Figure 6). Under the conditions used for polymerization on dsDNA (200 nM hRad51 in buffer A containing 100 µM ATP), the ssDNA molecule's length increased only slightly and continued fluctuating with an amplitude larger than that observed after length stabilization during polymerization experiments on dsDNA (Figure 6A, magenta curve, and Table 4). According to Equation (1), only 13 ± 5% of the ssDNA molecule was covered by hRad51, indicating less efficient hRad51 coverage of ssDNA than of dsDNA under these conditions. Even at higher hRad51 and ATP concentrations (3 µM hRad51 and 1 mM ATP), the molecule's length remained unstable and relatively short, with a percent coverage of 25 ± 5% (Figure 6A, red curve). It was reported previously that Ca2+ ions stabilize the interaction between hRad51 and ssDNA by reducing the rate of ATP hydrolysis and stimulating strand-exchange activity (32). We therefore investigated hRad51 polymerization on ssDNA in buffer B, which contains 5 mM Ca2+ (see ‘Materials and Methods’ section). At 200 nM hRad51 and 100 µM ATP, the coverage improved in the presence of Ca2+ compared to Mg2+ but it was still incomplete (36 ± 5%; Figure 6A, green curve). At 3 µM hRad51 and 1 mM ATP, however, the final extension reached 7 µm, corresponding to coverage of 88 ± 5% (Figure 6A, blue curve). In this case, we measured an effective persistence length ξ = 358 ± 30 nm (Figure 6B, blue curve). The persistence length is dramatically modified, as compared to that of ssDNA (black dots). This figure also plots the force versus length of nucleoprotein filaments assembled in the presence of ATPγS (discussed later).
Figure 6.
Kinetics of polymerization of hRad51 on ssDNA. (A) Polymerization of hRad51 on ssDNA in the presence of 5 mM Mg2+ (buffer A) or 5 mM Ca2+ (buffer B): 200 nM hRad51, 100 µM ATP, buffer A (magenta); 3 µM hRad51, 1 mM ATP, buffer A (red); 200 nM hRad51, 100 µM ATP, Buffer B (green), and 3 µM hRad51, 1 mM ATP, buffer B (blue). (B) Force versus extension curves for nucleoprotein filaments prepared under various conditions: naked ssDNA (black points); ssDNA–hRad51 filament polymerized with ATP (red); ssDNA–hRad51 filament polymerized with ATPγS (pink); ssDNA–RecA filament polymerized with ATP (blue), and ssDNA–RecA filament polymerized with ATPγS (green). Experimental data are fitted to the WLC model. (C) The influence of the stretching force on polymerization of 3 µM hRad51 on ssDNA in the presence of 1 mM ATP and 5 mM Ca2+ (buffer B): 6 pN applied force (green), 4 pN applied force (red) and 2 pN applied force (blue). (D) Polymerization and depolymerization curves for hRad51 on ssDNA using 3 µM hRad51 and buffer B in the presence of 1 mM ATP (red), 1 mM ATPγS (green) or 1 mM AMP-PNP (blue). The ssDNA was maintained at 6 pN throughout the experiment. In each experiment, the microchannel was rinsed for 10 min with buffer B containing the cofactor used during the polymerization.
Table 4.
The percentage of ssDNA length covered by hRad51 and the initial velocity of polymerization under different experimental conditions
| Buffer type | [hRad51] varies Force is fixed (6pN) | [hRad51] is fixed = 3 µM Forces varies | |||
|---|---|---|---|---|---|
| 200 nM | 3 µM | 2.0 ± 0.2 pN | 4.0 ± 0.4 pN | 6.0 ± 0.6 pN | |
| ATP, A buffer (2 mM Mg2+) | 13 ± 5% | 24 ± 5% | – | – | – |
| 7.4 ± 0.2 nm s−1 | 44.2 ± 0.2 nm s−1 | ||||
| 700±20 monomers min−1 | 4210 ± 20 monomers min−1 | ||||
| ATP, B buffer (5 mM Ca2+) | 36 ± 5% | 88 ± 5% | 24 ± 5% | 67 ± 5% | 88 ± 5% |
| 12.8 ± 0.2 nm s−1 | 80 ± 10 nm s−1 | 13 ± 1 nm s−1 | 80 ± 10 nm s−1 | 80 ± 10 nm s−1 | |
| 1220 ± 20 monomers min−1 | 8000 ± 1000 monomers min−1 | 1100 ± 90 monomers min−1 | 7400 ± 900 monomers min−1 | 8000 ± 1000 monomers min−1 | |
The error bars are given by the fit for each individual experiment. Each case of the table contains three lines: Line 1: percent coverage; Line 2: polymerization rate in nm/s; Line 3: Polymerization rate in monomers per min.
We next studied the effect of varying the force applied to a ssDNA molecule during polymerization in buffer B containing 3 µM hRad51 and 1 mM ATP. When the ssDNA was pulled at 2 or 4 pN instead of 6 pN during polymerization, the final length of the filament was shorter, with a coverage of 24 ± 5% and 67 ± 5%, respectively (Figure 6C, blue and red curves, and Table 4). Moreover, the initial polymerization velocity was directly proportional to the applied force (Table 4).
The polymerization rate of hRad51 on dsDNA, in contrast to ssDNA, was not stimulated by Ca2+ and the length versus time curves were not significantly different when calcium was used instead of magnesium (data not shown).
