Abstract
Summary: Bacteroides species are significant clinical pathogens and are found in most anaerobic infections, with an associated mortality of more than 19%. The bacteria maintain a complex and generally beneficial relationship with the host when retained in the gut, but when they escape this environment they can cause significant pathology, including bacteremia and abscess formation in multiple body sites. Genomic and proteomic analyses have vastly added to our understanding of the manner in which Bacteroides species adapt to, and thrive in, the human gut. A few examples are (i) complex systems to sense and adapt to nutrient availability, (ii) multiple pump systems to expel toxic substances, and (iii) the ability to influence the host immune system so that it controls other (competing) pathogens. B. fragilis, which accounts for only 0.5% of the human colonic flora, is the most commonly isolated anaerobic pathogen due, in part, to its potent virulence factors. Species of the genus Bacteroides have the most antibiotic resistance mechanisms and the highest resistance rates of all anaerobic pathogens. Clinically, Bacteroides species have exhibited increasing resistance to many antibiotics, including cefoxitin, clindamycin, metronidazole, carbapenems, and fluoroquinolones (e.g., gatifloxacin, levofloxacin, and moxifloxacin).
INTRODUCTION
By a variety of measures, the species Homo sapiens is more microbial than human. Microorganisms comprise only a small, albeit significant, percentage of the body weight (between 2 and 5 pounds of live bacteria). However, in terms of cell numbers, we are about 10% human and 90% bacterial (308)! Further, the number of genes in our microbiome may exceed the number of human genes by two orders of magnitude (264, 308), making us genetically 1% human and 99% bacterial! Consequently, bacteria play a major role in bodily functions, including immunity, digestion, and protection against disease (208). Colonization of the human body by microorganisms occurs at the very beginning of human life (208), and many of these organisms become truly indigenous to the host.
The human colon has the largest population of bacteria in the body (in excess of 1011 organisms per gram of wet weight), and the majority of these organisms are anaerobes; of these, ∼25% are species of Bacteroides (226), the bacterial genus that is focus of this review. This review will summarize the current state of knowledge about Bacteroides species, the most predominant anaerobes in the gut. The aspects of these organisms that will be covered will include their role as commensal organisms (The Good); their involvement in human disease (The Bad); and information about their physiology, metabolism, and resistance mechanisms as well as a brief overview of clinical characteristics (The Nitty-Gritty).
Bacteroidetes is one of the major lineages of bacteria and arose early during the evolutionary process (233). Bacteroides species are anaerobic, bile-resistant, non-spore-forming, gram-negative rods. The taxonomy of Bacteroides has undergone major revisions in the last few decades (see “Taxonomy” below), but the genus is now limited to species within the Bacteroides fragilis group, which now number >20. Names of species within the Bacteroides or Parabacteroides group to date are listed in Table 1 (146). Many of these species were isolated as single strains from human feces. The percentages of anaerobic infections that involve particular species of Bacteroides are indicated in Fig. 1 and were calculated from the Wadsworth Anaerobe Collection database, including more than 3,000 clinical specimens from which a Bacteroides species was isolated. The proportions of the most important species for the most common sites of isolation are indicated in Table 2. The numbers of B. fragilis isolates are 10- to 100-fold lower than those of other intestinal Bacteroides species, yet B. fragilis is the most frequent isolate from clinical specimens and is regarded as the most virulent Bacteroides species.
TABLE 1.
Species of the genera Bacteroides and Parabacteroides
| Species |
|---|
| Bacteroides |
| B. acidifaciens |
| B. caccae |
| B. coprocola |
| B. coprosuis |
| B. eggerthii |
| B. finegoldii |
| B. fragilis |
| B. helcogenes |
| B. intestinalis |
| B. massiliensis |
| B. nordii |
| B. ovatus |
| B. thetaiotaomicron |
| B. vulgatus |
| B. plebeius |
| B. uniformis |
| B. salyersai |
| B. pyogenes |
| B. finegoldii |
| B. goldsteinii |
| B. dorei |
| B. johnsonii |
| Parabacteroides |
| P. distasonis |
| P. merdae |
FIG. 1.
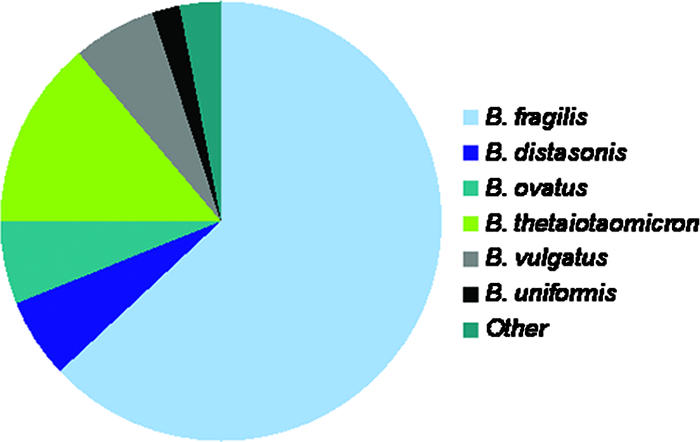
Proportions of Bacteroides species seen clinically.
TABLE 2.
Proportions of various species of the B. fragilis group found in anaerobic infections
| Site/specimen | No. of specimens | % of isolatesa
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B. caccae | B. capillosus | B. distasonis | B. fragilis | B. merdae | B. ovatus | B. putredinis | B. stercoris | B. thetaiotaomicron | B. uniformis | B. vulgatus | B. fragilis group | Totalb | ||
| Abdominal (peritoneal) | 713 | 2.8 | 0.8 | 10.0 | 31.0 | 0.4 | 10.9 | 0.4 | 1.8 | 15.4 | 4.9 | 9.4 | 7.0 | 95.0 |
| Appendix | 490 | 3.5 | 0.4 | 10.2 | 26.7 | 0.6 | 11.4 | 0.6 | 2.0 | 16.3 | 4.5 | 9.2 | 11.0 | 96.5 |
| Blood | 286 | 4.9 | 61.2 | 5.2 | 12.2 | 5.2 | 2.4 | 91.3 | ||||||
| Buttock, hip, or groin rectal | 195 | 8.7 | 35.9 | 9.2 | 15.4 | 10.3 | 5.1 | 84.6 | ||||||
| Miscellaneous wound ulcer | 182 | 44.0 | 11.0 | 14.8 | 8.2 | 11.0 | 89.0 | |||||||
| Foot abscess or ulcer | 179 | 41.3 | 8.9 | 9.5 | 5.6 | 22.3 | 87.7 | |||||||
| Leg | 86 | 2.3 | 5.8 | 36.0 | 4.7 | 23.3 | 2.3 | 7.0 | 17.4 | 98.8 | ||||
| Bone | 72 | 8.3 | 2.8 | 33.3 | 12.5 | 19.4 | 6.9 | 12.5 | 95.8 | |||||
| Bronchial specimen (including TTA)c | 66 | 4.5 | 39.4 | 3.0 | 7.6 | 7.6 | 31.8 | 93.9 | ||||||
| Chest | 63 | 3.2 | 11.1 | 9.5 | 4.8 | 61.9 | 90.5 | |||||||
| Above the neck | 53 | 3.8 | 3.8 | 22.6 | 3.8 | 13.2 | 5.7 | 39.6 | 92.5 | |||||
| Urinary tract infection | 21 | |||||||||||||
| Obstetric/gynecological | 19 | |||||||||||||
| Liver | 19 | |||||||||||||
| Bile duct | 14 | |||||||||||||
| Stump wound | 14 | |||||||||||||
| Penis | 11 | |||||||||||||
| Brain | 8 | |||||||||||||
| Skin | 7 | |||||||||||||
| Blind loop | 5 | |||||||||||||
| Hand | 5 | |||||||||||||
| Oral | 4 | |||||||||||||
| Cerebrospinal fluid | 3 | |||||||||||||
| Heart | 3 | |||||||||||||
| Arm | 3 | |||||||||||||
| Hemovac tube | 3 | |||||||||||||
| Vertebrae | 2 | |||||||||||||
| Prostrate | 2 | |||||||||||||
| Lymph | 1 | |||||||||||||
| Total | 2,529 | |||||||||||||
Proportions of species were calculated for sites for which >50 specimens were documented.
