Abstract
Selenium is incorporated into proteins as selenocysteine (Sec), which is dependent on its specific tRNA, designated tRNA[Ser]Sec. Targeted removal of the tRNA[Ser]Sec gene (Trsp) in mouse hepatocytes previously demonstrated the importance of selenoproteins in liver function. Herein, analysis of plasma proteins in this Trsp knockout mouse revealed increases in apolipoprotein E (ApoE) that was accompanied by elevated plasma cholesterol levels. The expression of genes involved in cholesterol biosynthesis, metabolism and transport were also altered in knockout mice. Additionally, in two transgenic Trsp mutant mouse lines (wherein only housekeeping selenoprotein synthesis was restored), the expression of ApoE, as well as genes involved in cholesterol biosynthesis, metabolism and transport were similar to those observed in wild type mice. These data correlate with reports that selenium deficiency results in increased levels of ApoE, indicating for the first time that housekeeping selenoproteins have a role in regulating lipoprotein biosynthesis and metabolism.
Keywords: apolipoprotein E, conditional-knockout, selenocysteine tRNA, housekeeping selenoproteins, stress-related selenoproteins
The importance of trace elements in human health can be assessed by the fact that their reduction in the diet may lead to various disorders. One important dietary trace element is selenium, which has potent cancer chemo-preventive properties [1]. It also has protective roles against viral infection [2], cardiovascular and muscular disorders along with roles in mammalian development, male reproduction and immune function [3]. Selenium is incorporated into a select group of proteins, selenoproteins, in the form of the amino acid, selenocysteine (Sec). The biological function of selenium is thought to be exerted primarily by these proteins [4]. Of the 24 and 25 selenoproteins identified in mice and humans, respectively [5], several are known to have roles in cellular redox regulation (e.g. methionine sulfoxide reductase B1 (Msrb1), glutathione peroxidases (Gpxs), and thioredoxin reductases (TRs)) [6]. Selenoprotein synthesis is dependent on the unique tRNA, designated tRNA[Ser]Sec [4] which is modified post-transcriptionally for proper functioning [7]. Two bases and one nucleoside modification occur within the anticodon loop of tRNA[Ser]Sec: A37 is modified to N6-isopentenyladenosine (i6A) and U34 is modified to methylcarboxyl-5′-methyluridine (mcm5U); mcm5U is further modified on its ribosyl moiety to Um34 [8]. Interestingly, the synthesis of Um34 on tRNA[Ser]Sec is responsive to selenium status [9].
Removing the gene for tRNA[Ser]Sec (designated Trsp) causes complete loss of selenoprotein expression and ΔTrsp is embryonic lethal [10; 11]. In addition, we generated transgenic mice in which ΔTrsp mice were rescued with a mutant Trsp transgene lacking i6A at position 37 (generated by an A37 → G37 mutation, designating the resulting transgenic mouse G37) [12] or mcm5U at position 34 (generated by an T34 → A34 mutation, designating the resulting transgenic mouse A34) in the Sec tRNA[Ser]Sec transgene product [13]. Interestingly, housekeeping, but not stress-related selenoproteins are synthesized by A34 and G37 Sec tRNAs[Ser]Sec that are transcribed and modified from the corresponding A34 and G37 transgenes [12].
We have also selectively removed Trsp in hepatocytes using loxP-Cre technology which demonstrated that proper liver function is dependent on selenoprotein expression [14]. The mean life span of the hepatocyte ΔTrsp knockout mice was significantly reduced compared with their wild type counterparts [14]. These mice died suddenly as a result of hepatocellular degeneration/necrosis. Most hepatocytes had vacuolated cytoplasm and mineralization with a majority of them being apoptotic. In contrast, the liver of mice carrying A34 or G37 transgenes appeared to be normal, with their lifespan being similar to that of the corresponding phenotypically normal litter mates (designated herein as wild type).