Depolymerization from ssDNA in the presence of ATP
We analysed the depolymerization kinetics of a hRad51–ATP–ssDNA nucleoprotein filament formed with 3 µM hRad51 and 1 mM ATP in buffer B as for the dsDNA nucleoprotein filament in Figure 4. After monitoring the polymerization on a ssDNA molecule held under an applied force of 6 pN, we rinsed out hRad51 monomers with buffer B and 1 mM ATP for 10 min. We then measured the filament length as a function of time, still pulling the filament at 6 pN (Figure 6D, red curve). In contrast to our findings with unconstrained dsDNA, the hRad51–ssDNA filament was much shorter after rinsing, indicating that a significant fraction of bound hRad51 dissociated from the ssDNA during the rinsing step. Moreover, the filament length continued to decrease sharply a few seconds after the end of the rinsing step and reached a value corresponding to the length of naked ssDNA molecule at this force. At this stage, we measured the force versus extension response of the molecule and obtained mechanical behaviour very similar to that of the initial ssDNA before polymerization (data not shown). Since the length and the mechanical behaviour of the molecule were the same before polymerization and after the rinsing step, we interpret the rapid shortening of the filament in the absence of hRad51 monomers as the consequence of rapid hRad51 depolymerization from the filament. By fitting the length versus time curve to a decreasing exponential function, we obtained a characteristic depolymerization time of 110 s, corresponding to an initial slope of 4.7 ± 0.5 nm s−1 i.e. 450 ± 50 monomers min−1 or 0.031 ± 0.003 monomer min−1 bp−1 (Table 3). For a hRad51 filament formed on dsDNA, we obtained a linear decrease with a velocity of 2.5 × 10−2 ± 2 × 10−3 nm s−1 (i.e. 3 ± 1 monomers min−1) when the filament was pulled at 6 pN (Table 3). Depolymerization from a hRad51–ATP–ssDNA filament is thus around 150 times faster than from a hRad51–ATP–dsDNA filament.
Effect of different nucleotide cofactors on the nucleoprotein filament formation and dissociation on ssDNA
We performed a similar experiment in which we replaced ATP with ATPγS and analysed the polymerization and depolymerization kinetics on ssDNA in buffer B (Figure 6D, green curve). The initial polymerization velocity was very similar to that obtained in the presence of ATP, the percentage of hRad51 coverage was 72 ± 5% and the effective persistence length was ξ = 640 ± 40 nm. In the presence of ATPγS, however, the nucleoprotein filament took around 12 times longer to reach an equilibrium length than in the presence of ATP, and the polymerization profile was irregular with some stepwise increases in length. Once equilibrium was achieved, we rinsed the flow cell of free hRad51 monomers with buffer B containing 1 mM ATPγS for 10 min and then measured the depolymerization of the hRad51–ATPγS–ssDNA filament held at 6 pN. In contrast to our findings with dsDNA, we observed no decrease in the nucleoprotein filament length, indicating no depolymerization. The same experiment was performed using AMP-PNP as a cofactor instead of ATPγS (Figure 6D, blue curve, depolymerization curve not shown). With this non-hydrolysable analogue, we obtained a final hRad51 percent coverage of 81 ± 5%, an effective persistence length of 460 ± 40 nm and a depolymerization velocity of less than 0.075 nm s−1 (Table 3).
DISCUSSION
Polymerization kinetics on dsDNA as a function of applied force
Figure 2A displays the polymerization kinetics of hRad51 on a torsionally unconstrained dsDNA molecule as a function of the force exerted on the DNA at a fixed concentration of hRad51 (200 nM) in buffer A (i.e. with Mg2+) with 0.1 mM ATP as the nucleotide cofactor. Data were fitted to a simple analytical model corresponding to random binding (black lines in Figure 2A). Since the depolymerization velocity was two orders of magnitude smaller than the initial polymerization velocity (see Figure 4 and Table 3), hRad51 depolymerization was not taken into account in this simple analysis. The number of hRad51 monomers, bound to the dsDNA molecule at a given time t is then given by:
| 2a |
| 2b |
where N is the total number of monomers that can polymerize on dsDNA, and ka is the hRad51 binding rate constant to dsDNA in s−1. Using Equation (2b), the DNA length is given by:
| 2c |
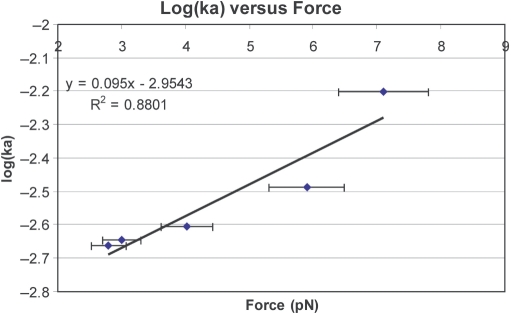
where li is the initial extension of the dsDNA before the injection of hRad51, and lf the final extension of the nucleoprotein filament measured at the end of polymerization. Figure 7 presents the binding constant ka extracted from the best fit, as a function of the force applied to the dsDNA molecule during polymerization. We observed that ka increases exponentially with this force. According to the Arrhenius law, we expect:
where Ea is the activation energy for protein binding. This exponential decrease suggests that Ea decreases linearly with the stretching force. Since a DNA molecule is stretched by a factor of 1.5 upon binding Rad51, the free energy of binding must contain a negative term linear in force (independent of the reaction path, since it is a purely mechanical term) and it is thus not surprising that this term be reflected by a linear dependence of the activation energy upon force. A semi-phenomenological microscopic model, however, suggests that only one-third of the activation energy is due to a direct mechanical contribution of the molecule's length to the free energy (see Supplementary Data, Section 3). Thus, some contribution may also arise from a deformation of the reaction path on a molecular scale.
Figure 7.
Graphic of the binding rate ka on a dsDNA versus the force applied during the polymerization.