The isolates enumerated account for >80% of infections.
TTA, transtracheal aspirate.
Bacteroides may be passed from mother to child during vaginal birth and thus become part of the human flora in the earliest stages of life (208). The bacteria maintain a complex and generally beneficial relationship with the host when retained in the gut, and their role as commensals has been extensively reviewed (308). A quote in a recent publication captured this attribute: “…with B. fragilis, as with real estate, it's location, location, location” (285). When the Bacteroides organisms escape the gut, usually resulting from rupture of the gastrointestinal (GI) tract or intestinal surgery, they can cause significant pathology, including abscess formation in multiple body sites (e.g., the abdomen, brain, liver, pelvis, and lungs) as well as bacteremia.
Recent genomic and proteomic advances have greatly facilitated our understanding of the uniquely adaptive nature of Bacteroides species. The completion of the sequencing projects for B. thetaiotaomicron in 2003 (306) and B. fragilis in 2004 to 2005 (54, 138) and subsequent proteomic analyses have vastly added to our understanding of the manner in which these organisms adapt to, and thrive in, the human gut. A few examples are (i) complex systems to sense the nutrient available and tailor nutrient-metabolizing systems accordingly, (ii) multiple pump systems to rid the bacteria of toxic substances, and (iii) the ability to control the environment by interacting with the host immune system so that it controls other (competing) pathogens. We have recently shown that the expression of the various resistance-nodulation-division (RND) pumps of B. fragilis depends upon the site of isolation, another indication that the bacterium can tailor its disposal system according to its habitat (198). Additionally, comparisons of sequence analyses of the genomes of these two species suggested important mechanisms to explain the respective niches and characteristics of these organisms.
A few interesting facts that are common to both genomes have been noted. First, there is an unusually low gene content for their genome size, which reflects a large number of proteins containing >1,000 amino acids (308); many of these predicted proteins were assigned putative functions based on homology with known bacterial proteins (∼60% in B. thetaiotaomicron). Second, in both B. fragilis and B. thetaiotaomicron, extensive DNA inversions may control expression of a large number of genes. Third, both species exhibit multiple paralogous groups of genes, i.e., genes that seem to have derived from a common ancestral gene and have since diverged from the parent copy by mutation and selection or drift. The reasons for this seemingly inefficient use of genetic space are not completely clear, but it would seem that Bacteroides species are genetic “pack rats” that prefer to have all possibly needed versions of relevant proteins at hand and therefore will not need to rely on unpredictable mutations.
THE GOOD
Bacteroides as Friendly Commensals
A recent review suggested that commensal is too mild a term for the relationship of Bacteroides to its human host. The term commensal implies that one partner benefits from the relationship and the other is unaffected. The authors suggested that mutualism is a more apt description, since both the bacteria and the human experience increased fitness as a result of the relationship (8). The intestinal microbiome endows us with many features that we have not had to evolve ourselves, and we provide the organisms with “bed and board.”
Appearance in the GI tract.
Bacteroides species in the neonate appear at approximately 10 days after birth (251). Breast-fed infants do not show appreciable numbers of Bacteroides organisms in their stool until after they are weaned; in these newborns, Bifidobacterium is the major genus (150). Bacterial interactions with the host intestinal cells are facilitated by the presence of cellular and stromal components, blood, mucins, and neurons in the intestinal mucosal layer (208).
Nutrient sources for intestinal bacteria.
Polysaccharides comprise the most abundant biological polymer and, as such, also the most abundant biological food source. Carbohydrate fermentation by Bacteroides and other intestinal bacteria results in the production of a pool of volatile fatty acids that are reabsorbed through the large intestine and utilized by the host as an energy source, providing a significant proportion of the host's daily energy requirement (118). Thus, gut flora provide nutrient sources for the host as well. Studies show that germfree animals lacking a gut flora need 30% more calories to maintain body mass than normal rats (104); the gut bacteria liberate and generate simplified carbohydrates, amino acids, and vitamins. Other organisms in the gut, without the array of sugar utilization enzymes that Bacteroides has, can benefit from the presence of Bacteroides by using sugars (generated by the glycosylhydrolases) that they would otherwise be unable to use (264). For example, Bifidobacterium longum has a better system for importing simple sugars than does B. thetaiotaomicron, but B. thetaiotaomicron can break down a large variety of glycosidic bonds, providing nutrients that B. longum can then use. Also, studies with mice indicate that B. thetaiotaomicron can redirect its carbohydrate-utilizing capability from dietary to host polysaccharides according to nutrient availability (265). In another study, the adaptation of B. thetaiotomicron to utilize different nutrients during the suckling and weaning periods was investigated (27). Transcriptome analysis indicated that B. thetaiotaomicron harvested from the ceca of suckling mice has increased expression of enzymes that can utilize host-derived polysaccharides (host glycans, hexoseamines, and sialic acids that are present in mucus and the underlying gut epithelium), as well as enzymes to aid in the catabolism of mono-and oligosaccharides present in mother's milk. After weaning, the repertoire of sugar-digesting metabolic enzymes was expanded so that plant-derived polysaccharides (which would now be present in the gut) could be utilized (27).
Adaptive survival in the GI tract.
Bacteroides species have a superb ability to utilize the nutrients at hand. In the large intestine, these bacteria utilize simple and complex sugars and polysaccharides for growth (118). At sites of infection, B. fragilis may utilize host cell surface glycoproteins and glycolipids as a nutrient source; these may include simple sugars such as galactose and mannose and more complex compounds (e.g., N-acetyl-d-glucosamine [NAG]) and N-acetylneuraminic acids). Indeed, the largest paralogous group of proteins in B. thetaiotaomicron are those involved in oligo- and polysaccharide uptake and degradation (2, 3, 58, 59, 72, 73, 156, 206, 207, 229, 247, 248, 276), capsular biosynthesis, and environmental sensing/signal transduction/DNA mobilization (306). The authors of the B. thetaiotaomicron genome sequence publication suggest that these expansions “reveal strategies used by B. thetaiotaomicron to survive and to dominate in the densely populated intestinal system” (306). The coupling of these paralogs with a variety of regulatory apparatus may explain the exquisitely tuned ability of Bacteroides to sense and adapt to environmental changes and stresses, such as would normally be encountered in the gut. Another system used by Bacteroides to adapt to the human gut is its ability to modulate its surface polysaccharides by “flipping” the promoters needed for their expression to an “on” or “off” position (137); this ability may allow it to evade a host immune response.