In the present study, we examined the plasma protein profile of hepatocyte ΔTrsp and the corresponding wild type mice and observed an elevation in the level of a protein, later identified as apolipoprotein E (ApoE). The elevated level of ApoE was accompanied by an increase in plasma cholesterol levels in hepatocyte ΔTrsp mice. A comparative gene expression analysis of hepatocyte ΔTrsp and wild type mice revealed an enhanced expression of genes involved in cholesterol biosynthesis and a decreased expression of genes involved in cholesterol metabolism or transport. Thus, the increase in plasma cholesterol levels accompanied by alteration of genes involved in cholesterol biosynthesis in the hepatocyte ΔTrsp mice reflects a link between selenoproteins and cholesterol biosynthesis. Interestingly, the levels of ApoE and cholesterol in the A34 and G37 transgenic and wild type mice were similar, as demonstrated by immunodetection and plasma lipid analysis, respectively. Since A34 and G37 transgenes restore housekeeping, but not stress-related selenoprotein synthesis, the data suggest a relationship between selenoproteins and lipoproteins, wherein housekeeping selenoproteins affect lipoprotein biosynthesis and metabolism.
Materials and methods
Materials
NuPage polyacrylamide gels, polyvinylidene difluoride membranes, Trizol reagent and Superscript II reverse transcriptase were purchased from Invitrogen (Carlsbad, CA, USA). SuperSignal West Dura substrate was obtained from Pierce (Rockford, IL, USA); goat polyclonal antibodies against ApoE and bovine anti-goat horseradish peroxidase-conjugated secondary antibodies were obtained from Santa Cruz Biotechnology, Inc (Santa Cruz, CA, USA). Fairplay® II Microarray Labeling kit was obtained from Stratagene (La Jolla, CA, USA), RNA Storage Solution from Ambion Inc. (Austin, TX, USA), MinElute column from Qiagen (Valencia, CA, USA) and SYBR green supermix from Bio Rad Laboratories (Hercules, CA, USA). All other reagents were commercially available products of the highest grade.
Mice and their genotyping
Control mice (genotype Trsp+/+-AlbCre+/+, designated wild type), liver ΔTrsp knockout mice (genotype Trspfl/fl-AlbCre+/+, designated ΔTrsp) and the A34 and G37 mutant Trsp transgenic mice (designated A34 and G37 mice and described above) were obtained as given [14]. Mice were either siblings or of similar ages, ranging between 8–9 weeks. The care of mice was in accordance with the National Institutes of Health institutional guidelines under the expert direction of Dr. Kyle Stump (NCI, National Institutes of Health, Bethesda, MD, USA). DNA was extracted from mouse tail clippings and genotyped by PCR with appropriate primers described earlier [14].
Microsequencing of protein
Plasma proteins were subjected to 10% SDS/PAGE and visualized by Coomassie Blue staining. The elevated protein band observed on gels in plasma from ΔTrsp mice was excised, digested with trypsin, and subjected to ESI-MS/MS analysis at the mass spectrometry core facility, Redox Biology Center, University of Nebraska. The resulting spectra were analyzed against mouse protein database and expressed sequence tag databases.
Analysis of plasma lipids and western blotting
The analysis of plasma lipids was carried out at the Diagnostic and Research Services Branch and Department of Laboratory Medicine, National Institutes of Health, Bethesda, MD, USA. Plasma samples from wild type, ΔTrsp, A34 and G37 mice were electrophoresed on 10% polyacrylamide gels, transferred to polyvinylidene difluoride membranes and immunoblotted with antibodies against ApoE (1:1,000 dilution). Following washes with TBST (Tween supplemented TBS), the membrane was incubated with bovine anti-goat horseradish peroxidase-conjugated secondary antibodies (1:20,000) and then washed with 0.1% TBST, incubated in SuperSignal West Dura substrate and exposed to X-ray film.