In previous studies, Prasad et al. (39) also analysed hRad51 polymerization on a single dsDNA molecule; they attached dsDNA molecules, labelled at one end with yoyo-1 fluorophores, to a coverslip and stretched the molecules at around 1 pN by means of a liquid flow. HRad51 protein was then injected into the flow cell in the presence of ATP. These authors did not present an analytic model to fit the polymerization kinetics, but the polymerization curve they obtained was close to a sigmoid in the presence of ATP. The difference in the kinetics they observed in their study and those in our present work is probably due to differences in the set-up. With magnetic tweezers, the stretching force exerted on a DNA molecule is uniform along the molecule, whereas in the case of a molecule stretched by a hydrodynamic force, the tension is non-uniform. Since nucleation is very sensitive to the force exerted on DNA, the polymerization kinetics might differ significantly between experiments involving uniform or non-uniform forces. In addition, with hydrodynamic stretching, the force varies with time, due to changes in the length and persistence length of the molecule upon hRad51 binding.
Polymerization on ssDNA
A similar kinetic model was applied to polymerization experiments carried out on ssDNA with 3 µM hRad51 in buffer B (i.e. with Ca2+) with ATP as the nucleotide cofactor (Figure 6C). To interpret these data properly, however, we must account for hRad51 depolymerization, which is around 150 times faster than from dsDNA nucleoprotein filaments (compare Figures 4 and 6D, and see Table 3). We therefore modelled the number of hRad51 monomers n(t) bound to the ssDNA as a function of time, as follows:
| 3a |
which leads to:
| 3b |
where ka is the hRad51 binding rate constant, kd is the dissociation rate constant and N is the maximum number of hRad51 monomers that can polymerize on a ssDNA molecule. The length versus time curves were fitted to the following equation:
| 3c |
Table 5 presents the binding and dissociation constant on ssDNA obtained by fitting the experimental data to Equation (3c). We observed that stretching a ssDNA molecule increases the binding constant of hRad51 from 4.3 × 10−3 ± 1 × 10−4 s−1 at 2 pN to 33 × 10−3 ± 4 × 10−3 s−1 at 6 pN.
Table 5.
The hRad51 binding rate constant, ka, and dissociation rate constant kd as a function of the force applied to the DNA molecule
| dsDNA, [hRad51] = 200 nM, buffer A | ssDNA, [hRad51] = 3 µM, buffer B | |||
|---|---|---|---|---|
| Force pN | ka (s−1) | Force (pN) | ka (s−1) | kd (s−1) |
| 2.8 ± 0.2 | 2.2 × 10−3 ± 2 × 10−4 | 2 ± 0.2 | 4.3 × 10−3 ± 1 × 10−4 | 13 × 10−3 ± 2 × 10−3 |
| 3.0 ± 0.3 | 2.3 × 10−3 ± 3 × 10−4 | – | – | – |
| 4.0 ± 0.4 | 2.5 × 10−3 ± 1 × 10−4 | 4 ± 0.4 | 30 × 10−3 ± 4 × 10−3 | 28 × 10−3 ± 7 × 10−3 |
| 6.0 ± 0.6 | 3.2 × 10−3 ± 4 × 10−4 | 6 ± 0.6 | 33 × 10−3 ± 4 × 10−3 | 4.6 × 10−3 ± 7 × 10−3 |
| 7.0 ± 0.6 | 6.7 × 10−3 ± 3 × 10−4 | – | – | – |
| dsDNA, [hRad51] = 3 µM, buffer B | ||||
| 6 pN | 19 × 10−3 ± 1 × 10−3 | |||
For dsDNA, the experimental curves were fitted using equation 2c, whereas for ssDNA we used equation 3c. The error bars are given by the fit for each individual experiment (note that, since hRad51 binding is not a simple bimolecular process, but involves cooperativity and different dynamic regimes, these constants are effective values at a fixed hRad51 concentration, and do not necessarily reflect directly microscopic rate constants).
Finally, we compared the binding constant of hRad51 on ssDNA with that on dsDNA under the same conditions (i.e. 3 µM hRad51, 1 mM ATP in buffer B at an applied force of 6 pN, which are the best conditions among those we tested for hRad51 to polymerize on ssDNA). We found that polymerization was faster on ssDNA than on dsDNA (Table 5). Thus, in these conditions, the hRad51 thermodynamic preference for dsDNA is due entirely to the fact that the depolymerization rate from dsDNA is 150 times slower than from ssDNA. As a consequence, in the presence of ATP, ssDNA nucleoprotein filaments are in dynamic equilibrium, but dsDNA ones are not.
Initial polymerization rate on dsDNA as a function of hRad51 concentration, and estimation of the size of the polymerization nucleus
In a previous study, Galetto et al. (40) observed RecA polymerization on dsDNA at protein concentrations varying from 86–238 nM. They measured the average number of protein clusters on DNA versus time at various protein concentrations, and applied a simple kinetic model involving a single multimolecular rate-limiting step:
| 4 |
They obtained a best-fit value for α equal to 4–5, and concluded that this corresponds to the number of protein monomers involved in the rate-limiting step. This result was also confirmed by Joo et al. (41).
Applying a similar approach, we plotted the initial slopes, measured from Figure 3A, as a function of hRad51 concentration (Figure 3B), and fitted them to Equation (4). The fit yields α = 5.5 ± 1.5, a value very similar to the value 4–5 obtained for RecA: this suggests a common nucleation mechanism for RecA and hRad51. Following the argument proposed by Galetto et al. (40), this would suggest that the rate-limiting step for hRad51 binding involves 5 or 6 monomers. Interestingly, this stoichiometry is very similar (within experimental error) to 6.4, the number of hRad51 monomers per helical turn in the filament. Structural information provides a simple rationale for this interpretation: extension of a helical nucleus by the docking of a new protein involves one protein–protein interaction if the nucleus has less than one turn, but it can involve two interactions (one along the helix spiral and one from one turn to the next), if the nucleus has one full turn or more. It is thus reasonable that the binding constant and the initial polymerization rate increases after the nucleus has reached the size of one helix turn.