Carbohydrate metabolism in B. thetaiotaomicron.
B. thetaiotaomicron has an extensive starch utilization system and multiple genes (sus genes) that are involved in starch binding and utilization. One hundred seventy-two glycosylhydrolases and 163 homologs of starch binding proteins (106 members homologous to SusC and 57 members homologous to SusD [306, 308]) enable the organisms to use the wide variety of dietary carbohydrates that might be available in the gut. Nearly half of the genes encoding the starch binding proteins (SusC homologs) are located next to glycosylhydrolase genes. In all, B. thetaiotaomicron contains more glycosylhydrolases than any sequenced prokaryote and appears to be able to cleave most of the glycosidic bonds found in nature (307). This ability to adapt to the use of different nutrient sources undoubtedly gives it an “edge” in its intestinal environment. These proteins may also be important in the attachment of the organism to mucus glycans.
Carbohydrate metabolism in B. fragilis.
The polysaccharide-utilizing ability of B. fragilis has not been as extensively studied, although analysis of the proteome of B. fragilis and comparison with B. thetaiotaomicron also suggests a tremendous capacity to use a wide range of dietary polysaccharides. A few years ago, we characterized a 200-kDa two-component protein (Omp200 [composed of Omp120 and Omp70, corresponding to their respective apparent molecular masses]). The intact two-component system had pore-forming ability in liposomes and black lipid bilayer membranes (two artificial systems that mimic the outer membrane of the cell and can measure pore formation). The 120-kDa component of this porin had significant homology to B. thetaiotaomicron SusC proteins (301). While Omp71 had no detectable similarity to SusD, it had homologs in the B. thetaiotomicron genome that are positioned next to a SusC homolog. Xu and Gordon speculated that the B. fragilis SusC-like component may be a conserved component of multifunctional outer membrane proteins. These multifunctional complexes may be divided into two groups: those with a downstream susD homolog that may affect acquisition/utilization of polysaccharides and those with homologs of omp71, encoding a protein whose function has not yet been defined (308).
The B. fragilis neuraminidase enzyme (product of the nanH gene) catalyzes the removal of terminal sialic acid from surface polysaccharides (105), and nanH mutants are often growth deficient. Because NAG is used in cell wall production, the ability to use extracytoplasmic NAG facilitates cell growth (181). Possibly, neuraminidase activity may render other carbon sources available when glucose levels are reduced (105), thus serving the nutritional requirements of the bacterium.
Miscellaneous enzymes used in sugar transport or utilization.
Many bacteria have transport-linked phosphorylation systems that allow sugars transported into the cells to be immediately utilized in pathways for energy metabolism or biosynthesis; any sugar transported across the cell membrane by these phosphotransfer systems can immediately enter metabolic or biosynthetic pathways. Genes for these systems were not found in the genome of either B. fragilis or B. thetaiotaomicron. Thus, they must have alternate ways of transporting sugars into the cell and attaching an active phosphate moiety. Recently, two broad-specificity hexokinases from B. fragilis were characterized, and their roles in hexose and NAG utilization were studied (31). These enzymes allow utilization of nutrients found in the gut (undigested dietary polysaccharides and host-derived glycoproteins) and at sites of infection (host cell surface antigens [including the Lewis antigen] and glycolipids) (31).
Association of levels of intestinal Bacteroides with obesity.
During the last 2 years, there have been a number of reports in prominent journals pointing out that the respective levels of the two main intestinal phyla, the Bacteroidetes and the Firmicutes, are linked to obesity, both in humans and in germfree mice (102, 143, 144, 280). The authors of the studies deduce that carbohydrate metabolism is the important factor. They observe that the microbiota of obese individuals are more heavily enriched with bacteria of the phylum Firmicutes and less with Bacteroidetes, and they surmise that this bacterial mix may be more efficient at extracting energy from a given diet than the microbiota of lean individuals (which have the opposite proportions) (280). In some studies, they found that the relative abundance of Bacteroidetes increases as obese individuals lose weight and, further, that when the microbiota of obese mice are transferred to germfree mice, these mice gain more fat than a control group that received microbiota from lean mice (280). Until very recently, reports in the literature agreed that B. thetaiotaomicron had more glycosylhydrolases than any sequenced prokaryote and appeared to be able to cleave most of the glycosidic bonds found in nature (307). However, the most recent genomic analysis found that environmental gene tags coding for many enzymes involved in the initial steps in breaking down otherwise indigestible dietary polysaccharides were enriched in obese mice (note that these were ob/ob homozygous mice with a defective leptin gene as well). The genome of Eubacterium rectale (a member of the Firmicutes division), which has not been completed, is significantly enriched for glycoside hydrolases compared to several completed genomes of Bacteroides species (280).
While it is not completely clear how significant these differences are or how well they will translate into human equivalents (9), they have, in fact, been extended to the human diet. A very recent study found that diets that are based on a high intake of protein but a low intake of fermentable carbohydrate (e.g., many of the popular diets, including Atkins, South Beach, etc.) may alter the gut flora. These workers found that proportions of Bacteroides and several clusters of Clostridia were not altered but that numbers of Roseburia, Eubacterium rectale, and bifidobacteria decreased significantly as carbohydrate intake decreased (79). Furthermore, human colonic butyrate-producing organisms that are related to Roseburia spp. and Butyrivibrio fibrisolvens showed an increased ability to use a variety of starches for growth compared to B. thetaiotaomicron (202).
Adaptation of B. thetaiotomicron and B. fragilis to their respective microenvironments.
While both B. thetaiotaomicron and B. fragilis contain large numbers of paralogous genes, comparison of the two genomes suggests that they are specifically tailored for their respective microenvironments. For example, B. fragilis has a pronounced capacity to create variable surface antigenicities by multiple DNA inversion systems (138). This surface-altering capability is more developed in B. fragilis, which is more frequently found at the mucosal surface (i.e., often the site of attack by host defenses) than is B. thetaiotaomicron. Also, the ability of B. fragilis to tolerate and use oxygen may account for the observation that it is found in greatest numbers at the mucosal surface, where the PO2 should be higher than it is within the intestinal lumen (13). The impressive capacity to utilize polysaccharides is more pronounced in B. thetaiotaomicron, which is more concentrated within the colon. The multiplicity of sensing systems of B. thetaiotaomicron, discussed below, also allow fine-tuned and efficient recruitment of the appropriate carbohydrate utilization systems. This was aptly illustrated by a very recent study that demonstrated adaptations in B. thetaiotomicron in the guts of mice during the suckling period and after weaning. By analyzing whole-genome transcriptional profiles of the bacterium harvested from the intestines of mice at different time points, the authors demonstrated that in sucking animals, glucose/galactose transporters and other glycosidases (enzymes that would be important in using host glycans sources) were expressed at higher levels, whereas B. thetaiotaomicron harvested from mice in the weaned stage showed increased expression of genes for enzymes that can liberate sugars from plant polysaccharides. Thus, during the suckling period, B. thetaiotomicron preferentially used host-derived polysaccharides as well as mono-and oligosaccharides present in mother's milk, and after weaning, this organism expanded its metabolism to exploit abundant polysaccharides of plant origin (27).