Sample preparation for microarray analysis
Total RNA from the liver of wild type, ΔTrsp, A34 and G37 mice was isolated using Trizol reagent according to manufacturer’s protocol and dissolved in the RNA Storage Solution. To generate cDNA, 10 μg RNA was incubated with 1 μl oligo dT at 70°C for 5 min and cooled on ice for 1 min. In a separate tube, 2 μl of 10x StrataScript reaction buffer, 1 μl of 0.1 M dithiothreitol, 1.0 μl of 20x aminoallyl dioxy-ribonocleotide triphosphate, 0.5 μl of 40U/μl RNAase inhibitor and 2 μl of 400U/μl StrataScriptII reverse transcriptase were mixed. This mixture was added to the RNA and oligo dT mix and reverse transcription was carried out at 48°C for 2 h. The cDNA was purified on MinElute columns and eluted from the column with 10 μl elution buffer and dried for 15 min using Speed-Vac. Finally, 5 μl of 2x coupling buffer and 5 μl Cy3 and Cy5 dye were mixed into control and experimental cDNAs, respectively, and incubated in dark at room temperature for 1 h. Following incubation, the labeled cDNA was purified on a MinElute column and eluted with 10 μl of elution buffer.
Gene expression analysis
Mouse oligonucleotide glass arrays were procured from the NCI microarray facility, Frederick, MD, USA. These high-quality oligonucleotide arrays were designated Mm-MEEBO-v1.3px, with each slide having 48 blocks containing 28 rows and 28 columns each. Each slide had 36960 oligonucleotide spots with a spacing of 155 μm. Arrays were prehybridized with 40 μl prehybridization buffer (5x SSC, 1% BSA and 0.1% SDS) under a coverslip for 1 h at 42°C. The slides were then washed with deionized water and isopropanol (each wash 2 min), spin-dried and kept at room temperature.
For hybridization, the Cy3 and Cy5 labeled cDNA were combined together and mixed with 1 μL [10 μg] COT-1 DNA, preheated at 100°C for 1 min to denature the target and cooled on ice for 2 min. This mixture was added to 20 μl of 2X F-hybridization buffer (50% formamide, 10X SSC, 0.2% SDS) and warmed to 42°C. The total cDNA probe (40 μl) was added to the prehybridized array and covered with a coverslip. The slides were placed in hybridization chambers and incubated at 42°C overnight (12–16 h). Following hybridization, the slides were washed for 2 min each in 2x SSC and 0.1% SDS, 1x SSC and 0.2x SSC and spin dried.
Microarray slides were scanned in both Cy3 (532 nm) and Cy5 (635 nm) channels using an Axon GenePix 4000B scanner (Axon Instruments, Foster City, CA, USA) with a 10 μM resolution. Scanned microarray images were exported as TIFF files to GenePix Pro 3.0 software for image analysis. For data analysis, data files (in gpr format) and images (in jpeg format) were imported into the microarray database (mAdb), and analyzed by software tools provided by NCBI.
Quantitative real-time PCR analysis
Gene expression was verified by real-time PCR, using the DNA Engine Opticon® 2 Real-Time PCR Detection System (MJ-Research/Bio Rad laboratories, Hercules, CA, USA) in combination with primer sequences outlined in Table 1. Two μg of total RNA from each sample was used to synthesize first strand cDNA in a 20 μl reaction mixture by using SuperScript II reverse transcriptase enzyme and random primers. Twenty ng of cDNA was utilized for the PCR reaction, using iQ™ SYBR green supermix and 500 nM of each primer, under the following conditions: initial denaturation for 5 min at 95°C, followed by 40 cycles consisting of 20 sec at 94°C, 20 sec at 55°C and 30 sec at 72°C. Amplifying known amounts of a PCR-product generated a standard curve at the same time as the samples. The expression of various mRNAs in each sample was normalized to the expression of 18S rRNA.
Table 1.