This interpretation is probably oversimplified, however, since Figure 3A also shows that the nucleoprotein filament polymerization curve does not have the same shape at all concentrations. This suggests complex kinetics involving several kinetic constants, which may not have the same concentration dependence (discussed further below). From a qualitative and phenomenological point of view, however, it probably remains safe to conclude that hRad51, as RecA, involves in its initial nucleation stage a nucleus of 5 to 6 monomers.
Mechanisms of hRad51 polymerization on dsDNA
The polymerization of hRad51 is more complex than that of RecA, and three qualitatively different regimes were observed when the hRad51 concentration was varied. At 50 nM and below, we observed no polymerization, even after several hours.
At 100 nM roughly linear growth of the filament and an abrupt saturation was observed (compare Figure 3A, green curve with Figure 1B of Ref. (37), or with Ref. (20)), reminiscent of the growth of RecA nucleoprotein filaments. This suggests a growth mechanism involving a single nucleation site, followed by polymerization at constant speed. We interpret the arrest of polymerization at a finite coverage in the present experiments on hRad51 as the consequence of sequence-specific nucleation and growth (see Supplementary Data).
At 125 nM and 150 nM, more complex behaviour was observed with definite stepwise growth and plateaus (Figure 3A, magenta and blue curves). This is consistent with a mechanism in which nucleation and growth occur at similar rates, so that the global increase in the molecule's length appears as a superposition of several individual events (nucleation, growth and stop). An analysis of the growth slopes revealed that they take discrete values (Supplementary Data Figures SF3 and SF4 and text), corresponding to discrete numbers of polymerization fronts growing in parallel. This allows an evaluation of the growth rate of a single front of 1 ± 0.4 nm s−1, roughly corresponding to 110 ± 40 monomers min−1. For comparison, the polymerization rate of a RecA nucleoprotein filament on dsDNA was 750 ± 12 monomers min−1 (20) in the presence of ATP, i.e. about seven times faster than hRad51.
At 200 nM hRad51 and higher concentrations, a smooth and fast exponential growth is observed in which nucleation is much faster than growth, and many discrete nucleation sites are simultaneously active so that no individual step can be discerned.
Structure and persistence length of dsDNA nucleoprotein filaments
Table 6 summarizes the data we obtained on hRad51 nucleoprotein filament assembly/disassembly and mechanical properties. Our polymerization experiments performed at high forces and at high hRad51 concentration (conditions in which polymerization is the fastest and the most reproducible), lead to a percent coverage of 82 ± 2% on dsDNA in the presence of ATP (Table 6). This was calculated assuming a rise per bp increased by a factor 1.5 ± 0.1 when compared to B-DNA. None led to a calculated coverage larger than 100%. Thus, our data are consistent with our initial assumption that hRad51 stretches both ssDNA and dsDNA by a factor 1.5 ± 0.1 when compared to B-DNA. This extension factor is close to that obtained with RecA, but some differences between the two proteins appear when the force versus length responses of the nucleoprotein filaments are compared. The persistence lengths obtained previously and under similar experimental conditions for RecA–dsDNA filaments were 450 ± 20 and 690 ± 40 nm with ATP and ATPγS, respectively (20,37,42). The effective persistence lengths of hRad51 filaments we obtained (390 ± 150 and 270 ± 80 nm for hRad51-ATP-dsDNA and hRad51-ATPγS-dsDNA nucleoprotein filaments, respectively, Table 6) are smaller than those of RecA (870 ± 130 and 960 ± 57 nm for RecA-ATP-dsDNA and RecA-ATPγS-dsDNA nucleoprotein filaments, respectively) (42). Moreover, we measured larger fluctuations from one experiment to another (Supplementary Data, Table ST1). This may be explained by differences in the polymerization mechanisms of the two proteins. RecA polymerizes by a slow nucleation followed by fast growth at all concentrations, whereas, as discussed above, hRad51 polymerization occurs by slow nucleation at low monomer concentrations (around 100 nM), and fast multiple nucleations at higher concentrations (200 nM and above), where a large coverage is obtained. Since one protein covers three base pairs at once, independent nucleation at many sites should lead to a large number of short independent nucleoprotein filaments along the DNA, nucleated at random and not in register. Thus, the hRad51 nucleoprotein filament should contain many defects (see Supplementary Data for more details). For RecA, by contrast, polymerization occurs through a small number of nucleation sites from which the protein polymerizes rapidly (20,40), and this high processivity should lead to a small number of long nucleoprotein domains. In addition, the fast depolymerization of RecA from dsDNA in the presence of ATP (120 monomers·min−1, (41)) may help to remove any remaining defects through a process of depolymerization and repolymerization. HRad51 polymerization from multiple nucleation sites and its slow depolymerization may contribute to the formation of nucleoprotein filaments with numerous and random discontinuities. This might explain why the percent coverage of DNA by hRad51 is less reproducible from one experiment to another than the same measurements with RecA, and why hRad51 filaments are less rigid than RecA filaments.
Table 6.