Environmental Sensing Systems
Beneficial symbiosis requires that the bacteria can sense changes in the environment so that they can adapt to alterations in their surroundings. The genome studies of B. thetaiotaomicron reveal that they have multiple genes encoding signal sensing systems; these include σ-factors and two-component regulatory systems. The function of these systems in Bacteroides is not understood to the extent that they are understood in aerobic bacteria, but indications are that they serve similar functions.
ECF σ-factors.
σ-factors are essential dissociable protein subunits of prokaryotic RNA polymerase that are necessary for initiation of transcription. These factors provide promoter recognition specificity to the polymerase and contribute to DNA strand separation; they then dissociate from the RNA polymerase core. In some cases the factor may regulate large numbers of prokaryotic genes, and in some cases the genes comprising a sigma factor regulon have a clearly defined function (131). One class of these factors, the extracytoplasmic function σ-factors, known as ECF-type σ-factors, are relatively small proteins (65). They are frequently associated with specific membrane-tethered cognates, known as anti-σ-factors. This cognate may receive a signal causing it to release its σ-factor; the released σ-factor can then interact with RNA polymerase to initiate transcription.
B. thetaiotaomicron contains the largest proportion and number of ECF σ-factors among the species of Bacteria and Archaea for which complete genome data are available (307). Approximately half of the ECF σ-factor genes are located next to open reading frames encoding putative anti-σ-factors. Further, all but one of these ECF σ-factor/anti σ-factor pairs is located upstream of open reading frames encoding homologs of the polysaccharide binding susC gene products. While the starch binding proteins are located on the cell surface, the starch-degrading enzymes are located in the periplasm, perhaps to allow Bacteroides exclusive access to the substrate (3). However, genomic analysis of B. thetaiotomicron identified a host of glycosyl hydrolases (α-galactosidases, β-galactosidases, α-glucosidases, β-glucosidases, β-glucuronidases, β-fructofuranosidases, α-mannosidases, amylases, and endo-1,2-β-xylanases, plus 14 other activities); 61% of these glycosylhydrolases are predicted to be in the periplasm or outer membrane or to be extracellular and may be important in shaping the nutrient availability of the intestinal ecosystem (306). Taken together, these results suggest a finely tuned regulatory system that allows B. thetaiotaomicron to sense the nutrients at hand and adjust its metabolism accordingly, thus benefiting (i) itself, (ii) other bacteria that cannot utilize the complex polysaccharide, and (iii) the human host, who obtains 10 to 15% of his/her caloric intake through microbial fermentation of oligosaccharides (307).
Two-component signal transduction systems.
The architecture and regulation of two component signal transduction systems have been extensively studied (268). These systems allow organisms to sense and respond to changes in many different environmental conditions. The prototype structure is well conserved and includes a histidine protein kinase that is regulated by environmental stimuli. In response to a stimulus, this protein autophosphorylates at a histidine residue, creating a high-energy phosphoryl group. The phosphoryl group is subsequently transferred to an aspartate residue in the response regulator protein; this induces a conformational change in the regulatory domain that results in activation of an associated downstream domain and causes the response (268). The majority of response regulators are transcription factors with DNA-binding effector domains, although some have C-terminal domains that function as enzymes. Examples of this system include regulation of the differential expression of ompF and ompC by the EnvZ-OmpR system in Escherichia coli and of the commitment to sporulation by the Spo system in Bacillus subtilis (268). While there are few functional studies of these systems in Bacteroides, it is reasonable to assume that they are similar to those described for other organisms. For example, expression of a two-component regulatory system gene from Bacteroides that was cloned into a multicopy plasmid vector in E. coli resulted in a decrease in the level of the outer membrane porin protein OmpF and an increase in the level of the outer membrane porin protein OmpC (204).
One Bacteroides two-component regulatory system which has been extensively studied is the RteA-RteB two-component system. The tetracycline resistance gene, tetQ, is part of the rteA-rteB-tetQ operon, which is located on a mobile element (CTnDot [see below]) found in many strains of Bacteroides. RteA is the sensor component, and RteB is the transcriptional regulator that controls the expression of a third downstream gene, rteC. The RteC product, in turn, controls the expression of a gene cluster (orf2C) that is important for excision (and therefore of transfer) of the CTnDot element. Tetracycline has a stimulatory effect on expression of the RteA-RteB system and, therefore, on both expression and transfer of tetracycline resistance. However, the rteA gene is not directly sensing tetracycline, and exactly what it is sensing is not yet clear (164).
In addition to the sizeable numbers of classical two-component systems (i.e., sensor kinases and response regulators), there is a family of 32 unique proteins in B. thetaiotaomicron that incorporate all of the domains found in the classical two-component system into a single polypeptide. In one system, nutrient sensing is coupled to regulation of monosaccharide metabolism (263). BT3172 belongs to this family of proteins, and its expression is regulated by polysaccharides in the environment. The presence of α-mannosides in the medium can induce expression of BT3172, which, in turn, will cause upregulated expression of secreted α-mannosidases. This system may also be important for capsular polysaccharide gene expression. Typically, expression of one of the capsular polysaccharide synthesis loci (cps3) is upregulated when polysaccharides are scarce. In a mutant deficient in BT3172, expression of cps3 is increased even in the presence of a medium rich in polysaccharides, suggesting that BT3172 is important for the bacterium to properly interpret its nutrient landscape in terms of adequate supply of polysaccharides (263).
Another feature of the ability of BT3172 to sense the neighboring polysaccharide landscape is that it can modulate “mimicry” so that the surface polysaccharide structure of the bacterium can be altered to match the surrounding landscape, possibly allowing the bacterium to avoid eliciting a host immune response (68).
Cross talk between Bacteroides and the intestinal cells.
Most of the characteristics of Bacteroides discussed to this point pertain to the adaptability of Bacteroides to its environment and changes that occur as a result of alterations in that environment. However, this is a two-way communication system, and studies have shown that the intestinal Bacteroides strains directly modulate gut function (94). Almost a decade ago, Hooper et al. demonstrated that B. thetaiotomicron can modify intestinal fucosylation in a complex interaction mediated by a fucose repressor gene and a signaling system (121). Subsequently, using transcriptional analysis, they demonstrated that B. thetaiotaomicron could modulate expression of a variety of host genes, including those involved in nutrient absorption, mucosal barrier fortification, and production of angiogenic factors (120). The line of communication from bacterium to intestinal cell can morph into a complete circle: B. thetaiotaomicron can stimulate production of RegIIIγ, a bactericidal lectin, which can then bind directly to bacterial peptidoglycan in gram-positive bacteria and result in bacterial killing (53).