Primers used for real-time PCR
| Gene | Forward Sequence | Reverse Sequence |
|---|---|---|
| Apoe | 5′-GAGGAACAGACCCAGCAAAT-3′ | 5′-GCCACAGAGGCCTGTATCTT-3′ |
| Cyb5r3 | 5′-CCCGACATCAAGTACCCTCT-3′ | 5′-GCCATCGATCCTAGTCGAG-3′ |
| Dhcr24 | 5′-TGCGAGTCGGAAAGTACAAG-3′ | 5′-TGGAGTTCAGCAAAGCTGTC-3′ |
| Ebp | 5′-TTGGCCTCTTCTCCATCTCT-3′ | 5′-CCTCGATCACAAGGTGAATG-3′ |
| Ldlr | 5′-TGGCCATCTATGAGGACAAA-3′ | 5′-GTGTGACCTTGTGGAACAGG-3′ |
| Pctp | 5′-TGGCATACTGGGAAGTGAAG-3′ | 5′-GACTTCTCGGGAAACTGAGG-3′ |
| Pmvk | 5′-AGGCTGAAGAGCAGACTTGG-3′ | 5′-CATGTCCCTCCGATAGGTCT-3′ |
| Star | 5′-CATTGGCCAAGAGCTCAAC-3′ | 5′-TGCTGGATGTAGGACAGCTC-3′ |
| Stard3 | 5′-CCCAGGAAGAGAACTGGAAG-3′ | 5′-CAGGATCACCTCCTGGTACA-3′ |
Results
Analysis of plasma proteins
Many proteins are synthesized in liver and transported to the plasma, and as we had shown previously the importance of selenoproteins in liver function [14], the plasma protein profiles of Δ Trsp and wild type mice were examined. Equal amounts of plasma proteins from both mouse lines were electrophoresed on polyacrylamide gels and stained with Coomassie Blue. Staining revealed elevated levels of a protein at ~35 kDa in the plasma of Δ Trsp mice (Fig. 1A). The corresponding band was excised from the gel and protein identity determined by tandem mass-spectrometry sequencing. This procedure revealed that the 35 kDa protein is ApoE (Fig. 1B).
Fig. 1.
Analysis of plasma proteins in wild type and ΔTrsp mice. (A) Plasma proteins from wild type and ΔTrsp mice were electrophoresed and stained with Coomassie Blue. The elevated protein (~35 kDa) in ΔTrsp mice is indicated by an arrow. (B) Mouse ApoE sequence. Peptides detected by MS/MS analysis are underlined and shown in bold.
ApoE protein and mRNA levels in knockout mice
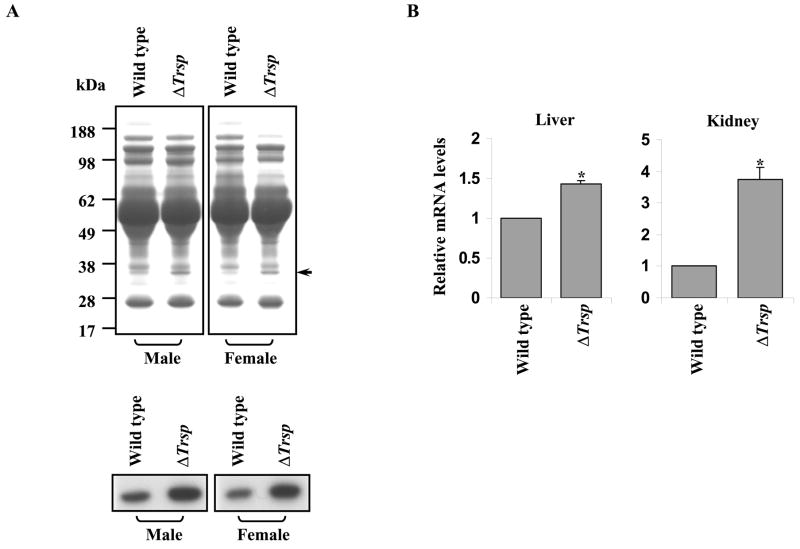
To verify that ApoE levels were increased in plasma of ΔTrsp mice, equal amounts of plasma protein from both male and female wild type and ΔTrsp mice were electrophoresed and stained with Coomassie brilliant blue (Fig. 2A). Separately, the samples were examined in immunoblot assays using polyclonal anti-ApoE antibodies (Fig. 2A, lower panel). Increased ApoE was detected in both male and female ΔTrsp mice compared to their corresponding wild type counterparts. In addition, real-time PCR revealed a 1.4 fold increase in the levels of Apoe mRNA in the liver of ΔTrsp mice (Fig. 2B). Interestingly, Apoe mRNA levels were also significantly higher (~ 3.75 fold) in kidneys of ΔTrsp mice as compared to the wild type mice.