Summary of the data on hRad51 nucleoprotein filament assembly/disassembly and mechanical properties
| hRad51 | ||
|---|---|---|
| ssDNA | DsDNA | |
| Persistence length with ATP (WLC) | 360 ± 30 nm | 390 ± 150 nm |
| Persistence length with ATPγS (WLC) | 670 ± 30 nm | 270 ± 80 nm |
| Average percent coverage in the presence of ATP | 88 ± 2% (3 µM hRad51, 6 pN) | 82 ± 2% (200 nM hRad51, 6 pN) |
| Average percent coverage in the presence of AMP-PNP | 72 ± 5% (3 µM hRad51, 6 pN) | 45 ± 16% (200 nM hRad51, 6 pN) |
| Average percent coverage in the presence of ATPγS | 72 ± 3% (3 µM hRad51, 6 pN) | 35 ± 4% (200 nM hRad51, 6 pN) |
| Initial polymerization velocity in the presence of ATP | 80 ± 10 nm s−1 | 7.8 ± 0.2 nm s−1 |
| (nm s−1 and monomers min−1) | 8000 ± 1000 monomers min−1 (3 µM hRad51, 6 pN) | 840 ± 20 monomers min−1 (200 nM hRad51, 6pN) |
| Depolymerization velocity in the presence of ATP | 450 monomers min−1 (6 pN) | 3 monomers min−1 (6 pN) |
| Cooperativity during polymerization, with ATP (number of monomers involved in a nucleus) | – | 5.5 ± 1.5 |
Comparison with AFM experiments
Ristic et al. (34) recently reported a study of nucleoprotein hRad51 filaments assembled on dsDNA and ssDNA with various cofactors. Apart from very preliminary real-time measurements of the hRad51 polymerization by magnetic tweezers, most of this study focused on images obtained by AFM after immobilization of the DNA on a surface. Essentially, two types of filaments were observed: ‘regular’ filaments, resembling those previously reported for RecA–DNA nucleoprotein filaments, and ‘irregular’ filaments, involving proteins as individual spots distributed around a curvilinear path, on a width of typically more than 100 nm. If such a spatial distribution reflects the actual distribution of proteins along a nucleoprotein filament in solution, we might expect spectacular differences in the nucleoprotein filament length and persistence length. However, we saw no strong correlation between filament length and persistence length measured in our experiments (Table 4), or between the persistence length of the filaments we obtained in a given buffer, and the regularity of the filaments as observed in (34) with a comparable buffers.
We observed higher coverage of ssDNA by hRad51 when the filaments were assembled in the presence of Ca2+ than in the presence of Mg2+ (Table 4): this correlates with the report that filaments assembled in the presence of Ca2+ were more regular than those assembled in Mg2+ (34). For hRad51 coverage of dsDNA, however, there is less agreement between the two studies. For instance, we measured a shorter persistence length and percent coverage for nucleoprotein filaments assembled with AMP-PNP than for those assembled with ATP, whereas Ristic et al. (34) report more regular filaments when assembled in AMP-PNP rather than ATP. It is very unlikely that a spreading of hRad51 proteins as a ‘cloud’ extending over several tens of nanometres reflects the actual distribution of the proteins in the filament in solution, since this would yield very weak and unstable protein–protein interactions. More probably, these clouds result from a dissociation of proteins from the nucleoprotein filament during the immobilization process. Such artefacts can arise from numerous sources and have been reported by other groups (E. Le Cam, personal communication).
SsDNA, dsDNA and the role of ATP hydrolysis and partner proteins
A major difference between RecA and hRad51, reported widely in the literature (6,21,22), is that the RecA prefers to polymerize on ssDNA whereas hRad51 has a higher affinity for dsDNA. Our data reported here agree with this general interpretation, however, we demonstrate that hRad51 actually polymerizes faster on ssDNA than on dsDNA (Table 6, see ‘initial polymerization velocity in the presence of ATP’). Its thermodynamic preference for dsDNA is entirely due to a depolymerization rate 150 times slower from dsDNA than from ssDNA (Table 6, see ‘depolymerization velocity in the presence of ATP’). Then two questions arise: first, can we understand the reasons for this slow depolymerization on a molecular basis, and second, what consequences can it have on the strand-exchange process?
For RecA, it was shown that depolymerization is triggered by ATP hydrolysis (18), so we first investigated the effect of ATP hydrolysis on hRad51 depolymerization using poorly hydrolysable ATP analogues. We showed that in the presence of ATP, hRad51 monomers depolymerize from ssDNA with a dissociation rate of 13 × 10−3 ± 2 × 10−3 s−1 at the lowest force (2 pN, Table 5), whereas in the presence of ATPγS or AMP-PNP, depolymerization is extremely slow. Moreover, the hRad51 catalytic constant for ATP hydrolysis, kcat obtained in bulk experiment in the presence of ssDNA (6) (0.28 min−1) and the dissociation constant obtained in this study (13 × 10−3 ± 2 × 10−3 s−1 i.e. 0.8 ± 0.1 min−1) are in the same order of magnitude (note that, under our experimental conditions, the ATP hydrolysis rate is given by kcat because the ATP concentration is much larger than Km, the Michaelis constant (6)). Correlating depolymerization of hRad51 from ssDNA with ATP hydrolysis is also consistent with the results of a previous study (22) using a hRad51 mutant (Rad51K133R) that binds to ssDNA but does not hydrolyse ATP. Thus, our results are fully consistent with the suggestion, already made on the basis of bulk experiments that hRad51 depolymerization from ssDNA correlates with ATP hydrolysis.