Interactions with the Immune System
There are numerous studies detailing the host immune response to bacterial virulence factors. However, the vast majority of human-bacterial interactions are benign and commensal or mutualistic in nature. Intestinal bacteria are important in the development of gut-associated lymphoid tissues (GALT): in the absence of bacterial colonization, development of GALT is defective (117). In rabbits, the combination of B. fragilis and Bacillus subtilis consistently promoted GALT development and led to development of the preimmune antibody repertoire (212). Mazmanian and Kasper reviewed the factors that allow the GI tract—an environment with multiple immune capabilities—to coexist with the huge numbers of bacteria found there (154) and proposed a model whereby the immune capabilities of the GI tract are profoundly affected by some of those bacteria. The general outline of this model is described below.
Polysaccharides produced by B. fragilis are important in the activation of the T-cell-dependent immune response.
A zwitterionic polysaccharide (ZPS) produced by B. fragilis can activate CD4+ T cells (i.e., T helper cells expressing the CD4 [cluster of differentiation 4] glycoprotein). Polysaccharides A and B (PS-A and PS-B) of the B. fragilis capsular polysaccharide complex are both ZPSs. Normally, polysaccharides (which almost never carry a positive charge) are considered activators of B cells, and they promote increased immunoglobulin M (IgM) production but without IgG production and without a memory response. However, the unusually structured ZPSs can bind onto the borders of the peptide-binding groove on major histocompatibility complex class II molecules of the antigen-presenting cells (APCs) and dock in this groove, and they can thus be presented to the T-cell receptors of CD4+ T cells in the same way that a peptide or glycopeptide conjugate would be presented. Experimental data indicate that these ZPSs are internalized by the APCs, processed by chemical oxidation into smaller fragments, and then presented to the T cell at the surface of the APC. Indeed, these ZPSs appear to be important in the development of CD4+ T cells. Splenocytes from germfree mice showed a lower proportion of CD4+ cells than splenocytes from conventionally colonized mice, and colonization with B. fragilis (even in the absence of all the other gut microflora) could correct this proportion in the germfree animals. Moreover, colonization with a mutant that could not produce PS-A was not able to correct the defect, whereas PS-A alone could also correct the defect (154, 155).
The CD4+ T cells stimulated by PS-A produce interleukin-10 (IL-10), which can act to prevent abscess formation and other inflammatory responses. Also, in either in vivo or in vitro experiments, these PS-A-stimulated cells also produce gamma interferon, IL-2, and IL-12. The studies accomplished by this group reveal an intricate “dance” between the microbe and certain components of the host immune system, and the authors conclude that the PS-A of B. fragilis “is necessary and sufficient to mediate the generation of a normal mature immune system” (154).
Gut bacteria and the “hygiene hypothesis.”
There have been reports that modulation of the immune system by the commensal gut bacteria is important in allergy development. One scenario is that increases in vaccination, antibiotic usage, and disinfectant use decrease the gut flora at an important point in the development of the immune system, which results in a skewing of the immune system toward TH2 cell response and overproduction of TH2 cytokines and IgE; the innate immune mechanisms and TH1, TH2, and regulatory T cells are all part of this fine balancing act (210). The specific bacterial determinants of this phenomenon are not clear. Mazmanian and Kasper suggest that B. fragilis PS-A may be involved (154). Others suggest that lipopolysaccharide (LPS) (not necessarily Bacteroides LPS), which at low levels is an inducer of IL-12 and gamma interferon (cytokines that stimulate TH1-mediated immunity and decreases the production of TH2 inflammatory cytokines such as IL-4, IL-5, and IL-13), is part of this process (293).
Bacteroides can affect expression of Paneth cell proteins.
Small intestinal crypts house stem cells that serve to constantly replenish epithelial cells that die and are lost from the villi. Paneth cells (immune systems cells similar to neutrophils), located adjacent to these stem cells, protect them against microbes by secreting a number of antimicrobial molecules (defensins) into the lumen of the crypt (97), and it is possible that their protective effect even extends to the mature cells that have migrated onto the villi (97). In animal models, B. thetaiotaomicron can stimulate production of an antibiotic Paneth cell protein (Ang4) that can kill certain pathogenic organisms (e.g., Listeria monocytogenes) (119). In newborn mice, B. thetaiotaomicron promotes angiogenesis and postnatal development (266).
Limiting Colonization of the GI Tract by Pathogens
Studies by Wells and colleagues indicate that anaerobic bacteria play a pivotal role in limiting the translocation of normal intestinal bacteria but that other bacterial groups also have a role in preventing the intestinal colonization and translocation of potential pathogens (295). Recent studies suggest that Bacteroides, and possibly specific species of Bacteroides, have a role in preventing infection with Clostridium difficile (122, 123). As detailed above, the development of the immune response that limits entry and proliferation of potential pathogens is profoundly dependent upon B. fragilis. Also as mentioned, Paneth cell proteins may produce antibacterial peptides in response to stimulation by B. thetaiotomicron (119), and these molecules may prevent pathogens from colonizing the space. In addition, B. thetaiotomicron can induce Paneth cells to produce a bactericidal lectin, RegIIIγ, which exerts its antimicrobial effect by binding to the peptidoglycan of gram-positive organisms (53).
TRANSITION: FROM COMMENSAL TO PATHOGEN
As outlined above, Bacteroides species are normally commensals in the gut flora. However, these organisms can also be responsible for infections with significant morbidity and mortality. A similar scenario is found with the “commensal gone bad” (186), i.e., Enterococcus faecalis. E. faecalis is normally a benign resident of the gut flora. However, the first vancomycin-resistant enterococcal strain had a number of DNA elements, apparently of foreign origin, which comprised a quarter of the genome of that strain (186). These elements included a variety of resistance determinants as well as a pathogenicity island carrying a number of virulence-associated genes. Thus, there may be considerable “sharing” of genes within the crowded neighborhood of the gut flora. Acquiring genes that favor the “new” resident (e.g., genes that code for improved adhesion, new nutrition pathways, antibiotic resistance, and inhibition of host defenses) will give these organisms an edge in establishing a niche for themselves. Indeed, some bacteria may not even need to acquire new genes. Organisms such as Bacteroides with such a large genome bank at their disposal may simply need to turn on certain genes (such as those involving new nutrition pathways, efflux pumps to rid the cell of toxic substrates, or new surface epitopes) to change from friendly commensal to dangerous threat (104).
Additionally, the capsular polysaccharide of B. fragilis, which is so important in development of the host immune system, is also responsible for abscess formation; it is thus one of the most important virulence determinants in this bacterium and is the most obvious bacterial element that is both “friend and foe.”
The proportions of various species of the B. fragilis group found in anaerobic infections are given in Table 2.