Fig. 2.
Analysis of ApoE and Apoe mRNA in wild type and ΔTrsp mice. (A) Coomassie Blue staining of plasma proteins from male and female wild type and ΔTrsp mice (upper panel) and immunodetection of ApoE using polyclonal anti-ApoE antibodies in plasma from the same mice (lower panel). (B) The relative Apoe mRNA levels in liver and kidney samples of wild type and ΔTrsp mice, determined by real-time PCR. The level of Apoe mRNA in each sample was normalized to that of 18S rRNA and the normalized value for Apoe mRNA in ΔTrsp mice was then plotted relative to that of wild type mice along with the error bars. The results are representations of 4 independent experiments, each carried out in triplicate (*, p < 0.0005 versus wild type mice).
Analysis of cholesterol levels and genes involved in cholesterol biosynthesis
Cholesterol levels in plasma were examined in male and female wild type and ΔTrsp mice (Fig. 3A). The elevated level of ApoE was also reflected in plasma lipids with an increase in plasma cholesterol levels of ΔTrsp mice. Total plasma cholesterol was increased by 38.9% in ΔTrsp males and 35.5% in ΔTrsp females. We further focused on male mice as they had a lower mean lifespan than their female counterparts [14].
Fig. 3.
Cholesterol levels and analysis of genes involved in cholesterol metabolism in wild type and ΔTrsp mice. (A) Examination of plasma cholesterol levels in wild type and ΔTrsp male and female mice. The values represent means ± SE, n = 5 mice/sex · genotype−1 (p < 0.01 for both genders). (B) Microarray analysis, using total RNA from liver of wild type and ΔTrsp mice showing genes with altered expression involved in cholesterol biosynthesis, metabolism or transport. Results from four independent hybridizations are shown in the column designated ΔFOLD wherein an arrow pointing up indicates a relative increase, and an arrow pointing down, a relative decrease in mRNA expression; and the standard error is shown in the column designated SE. (C) Real-time PCR analysis of mRNA levels of some of the genes tabulated in Fig. 3B. The results represent 3–4 independent experiments, each carried out in triplicate and shown along with the error bars (*, p < 0.005; **, p < 0.1)
The elevation of cholesterol in the plasma of ΔTrsp mice prompted us to examine the expression profile of genes associated with cholesterol biosynthesis. Comparative analyses of gene expression using microarrays revealed altered expression of genes involved in cholesterol biosynthesis, metabolism and transport (Fig. 3B). Genes displaying a change of two-fold or more were placed into a tabular form. The loss of Trsp in liver was associated with an increased expression of several genes involved in cholesterol biosynthesis (e.g., Cyb5r3, Dhcr24, Ebp and Pmvk) and a decreased expression of genes involved in cholesterol metabolism or transport (e.g., Ldlr, Pctp, Star and Stard3). A quantitative analysis of these genes confirmed the microarray data for Cyb5r3, Dhcr24, Ebp, Pmvk and Ldlr (Fig. 3C). The transcript levels of Pctp, Star and Stard3 were too low to be detected under these experimental conditions and were not further examined.