In the case of hRad51 depolymerization from dsDNA, the situation is quite different. The kcat in the presence of dsDNA, as reported in the literature (6), is not very different from that measured in the presence of ssDNA (0.12 min−1 for dsDNA and 0.28 min−1 for ssDNA), yet we observed that depolymerization from dsDNA was 150-fold slower than from ssDNA. This suggests that ATP hydrolysis does not trigger hRad51 depolymerization from dsDNA, but that the protein remains bound to DNA in an ADP-bound form. Assuming that the rate of conversion from a hRad51–ATP–dsDNA filament to a hRad51–ADP–dsDNA filament is similar to the rate of ATP hydrolysis reported in (6) (0.28 min−1), and comparing this rate to the dissociation rate of hRad51 from dsDNA measured here, one might expect that, in the steady state, most hRad51–dsDNA nucleoprotein filaments prepared with ATP in vitro are in the hRad51–ADP–dsDNA form.
In summary, ATP hydrolysis and its consequences for nucleoprotein filament depolymerization are very different for hRad51 and RecA. First, RecA hydrolyses ATP much more rapidly than hRad51 hydrolyses ATP, and second, dissociation of RecA monomers from both ssDNA and dsDNA correlates with ATP hydrolysis (6,18–20). In contrast, dissociation of hRad51 monomers from ssDNA, correlates with ATP hydrolysis, but hRad51 dissociation from dsDNA is independent of the rate of ATP hydrolysis.
The high affinity of hRad51 for dsDNA and its slow dissociation seems counterintuitive for a DNA repair process that we might imagine would initiate on ssDNA. It suggests that, in vivo, other partners and mechanisms must come into play to restore the preference of hRad51 for ssDNA. Rad54 may be one such protein partner: hRad54 interacts with hRad51 to stabilize ssDNA nucleoprotein filaments (43).
The slow rate of hRad51 dissociation from dsDNA is also intriguing in the light of recent findings about RecA-mediated strand exchange, in which depolymerization of RecA was associated with ATP hydrolysis and shown to be essential for the release of the exchanged nucleoprotein filament after strand exchange (44–47). The detailed mechanisms involved are still unclear, but the energy from ATP hydrolysis is thought to fuel irreversible strand exchange and to bypass sequence heterologies in the RecA-mediated homologous recombination reaction. Our data show that, in the presence of ATP, hRad51 remains attached to dsDNA and depolymerization is slow. Thus, in contrast with the behaviour of RecA, spontaneous hRad51 depolymerization from heteroduplex dsDNA is probably not an efficient mechanism for terminating strand exchange and releasing the exchanged pair as a free dsDNA molecule. Another mechanism must be at work. This could be an other important role of hRad54 (beyond its stabilization effect on hRad51-ssDNA nucleoprotein filaments), since it was recently shown that Rad54 can dissociate Rad51 from dsDNA in an ATP-dependent manner (9–11). This view is also supported by recent work by Kiianitsa et al. (10) showing that Rad51 bound to heteroduplex dsDNA, the product of homologous recombination after DNA strand exchange, stimulates the Rad54 ATPase activity up to six-fold, and leads to a faster turnover of Rad51 in the product complex.
This brief comparison between hRad51 and RecA is consistent with the fact that RecA catalyses homologous pairing and strand exchange efficiently in the absence of other proteins (5) whereas hRad51 protein seems to require the assistance of numerous partners to perform the same activities. Besides Rad54 (9,10,11,43), other factors such as RPA, Rad52 (48–53) and Brca2 (54,55), have been proposed as putative partners in homologous recombination in eukaryotes. In vivo, RPA protein (the counterpart of single strand-binding protein, SSB in higher eukaryotes) removes secondary structures from ssDNA; hRad52 and Brca2 enhance the ability of hRad51 to displace RPA from ssDNA and to form extended nucleoprotein filaments on ssDNA (53–55). Our study of hRad51 nucleation, growth and dissociation shows that hRad51 lacks some of the essential functions of RecA; overall, these functions seem to correspond well to those accomplished by hRad51 partner proteins.
CONCLUSIONS
We observed in real time the formation of hRad51 nucleoprotein filaments on topologically unconstrained dsDNA and ssDNA, under various conditions and in real time. In each case, we analysed the assembly and disassembly dynamics of the DNA–hRad51 nucleoprotein filaments and measured their persistence lengths.
The polymerization kinetics of hRad51 are more complex than those of RecA and present several regimes depending on protein concentration, probably reflecting a different balance between nucleation and growth. The initial growth rate scales as the fifth or sixth power of the concentration. Following the method proposed by Galletto et al. (40) for RecA, we deduce from this power that the nucleus necessary for triggering cooperative growth of a filament may involve five or six monomers of hRad51. We also suggest that this local cooperative growth might arise from specific, protein–protein interactions that occur when a full turn of the protein helix has been assembled.
Our data also suggest that polymerization occurs by slow nucleation and fast growth at low hRad51 concentrations, leading to a high cooperativity, and by fast nucleation at multiple sites at high hRad51 concentrations, leading to a low cooperativity. This contrasts with RecA, whose polymerization at all concentrations involves a slow nucleation step followed by extensive growth.