THE BAD
Virulence
Although B. fragilis accounts for only 0.5% of the human colonic flora (190), it is the most commonly isolated anaerobic pathogen, due in part to its potent virulence factors. Virulence factors can generally be subdivided into three broad categories: those involved in (i) adherence to tissues, (ii) protection from the host immune response (such as oxygen toxicity and phagocytosis), or (iii) destruction of tissues. Bacteroides strains may possess all of these characteristics. The fimbriae and agglutinins of B. fragilis function as adhesins, allowing them to be established in the host tissue. The polysaccharide capsule, LPS, and a variety of enzymes protect it from the host immune response. The capsule is responsible for abscess formation, and histolytic enzymes found in B. fragilis can mediate tissue destruction.
The bacterial capsule.
The capsule of B. fragilis initiates a unique immune response in the host: abscess formation. The actual formation of the abscess is an example of a pathological host response to the invading bacterium: a fibrous membrane localizes invading bacteria and surrounds a mass of cellular debris, dead polymorphonuclear leukocytes, and a mixed population of bacteria (282). Abscesses left untreated can expand and can even cause intestinal obstruction, erosion of resident blood vessels, and ultimately fistula formation. Abscesses may also rupture and result in bacteremia and disseminated infection.
B. fragilis is the only bacterium that has been shown to induce abscess formation as the sole infecting organism. Abscess formation has been clearly linked to the B. fragilis capsule in an animal model (282). Injection of capsules alone was sufficient to induce abscess formation (67), while systemic injection prevented abscess formation in rats, presumably due to antibody development and subsequent protection (281). Responses to most other polysaccharide antigens are T-cell independent, but abscess formation induced by B. fragilis is dependent on T cells (243-245, 310).
The B. fragilis capsule was first analyzed with a prototype strain. Two distinct high-molecular-weight polysaccharides (PS-A and PS-B) that are coexpressed were described (179, 283), and the structures of these two polysaccharides were elucidated (14). PS-A is made up of repeating tetrasaccharide units, and PS-B is made up of repeating hexasaccharide units (283, 289). Other strains of B. fragilis were subsequently analyzed, and all possessed a complex capsular polysaccharide composed of at least two different polysaccharides; these polysaccharides were antigenically diverse, although some cross-reactivity with the prototype capsular polysaccharide was seen (180). A third capsular polysaccharide (PS-C) was also found, and the biosynthetic loci involved were cloned and sequenced (67, 129).
The assignment of specific biosynthetic loci (involving up to 22 genes/locus) to specific polysaccharides has been amended since first described (67), but the basic features of the complex polysaccharides remain. The most predominant feature of these polysaccharides is the presence of both positively and negatively charged groups on each repeating unit. The two polysaccharides have very different net charges at physiological pH and exhibit variable expression on the bacterial cell surface (180). The zwitterionic motif is necessary for the activities of this group of molecules, including promoting the formation of abscesses (282). The structural basis of the abscess-modulating activity of the polysaccharide has been extensively studied; one model suggests that grooves in the polysaccharide may serve as “docking sites” for α-helices of specific molecules (e.g., immunomodulating molecules such as major histocompatibility/antigen molecular complexes) and thus trigger specific T-cell responses which then lead to abscess formation (289).
There are various opinions concerning the prevalence of capsule among clinical isolates of B. fragilis (48, 180, 193); one possible explanation is that different staining techniques will detect capsule to different extents (180). Electron micrographs reveal that even within an individual strain of B. fragilis, one might observe a large capsule, a small capsule, and noncapsulate variants. The large capsule and unencapsulated strains share antigenic epitopes, but the bacteria with small capsules are different. Intra- and interstrain antigenic variation was noted (184), and this variation has been observed in clinical isolates from a variety of anatomical sites and different geographical locations and also in bacteria grown in an in vivo model of peritoneal infection (184). Expression of the different capsular types is inheritable, since populations can be enriched for their particular type by subculture from different layers of density gradients. In some bacteria that appear noncapsulate, an additional electron-dense layer might be visible adjacent to the outer membrane.
Evasion of host immune response.
The ability to evade the host immune response certainly contributes to the virulence of a bacterium. The B. fragilis capsule can mediate resistance to complement-mediated killing and to phagocytic uptake and killing (98, 209, 252). Recent studies indicate that B. fragilis may interfere with the peritoneal macrophages, the first host immunologic defense response to rupture of the intestine or other compromise of the peritoneal cavity (287). Macrophages are important for early immune responses to invading microorganisms, and the production of nitric oxide (NO) is central to this function. NO is generated by inducible nitric oxide synthase (iNOS) following exposure to certain cytokines (e.g., gamma interferon). These cytotoxic radicals enhance microbicidal function but can also act on host cells to produce cell necrosis or death. In the study mentioned above, macrophages activated by interaction with B. fragilis showed decreased NO production, decreased iNOS activity, and colocalization of iNOS and actin filaments in the macrophage cytoskeleton, along with pore formations not seen in the control cells. The authors concluded that the infection of macrophages with B. fragilis leads to actin filaments and iNOS extrusion through the pore formations, thus allowing the bacteria to evade killing by the macrophages.
Another remarkable feature of Bacteroides is its ability to modulate its surface polysaccharides. The production of these polysaccharides is regulated by a reversible inversion of the DNA segment containing the promoter needed for their expression to an “on” or “off” position (137). These inversions are mediated by invertase genes; mpi, the best known of these genes, codes for a global DNA invertase that is involved in inverting 13 distinct DNA regions, including the promoters of seven of the capsular polysaccharide biosynthesis regions (69). By changing its surface architecture, Bacteroides may avoid the host immune response; other potential effects of surface modification would include the ability to colonize host tissue or form biofilms.
Enzymes implicated in virulence.
Proteases of B. fragilis have been implicated in destroying brush border enzymes (214); these enzymes on the microvillus membranes aid in the final digestion of food and in mechanisms that provide for the selective absorption of nutrients. The most widespread histolytic enzymes in B. fragilis include hyaluronidase and chondroitin sulfatase, which attack the host extracellular matrix (222). Some strains produce other histolytic (e.g., fibrinogenolytic) enzymes (57). Two hemolysins (HlyA and HlyB) have been characterized in B. fragilis; these are two-component cytolysins that act together in hemolysis of erythrocytes (216).
Neuraminidase, the product of the nanH gene in Bacteroides species, cleaves mucin polysaccharides and enhances growth of the bacterium by generating available glucose (105). This enzyme is found in many pathogenic bacteria and is generally considered a virulence factor (223), and many strains of B. fragilis produce neuraminidase (22, 274). Neuraminidase can catalyze the removal of the sialic acid from host cell surfaces and from important immunoactive proteins such as IgG and some components of complement and may consequently disrupt important host functions (237).
Enterotoxin.
The B. fragilis enterotoxin (BFT) is a zinc metalloprotease (136, 162) and may destroy the zonula adherens tight junctions in intestinal epithelium by cleaving E-cadherin (303), resulting in rearrangements of the actin cytoskeleton of the epithelial cells and loss of tight junctions. The result is that this barrier leaks and results in diarrhea (303). More recent evidence indicates that this action is initiated when BFT binds to a specific receptor other than E-cadherin (305). BFT is secreted by enterotoxigenic B. fragilis (ETBF) strains, which encode three isotypes of BFT on distinct bft loci, carried on a 6-kb genome segment unique to these strains, called the B. fragilis pathogenicity island (238).