Analysis of ApoE levels and cholesterol levels in selenoprotein replacement mice
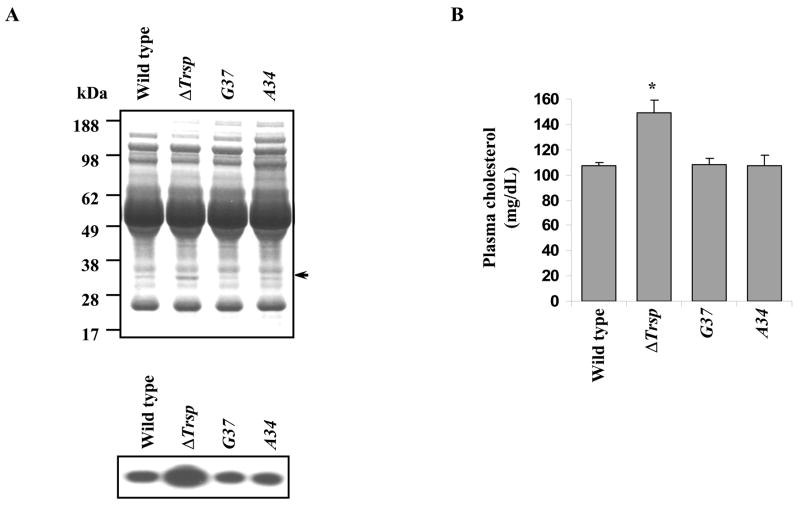
To assess whether the elevation in ApoE and cholesterol levels was due to altered expression of housekeeping (e.g. TR1 and GPX4) or stress-related (e.g. GPX1 and SELT) selenoproteins, the plasma protein profiles of wild type and ΔTrsp mice were compared to the corresponding levels in A34 and G37 transgenic mice, which are known to express the former class of selenoproteins, but express the latter class poorly [14]. Equal amounts of plasma proteins from each mouse line were electrophoresed and stained with Coomassie brilliant blue (Fig. 4A).Western blot analysis detected ApoE levels in these mice and we found that A34 and G37 mice had normal levels of ApoE (Fig. 4A, lower panel).
Fig. 4.
ApoE and cholesterol levels in A34 and G37 mice. ApoE was assessed by western blotting and cholesterol levels were measured in plasma lipids of wild type, ΔTrsp, A34 and G37 mice. (A) Coomassie Blue staining of plasma proteins from wild type, ΔTrsp, A34 and G37 male mice (upper panel) and western blotting carried out using polyclonal anti-ApoE antibodies (lower panel). (B) Plasma cholesterol levels. Values represent means ± SE, n = 5 mice/genotype for wild type and ΔTrsp mice and n = 4 mice/genotype for A34 and G37 mice (*, p < 0.01).
Plasma lipid analysis showed that although the levels of cholesterol were elevated in ΔTrsp mice with respect to wild type mice, they were virtually identical in wild type and the A34 and G37 mice (Fig. 4B). Furthermore, expression of genes associated with cholesterol biosynthesis was restored in selenoprotein replacement mice and was more in line with that in wild type mice (data not shown).
Discussion
Selenoproteins are critical for proper liver function as their loss in this organ leads to severe necrosis and hepatocellular degeneration [14]. This observation suggested that the absence of selenoproteins in liver may influence the function of other proteins. To examine whether the levels of secreted proteins are altered in the mouse model of hepatic selenoprotein deficiency, we compared the levels of major plasma proteins from ΔTrsp and wild type mice and observed an increase in a ~35 kDa protein in liver knockout mice. Microsequencing and western blotting identified this protein as ApoE. The increase in ApoE was accompanied by an increase in cholesterol levels in the plasma of ΔTrsp mice, which did not appear to be gender specific. A comparative gene expression analysis of in livers of ΔTrsp and wild type mice revealed an enhanced expression of genes involved in cholesterol biosynthesis and a decreased expression of genes involved in cholesterol metabolism and transport in ΔTrsp mice. The Apoe mRNA levels were also significantly higher in the kidney of ΔTrsp mice as compared with the kidney from wild type mice. Earlier reports suggested that Apoe is synthesized in both liver and kidney [15], and the increased mRNA levels observed in kidney could also account for elevated levels of the protein in plasma.