The extensive literature on homologous recombination in both prokaryotes and eukaryotes suggests that during the evolution of higher eukaryotes, several functions accomplished solely by a RecA-like ancestral protein were divided between Rad51 and its paralogues. This greater complexity of the homologous recombination process in eukaryotes may well allow for diverse regulatory mechanisms depending on, for example, the specialization of particular cell types or their state of differentiation. We are far from a complete understanding of the associated pathways and regulation mechanisms, however. For instance, we do not understand why, during evolution, the main actor of homologous recombination has ‘lost’ its preference for ssDNA compared to dsDNA, which seemed a very efficient way of avoiding dispersal of the protein on the vast excess of dsDNA not requiring repair. Our study provides a first clue to this apparent paradox: we confirm that, from a thermodynamic point of view, the favourite substrate of hRad51 is dsDNA, however, we demonstrate that hRad51 polymerizes faster on ssDNA than on dsDNA, and that its thermodynamic preference for dsDNA is entirely due to a depolymerization rate 150 times slower from dsDNA than from ssDNA. We also confirm that hRad51 dissociation correlates with ATP hydrolysis for ssDNA but not for dsDNA. Our data suggest that hRad51–ATP–dsDNA is converted into a relatively stable hRad51–ADP–dsDNA nucleofilament, before significant depolymerization occurs. This very slow depolymerization of hRad51 from dsDNA raises intriguing questions about the mechanisms used by higher eukaryotes to free the exchanged duplex rapidly after strand exchange and to reassemble it into chromatin. In E. coli, RecA dissociation is spontaneous and associated with ATP hydrolysis but another mechanism must be at play in higher eukaryotes. This may be the role of one or several partner proteins of hRad51. In particular, recent data highlight the probable importance of Rad54, which can dissociate dsDNA–Rad51 filaments by an active, ATP-dependent translocation mechanism (10,11). Our data show that the mechanisms responsible for hRad51 dissociation in vivo and the ‘dsDNA preference paradox’ are closely linked. This makes investigation of the role of partner proteins, such as RPA, Rad52 and Rad54, all the more timely and desirable.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This study was funded by Joint Programmes « Nanoscience-(#04 2-112) » and « Informatique, Mathématique, et Physique en Biologie Moléculaire (IMPBio 04-5-238) » from « Centre National de Recherche Scientifique » and « Ministère de l’Education Nationale et de la Recherche et de la Technologie »; Agence National de la Recherche ‘PNANO’ (ANR-05-NANO-062-03). We thank A. Bancaud for the construction of the magnetic tweezers set-up and advice on the experiments. We thank H. Kurumizaka (Waseda University School of Science and Engineering, Tokyo, Japan) for providing the proteins used in preliminary experiments. We are also indebted to V. Croquette and J.F. Allemand for fruitful discussions and comments and to S. Kowalczykowski and P. Bianco for their enlightening comments. Funding to pay the Open Access publication charges for this article was provided by Agence Nationale de la Recherche « PNANO » (ANR-05-NANO-062-03).
Conflict of interest statement. None declared.
REFERENCES
- 1.Critchlow SE, Jackson SP. DNA end-joining: from yeast to man. Trends Biochem. Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 2.Krejci L, Chen L, Van Komen S, Sung P, Tomkinson A. Mending the break: two DNA double-strand break repair machines in eukaryotes. Prog. Nucleic Acids Res. Mol. Biol. 2003;74:159–201. doi: 10.1016/s0079-6603(03)01013-4. [DOI] [PubMed] [Google Scholar]
- 3.Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 4.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:1–11. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 5.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 1998;3:570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 6.Tombline G, Fishel R. Biochemical characterization of the human Rad51 protein: I. ATP hydrolysis. J. Biol. Chem. 2002;277:14417–14425. doi: 10.1074/jbc.M109915200. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Stasiak A, Masson J.-Y, Mcllwraith MJ, Stasiak A, West SC. Conformational changes modulate the activity of human Rad51 protein. J. Mol. Biol. 2004;337:10855–10860. doi: 10.1016/j.jmb.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 8.Baumann P, West SC. Role of the human Rad51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 9.Wolner B, Peterson CL. ATP-dependent and ATP-independent role for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. J. Biol. Chem. 2005;280:10855–108860. doi: 10.1074/jbc.M414388200. [DOI] [PubMed] [Google Scholar]
- 10.Kiianitsa K, Solinger JA, Heyer WD. Terminal association of Rad54 protein with the Rad51-dsDNA filament. Proc. Natl Acad. Sci. USA. 2006;103:9767–9772. doi: 10.1073/pnas.0604240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Zhang XP, Solinger JA, Kiianitsa K, Yu X, Egelman EH, Heyer WD. Rad51 and Rad54 ATPase activities are both required to modulate Rad51-dsDNA filament dynamics. Nucleic Acids Res. 2007;35:4124–4140. doi: 10.1093/nar/gkm412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shibata T, Nishinaka T, Mikawa T, Aihara H, Kurumizaka H, Yokoyama S, Ito Y. Homologous genetic recombination as an intrinsic dynamic property of a DNA structure induced by RecA/Rad51-family proteins: a possible advantage of DNA over RNA as genomic material. Proc. Natl Acad. Sci. USA. 2001;17:8425–8432. doi: 10.1073/pnas.111005198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 2004;8:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 14.Yu X, Jocabs SA, West SC, Ogawa T, Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc. Natl Acad. Sci. USA. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egelman EH. Does a stretched DNA structure dictate the helical geometry of RecA-like filaments? J. Mol. Biol. 2001;309:539–542. doi: 10.1006/jmbi.2001.4686. [DOI] [PubMed] [Google Scholar]
- 17.Roca AI, Cox MM. RecA protein: structure, function, and role in recombinational DNA repair. Prog. Nucleic Acids Res. Mol. Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 18.Arenson TA, Tsodikov OV, Cox MM. Quantitative analysis of the kinetics of end-dependent disassembly of RecA filaments from ssDNA. J. Mol. Biol. 1999;288:391–401. doi: 10.1006/jmbi.1999.2705. [DOI] [PubMed] [Google Scholar]