There is evidence that the enterotoxin pathogenicity island is contained within a novel conjugative transposon (91). This pathogenicity island is flanked by genes encoding mobilization proteins (92) and may thus be transmissible to nontoxigenic strains. A recent study found that 57% of blood culture isolates contained the pathogenicity island and/or its flanking segments (19% had both and 38% had just the flanking segments). Comparatively, in B. fragilis isolates from other clinical sources, 10% had both the pathogenicity island and flanking segments, 43% had only the flanking segments, and 47% had neither. The authors deduced that the pathogenicity island and the flanking elements may be general virulence factors of B. fragilis (61). BFT also induces cyclooxygenase 2 and fluid secretion in intestinal epithelial cells (133). Finally, BFT has a possible role as a carcinogen in colorectal cancer (277).
The presence of the BFT gene is generally detected by PCR techniques (246). In a study of strains from Germany and from southern California, blood culture isolates were more likely to carry the enterotoxin gene than were other isolates (62). There is some association of ETBF and inflammatory bowel disease (IBD), although rigorous, clear-cut correlation has not been demonstrated (12, 192). While the enterotoxin gene was not found in patients with inactive IBD, 13% of patients with IBD and 19% of patients with active disease were toxin positive. For an exhaustive description of ETBF, see the review by Sears (238).
Endotoxin/LPS.
LPS in B. fragilis has an unusual structure (291) and is 10 to 1,000 times less toxic than that of E. coli. Thus, it is generally not referred to as “endotoxin,” although it does have a demonstrable toxicity (71). The induction of endotoxin liberation on exposure to antibiotics was many times higher with B. fragilis than with the other species of the B. fragilis group, which may also help to explain why this species is particularly associated with clinical infections and higher mortality (221). Both LPS and capsule may also function as adhesins that allow the bacterium to become established at the site of infection (15).
Aerotolerance of Bacteroides.
Aerotolerance is not an obvious virulence factor, but it is likely that the ability to survive oxidative stresses plays a role in its ability to initiate or persist in infection (253). Further discussion of the oxidative stress response in Bacteroides is found later in this review.
Infections in Adults
Anaerobic infections are usually polymicrobial, and Bacteroides fragilis is found in most of these infections, with an associated mortality of more than 19% (107). If a documented B. fragilis infection is left untreated, the mortality rate is reported to be about 60% (107). This mortality rate can be greatly improved, however, with use of appropriate antimicrobial therapy (107). Therefore, therapeutic regimens are normally designed to cover this species.
Intra-abdominal sepsis.
Intra-abdominal sepsis is the most common infection caused by Bacteroides. After disruption of the intestinal wall, rupture of the diverticula, or other perforations due to a surgical wound, malignancies, or appendicitis, members of the normal flora infiltrate the normally sterile peritoneal cavity, and the resultant infections reflect the gut flora composition. During the early, acute stage of infection (approximately 20 h), the aerobes, such as E. coli, are the most active members of infection, establishing preliminary tissue destruction and reducing the oxidation-reduction potential of the oxygenated tissue. Once sufficient oxygen has been removed to allow the anaerobic Bacteroides species to replicate, these bacteria begin to predominate during the second, chronic stage of infection.
Perforated and gangrenous appendicitis.
Detailed bacteriologic studies performed in our laboratory recovered more than 20 genera and 40 species of organisms from specimens taken from patients with gangrenous and perforated appendicitis (21); B. fragilis and E. coli were the most frequently recovered anaerobic and aerobic species, respectively. B. thetaiotaomicron was also frequently recovered (in more than 70% of the specimens). The other species of the B. fragilis group were also found but in lower percentages of specimens.
Gynecological infections.
Bacteroides species are not part of the normal flora of the vagina but are occasionally isolated from vaginal cultures. The rates of vaginal carriage of Bacteroides in healthy women (both pregnant and nonpregnant) were estimated to be between 0 and 6% (142, 145), but this rose to 16% in women in labor (141) and to 27 to 28% in patients with cervicitis (145). In a study of 120 pregnant women attending a hospital in Warsaw, Poland, several distinct subgroups of B. fragilis were found, including one ETBF strain, which was genetically different than ETBF strains obtained from other sources (142).
Pelvic infections in which B. fragilis is likely to be involved are often characterized by the presence of an abscess (175); B. fragilis has been isolated from Bartholin's abscess (an abscess in the glands at the side of the vaginal opening) (45) as well as abscesses in the ovaries or fallopian tubes (23). In a large study assessing risk factors for intrauterine growth retardation, colonization of the cervix and/or vagina with Bacteroides, Porphyromonas, and Prevotella was significantly associated with intrauterine growth retardation (100). In a study of 39 women with mild to severe pelvic inflammatory disease, B. thetaiotaomicron was recovered from the endometria or Fallopian tubes of several women with moderate to severe pelvic inflammatory disease (116).
Skin and soft tissue infections.
Necrotizing soft tissue infections are typically polymicrobic. In one retrospective study of 196 patients, nearly half had mixed aerobic and anaerobic growth, and Bacteroides species were the most common organisms isolated (84). In our studies, Bacteroides was not the most common anaerobic organism isolated; nevertheless, 7% of the anaerobes belonged to the B. fragilis group (299). In general, the organisms found in soft tissue infections reflect the normal flora found in the adjacent region. A comprehensive study of the bacteriology of human bite wounds (which include clenched fist injuries) included multiple anaerobes, basically reflecting the oral flora (e.g., Prevotella, Fusobacterium, and Peptostreptococcus); Bacteroides was not reported in these specimens (158). In a study of infected dog and cat bites, 56% of specimens (50 dog bites and 57 cat bites) yielded both aerobes and anaerobes. B. fragilis was isolated from one patient with a dog bite and from one patient with a cat bite, and B. ovatus was isolated from one patient with a dog bite (272).
Endocarditis and pericarditis.
Involvement of anaerobic bacteria in endocarditis is unusual (26), but when it does occur it may have serious consequences (including valvular destruction, dysrhythmias, and cardiogenic shock), with a mortality rate of 21 to 43% (42). The predominant anaerobes in pericarditis are the B. fragilis group and probably occur from hematogenous spread (40). If B. fragilis is found, the most likely source is the GI tract; a literature review of endocarditis due to anaerobes reported 53 cases, and a variety of sources of the infecting organism, including a GI malignancy, liver abscess, ruptured appendix, and decubitus ulcer (26).
Bacteremia.
The incidence of anaerobic bacteremia decreased in the 1980s but has been steadily increasing since the early 1990s. At the Mayo Clinic, 91 cases/year during 2001 to 2004 were seen (a 74% increase over that in the 1980s) (139). Increasing numbers of compromised and/or elderly patients may be one reason for this increase; also, improved survival rates among cancer patients may be another reason: chemotherapy may cause damage of GI mucosal barriers, allowing anaerobic bacteria to pass through and ultimately enter the bloodstream (139).