It was previously reported that selenium deficiency results in an increased plasma cholesterol concentration [16; 17] along with an increase in ApoE levels [18]. It was speculated that this increase was related to an increase in the HDL1 fraction [18] which is rich in ApoE. The present study analyzed the effects of the targeted removal of Trsp in liver of mice fed a selenium sufficient diet. Interestingly, the findings of increased cholesterol and elevated levels of ApoE in the ΔTrsp mice are similar to those reported earlier in rats maintained on selenium deficient diets [19]. Thus, these observations could be attributed to the absence of selenoproteins in liver, even though our mice were fed selenium sufficient diets. Since stress-related selenoproteins are more susceptible to selenium status than housekeeping selenoproteins, one might expect that the former subclass of selenoproteins is responsible for the observed effect. To test this possibility we examined levels of ApoE and cholesterol in the A34 and G37 transgenic mice. The housekeeping, but not stress-related selenoprotein population was replaced in these transgenic mice, yet the levels of ApoE and plasma cholesterol were restored to those observed in the corresponding wild type mice. Thus, the observed changes in ApoE and cholesterol levels and their restoration in mice in which housekeeping selenoproteins were expressed, suggests that stress-related selenoproteins could not account for this effect.
Several selenoprotein mRNAs were restored in A34 and G37 transgenic mice, such as Dio1, Selk, Sepp1 and Sep15 [13], while others like TR1 and TR3 were restored partially. Although mRNA levels might not necessarily match protein expression levels, our data suggest that some of these or other restored selenoproteins are responsible for the role of selenium in ApoE and cholesterol metabolism.
Earlier studies have suggested a role of selenium [20] and/or selenoproteins in cardiovascular disorders [21] and reports indicate that cholesterol levels increase in selenium deficient animals compared to adequate selenium fed animals [22]. Our studies suggest that these effects are most likely executed through a select group of selenoproteins as our mouse models have shown that the loss of Sec tRNA[Ser]Sec elicits similar conditions, even under selenium sufficient diets. It is of interest to note that the selenium content of liver from ΔTrsp and the mutant Trsp transgenic mice are similar and only about 30% of wild type. This observation provides further evidence that the selenium effect is mediated through (a) housekeeping selenoprotein(s) providing evidence for a novel role of this subclass in human health through modulation of lipoprotein and cholesterol metabolism.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research and by NIH grants to V.N.G.
Abbreviations
- ApoE
apolipoprotein E
- Sec
selenocysteine
- TBS
tris-buffered saline
- Trsp
selenocysteine tRNA gene
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Whanger PD. Selenium and its relationship to cancer: an update dagger. Br J Nutr. 2004;91:11–28. doi: 10.1079/bjn20031015. [DOI] [PubMed] [Google Scholar]
- 2.Rayman MP. The importance of selenium to human health. Lancet. 2000;356:233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 3.Hatfield DL. Selenium: Its Molecular Biology and Role in Human Health. Kluwer Academic Publishers; Norwell, MA: 2001. [Google Scholar]
- 4.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 6.Kohrl J, Brigelius-Flohe R, Bock A, Gartner R, Meyer O, Flohe L. Selenium in biology: facts and medical perspectives. Biol Chem. 2000;381:849–864. doi: 10.1515/BC.2000.107. [DOI] [PubMed] [Google Scholar]
- 7.Choi IS, Diamond AM, Crain PF, Kolker JD, McCloskey JA, Hatfield DL. Reconstitution of the biosynthetic pathway of selenocysteine tRNAs in Xenopus oocytes. Biochemistry. 1994;33:601–605. doi: 10.1021/bi00168a027. [DOI] [PubMed] [Google Scholar]