- 19.Lindsley JE, Cox MM. Dissociation pathway for RecA nucleoprotein filaments formed on linear duplex DNA. J. Mol. Biol. 1989;205:695–711. doi: 10.1016/0022-2836(89)90315-x. [DOI] [PubMed] [Google Scholar]
- 20.Shivashankar GV, Feingold M, Krichevsky O, Libchaber A. RecA polymerization on double-stranded DNA by using single-molecule manipulation: the role of ATP hydrolysis. Proc. Natl Acad. Sci. USA. 1999;96:7916–7921. doi: 10.1073/pnas.96.14.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazin A, Zaitseva E, Sung P, Kowalczykowski SC. Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 2000;19:1148–1156. doi: 10.1093/emboj/19.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sun P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair. 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Tombline G, Heinen CD, Shim KS, Fishel R. Biochemical characterization of the human Rad51 protein. III. Modulation of DNA binding by adenosine nucleotides. J. Biol. Chem. 2002;17:14434–14442. doi: 10.1074/jbc.M109917200. [DOI] [PubMed] [Google Scholar]
- 24.Amitani I, Baskin RJ, Kowalczykowski SC. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Alexeev A, Mazin A, Kowalczykowski SC. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 26.New JH, Kowalczykowski SC. Rad52 protein has a second stimulatory role in DNA strand exchange that complements replication protein-A function. J. Biol. Chem. 2002;277:26171–26176. doi: 10.1074/jbc.M203670200. [DOI] [PubMed] [Google Scholar]
- 27.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 28.San Filippo J, Chi P, Sehorn MG, Etchin J, Krejci L, Sung P. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J. Biol. Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stark JM, Hu P, Pierce AJ, Moynahan PE, Ellis N, Jasin M. ATP hydrolysis by mammalian Rad51 has a key role during homology-directed DNA repair. J. Biol. Chem. 2002;277:20185–20194. doi: 10.1074/jbc.M112132200. [DOI] [PubMed] [Google Scholar]
- 30.Petalcorin MI, Sansall J, Wigley DB, Boulton SJ. CeBRC-2 stimulates D-loop formation by Rad-51 and promotes DNA single-strand annealing. J. Mol. Biol. 2006;361:231–242. doi: 10.1016/j.jmb.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 31.Shim KS, Tombline G, Heinen CD, Charbonneau N, Schmute C, Fishel R. Magnesium influences the discrimination and release of ADP by human Rad51. DNA Repair. 2006;10:704–717. doi: 10.1016/j.dnarep.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 32.Bugreev DV, Mazin A. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl Acad. Sci. USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Namsaraev EA, Berg P. Interaction of Rad51 with ATP and Mg2+ induces a conformational change in Rad51. Biochemistry. 1998;37:11932–11939. doi: 10.1021/bi9810297. [DOI] [PubMed] [Google Scholar]
- 34.Ristic D, Modesti M, Van der Heijden T, Van Noort J, Dekker C, Kanaar R, Wyman C. Human Rad51 filaments on double-and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strick T, Allemand J.-F, Bensimon D, Croquette V. Behavior of supercoiled DNA. Biophys. J. 1996;74:2016–2028. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strick T, Allemand J.-F, Croquette V, Bensimon D. Twisting and stretching single DNA molecules. Prog. Biophys. Mol. Biol. 2000;74:115–140. doi: 10.1016/s0079-6107(00)00018-3. [DOI] [PubMed] [Google Scholar]
- 37.Fulconis R, Bancaud A, Allemand J.-F, Croquette V, Dutreix M, Viovy J.-L. Twisting and untwisting a single DNA molecule covered by RecA protein. Biophys. J. 2004;87:2552–2563. doi: 10.1529/biophysj.104.043059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouchiat C, Wang MD, Allemand J.-F, Strick T, Block SM, Croquette V. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys. J. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prasad TK, Yeykal CC, Greene E. Visualizing the assembly of human Rad51 filaments on double-stranded DNA. J. Mol. Biol. 2006;363:713–728. doi: 10.1016/j.jmb.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 40.Galetto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 41.Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Real time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 42.Hegner M, Smith SB, Bustamante C. Polymerization and mechanical properties of single RecA-DNA filaments. Proc. Natl Acad. Sci. USA. 1999;96:10109–10114. doi: 10.1073/pnas.96.18.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazin A, Alexeev AA, Kowalczykowski SC. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 2003;278:14029–14036. doi: 10.1074/jbc.M212779200. [DOI] [PubMed] [Google Scholar]
- 44.Fulconis R, Mine J, Bancaud A, Dutreix M, Viovv J.-L. Mechanism of RecA-mediated homologous recombination revisited by single molecule nanomanipulation. EMBO J. 2006;25:4293–4304. doi: 10.1038/sj.emboj.7601260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacFarland KJ, Shan Q, Inman RB, Cox MM. RecA as a motor protein. J. Biol. Chem. 1997;272:17675–17685. doi: 10.1074/jbc.272.28.17675. [DOI] [PubMed] [Google Scholar]
- 46.Spengler SJ, Stasiak A, Cozzarelli NR. Quantitative analysis of the contributions of enzyme and DNA to the structure of lambda integrative recombinants. Cold Spring Harb. Symp. Quant. Biol. 1984;49:745–749. doi: 10.1101/sqb.1984.049.01.084. [DOI] [PubMed] [Google Scholar]
- 47.Howard-Flanders P, West SC, Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984;309:215–220. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- 48.Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1997;12:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 49.Navadgi VN, Shukla A, Vempati RK, Rao BJ. DNA mediated disassembly of hRad51 and hRad52 proteins and recruitment of hRad51 to ssDNA by hRad52. FEBS J. 2006;273:199–207. doi: 10.1111/j.1742-4658.2005.05058.x. [DOI] [PubMed] [Google Scholar]
- 50.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 51.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;22:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 52.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast Rad51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 53.Sugiyama T, Kowalczykowsi SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 54.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates Rad51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 55.Kowalczykowski SC. Catalyst of a catalyst. Nature. 2005;433:591–592. doi: 10.1038/433591a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.