- 8.Kim LK, Matsufuji T, Matsufuji S, Carlson BA, Kim SS, Hatfield DL, Lee BJ. Methylation of the ribosyl moiety at position 34 of selenocysteine tRNA[Ser]Sec is governed by both primary and tertiary structure. RNA. 2000;6:1306–1315. doi: 10.1017/s1355838200000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatfield D, Lee BJ, Hampton L, Diamond AM. Selenium induces changes in the selenocysteine tRNA[Ser]Sec population in mammalian cells. Nucleic Acids Res. 1991;19:939–943. doi: 10.1093/nar/19.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosl MR, Takaku K, Oshima M, Nishimura S, Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc Natl Acad Sci USA. 1997;94:5531–5534. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumaraswamy E, Carlson BA, Morgan F, Miyoshi K, Robinson GW, Su D, Wang S, Southon E, Tessarollo L, Lee BJ, Gladyshev VN, Hennighausen L, Hatfield DL. Selective removal of the selenocysteine tRNA [Ser]Sec gene (Trsp) in mouse mammary epithelium. Mol Cell Biol. 2003;23:1477–1488. doi: 10.1128/MCB.23.5.1477-1488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlson BA, Xu XM, Gladyshev VN, Hatfield DL. Selective rescue of selenoprotein expression in mice lacking a highly specialized methyl group in selenocysteine tRNA. J Biol Chem. 2005;280:5542–5548. doi: 10.1074/jbc.M411725200. [DOI] [PubMed] [Google Scholar]
- 13.Carlson BA, Moustafa ME, Sengupta A, Schweizer U, Shrimali R, Rao M, Zhong N, Wang S, Feigenbaum L, Lee BJ, Gladyshev VN, Hatfield DL. Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34. J Biol Chem. doi: 10.1074/jbc.M707036200. (In press) [DOI] [PubMed] [Google Scholar]
- 14.Carlson BA, Novoselov SV, Kumaraswamy E, Lee BJ, Anver MR, Gladyshev VN, Hatfield DL. Specific excision of the selenocysteine tRNA[Ser]Sec (Trsp) gene in mouse liver demonstrates an essential role of selenoproteins in liver function. J Biol Chem. 2004;279:8011–8017. doi: 10.1074/jbc.M310470200. [DOI] [PubMed] [Google Scholar]
- 15.Blue ML, Williams DL, Zucker S, Khan SA, Blum CB. Apolipoprotein E synthesis in human kidney, adrenal gland, and liver. Proc Natl Acad Sci USA. 1983;80:283–287. doi: 10.1073/pnas.80.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stone WL, Stewart ME, Nicholas C, Pavuluri S. Effects of dietary selenium and vitamin E on plasma lipoprotein cholesterol levels in male rats. Ann Nutr Metab. 1986;30:94–103. doi: 10.1159/000177181. [DOI] [PubMed] [Google Scholar]
- 17.Stone WL, Scott RL, Stewart EM, Kheshti A. Lipoprotein alterations in the spontaneously hypertensive rat fed diets deficient in selenium and vitamin E. Proc Soc Exp Biol Med. 1994;206:130–137. doi: 10.3181/00379727-206-43731. [DOI] [PubMed] [Google Scholar]
- 18.Mazur A, Nassir F, Gueux E, Moundras C, Bellanger J, Grolier P, Rock E, Rayssiguier Y. Diets deficient in selenium and vitamin E affect plasma lipoprotein and apolipoprotein concentrations in the rat. Br J Nutr. 1996;76:899–907. doi: 10.1079/bjn19960096. [DOI] [PubMed] [Google Scholar]
- 19.Dhingra S, Bansal MP. Attenuation of LDL receptor gene expression by selenium deficiency during hypercholesterolemia. Mol Cell Biochem. 2006;282:75–82. doi: 10.1007/s11010-006-1266-1. [DOI] [PubMed] [Google Scholar]
- 20.Wojcicki J, Rozewicka L, Barcew-Wiszniewska B, Samochowiec L, Juzwiak S, Kadlubowska D, Tustanowski S, Juzyszyn Z. Effect of selenium and vitamin E on the development of experimental atherosclerosis in rabbits. Atherosclerosis. 1991;87:9–16. doi: 10.1016/0021-9150(91)90227-t. [DOI] [PubMed] [Google Scholar]
- 21.Shrimali RK, Weaver JA, Miller GF, Starost MF, Carlson BA, Novoselov SV, Kumaraswamy E, Gladyshev VN, Hatfield DL. Selenoprotein expression is essential in endothelial cell development and cardiac muscle function. Neuromuscul Disord. 2007;17:135–142. doi: 10.1016/j.nmd.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhingra S, Bansal MP. Hypercholesterolemia and apolipoprotein B expression: regulation by selenium status. Lipids Health Dis. 2005;4:28. doi: 10.1186/1476-511X-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]