In one study of Bacteroides bacteremia, 44% of 128 patients had a surgical procedure within 4 weeks of the bacteremia and 30% had a malignancy (169). Twenty-eight percent of the patients had a polymicrobial bacteremia, and nine of the patients were infected with two different Bacteroides species. The mortality rate for all patients was 16% if active therapy was instituted and 45% if inappropriate therapy was given. In another study, bacteremia occurred in 27% of patients with necrotizing soft tissue infections; Bacteroides isolates were the most common species recovered (84). In this study, the presence of bacteremia was the only microbiological variable known to affect mortality. In a medical center setting in Taiwan, 48% of the systemic infections could be traced to a GI source, and 27% of patients with community-acquired anaerobic bacteria had an underlying malignancy. Strains of the Bacteroides fragilis group species were the most common anaerobic isolates, occurring in 45% of the cases (124).
According to a review by Brook, the mortality rate for Bacteroides bacteremia is up to 50% and is somewhat dependent on the species recovered (B. thetaiotaomicron > B. distasonis > B. fragilis); whether this is due to differences in virulence factors or to differences in antimicrobial susceptibility is not known (44).
Septic arthritis.
Bacteroides fragilis is a rare cause of septic arthritis. Most patients with B. fragilis septic arthritis have a chronic joint disease, particularly rheumatoid arthritis, and sources of infection include lesions of the GI tract and the skin (1, 29, 76, 112, 157, 220, 309). There is one report of a case of hip septic arthritis in an alcoholic patient (157). In that paper, the authors reported that about 9% of cases of anaerobic septic arthritis are attributed to B. fragilis, but the references for these data were two decades old, and the taxonomic changes that have occurred since then would render that data misleading. A 2006 report reviewed cases of infection in prosthetic joints and did find reports of infection with anaerobes, including Clostridium difficile, Clostridium perfringens, Porphyromonas melaninogenica, and Veilonella species, but not Bacteroides species (151). However, one recent report of an improved method of culturing these infections found one isolate of B. fragilis in intraoperative specimens from 72 patients with prosthetic joint revision (a total of 155 isolates were recovered) (239).
Brain abscess and meningitis.
While not common, cases of meningitis due to B. fragilis have been reported (4, 88, 93, 163, 168, 172, 183). If these organisms are isolated, a predisposing source of infection should be sought (168). Ventriculoperitoneal shunts that perforate the gut may lead to a shunt infection with Bacteroides and ultimately to meningitis (50).
IBD.
ETBF has been implicated in IBD, but the correlation is not straightforward (12). In one study, the rate of ETBF was high in patients with or without disease. The prevalence of the enterotoxin gene was higher in luminal washings of patients with diarrhea in the control group than in patients without diarrhea, but overall no difference was seen in the prevalences of the toxin gene in patients with and IBD than in the control group. One hypothesis to explain a potential pathogenic mechanism is that colonization with ETBF leads to acute or chronic intestinal inflammation (304); also, the enterotoxin may cleave E-cadherin, an intercellular adhesion protein forming the zonula adherens of intestinal epithelial cells (303) (which limits the ability of water or larger molecules to pass between cells), thus leading to the increased permeability of intestinal epithelial cells. In polarized cell monolayers, BFT alters the apical F-actin structure, resulting in disruption of the epithelial barrier function (39), which may consequently contribute to the diarrhea1 disease associated with B. fragilis infection (28).
Gut bacteria have been implicated as environmental factors in the inflammatory process of ulcerative colitis, a chronic inflammatory mucosal disease. The pANCA (perinuclear antineutrophil cytoplasmic antibody) autoantibody, which is directed against neutrophil proteins, cross-reacted with a microbial antigen epitope in E. coli and Bacteroides (64). In E. coli, the epitope was located on the OmpC protein, one of the well-characterized porin proteins. In B. thetaiotaomicron and B. caccae, the epitope was found on ∼80-kDa and ∼100-kDa proteins, respectively.
Crohn's disease is a subacute or chronic inflammation of the GI tract that may include ulcers and granulomas (95). The role of the commensal bacteria in Crohn's disease and ulcerative colitis is currently being studied intensively (11, 269, 270). Some studies have implicated E. coli and B. vulgatus in the development of this disease. High titers of antisera to a 26-kDa antigen on the surface of B. vulgatus was found in patients with Crohn's disease (10), but the association of these bacteria with the disease process remains somewhat unclear (95). In a very recent report, two hypotheses of the nature of the bacterial role in IBD (including ulcerative colitis and Crohn's disease) are discussed (270). One theory attributes an excessive immunologic response to normal microflora to a malfunction in the immune system. The second theory suggests that changes in the composition of gut microflora and/or deranged epithelial barrier function elicits pathological responses from the normal mucosal immune system. The authors conclude that IBD is characterized by an abnormal mucosal immune response but that microbial factors and epithelial cell abnormalities are implicated in this response. Paneth cells, for example, produce defensins, which are small cationic peptides with antimicrobial activity. Lack of production of these peptides may allow higher bacterial concentrations in the ileal intestinal crypt, which could ultimately contribute to inflammation. Several studies indicated that the gut flora could drive mucosal inflammation, perhaps due to a lack of immune tolerance to the antigens in this flora.
Anaerobes in Pediatric Infections
Infections with intra-abdominal origin.
Sites of Bacteroides infections in children mirror those found in adults. As in adults, Bacteroides isolates are most predominant in infections that have an intra-abdominal origin; normally present in the GI tract, these organisms may enter the peritoneal cavity due to a disturbance such as perforation, obstruction, or direct trauma. A few studies evaluating the microbiology of the peritoneal cavity and postoperative wounds in children following perforated appendix in pediatric patients found that Bacteroides species were recovered from 93% of peritoneal fluids, along with enteric gram-negative bacteria and enterococci (38). Complications following peritonitis may include subphrenic, hepatic, splenic, and retroperitoneal abscesses (39) (which may occur secondary to appendicitis), necrotizing enterocolitis, pelvic inflammatory disease, tubo-ovarian infection, surgery, or trauma (39). B. fragilis was the most common anaerobe found in postsurgical wound infections in wounds relating to the gut flora (37). As expected, wounds and other subcutaneous tissue infections in the rectal area, or that otherwise originated from the gut flora, are typically polymicrobial and often included Bacteroides species. A few studies found that ETBF was associated with diarrheal disease in young children 1 to 5 years of age (235).
Bone and joint infections in children.
Anaerobes have rarely been reported as a cause of joint and bone infections in children. If found, anaerobic infections in arthritis typically involve a single isolate; the isolates found include anaerobic gram-negative bacilli (both B. fragilis group and Fusobacterium species), Clostridium spp., and Peptostreptococcus spp. Anaerobic arthritis is generally secondary to hematogenous spread. Anaerobic osteomyelitis will usually occur due to an anaerobic infection elsewhere in the body and may involve more than one organism (36). Some of these infections may result in positive blood cultures, and the organisms recovered are similar to those from the infected sites (46). Osteomyelitis involving long bones, which may occur after trauma or fracture, may also involve Bacteroides (35). A fairly recent review by Brook comments on the reports of the recovery of anaerobic organisms from infected bones in children (36).
Bacteremia in children.
The incidence of anaerobic bacteremia in children appears to be lower than it is in adults (∼1 to 8% of blood cultures) The particular anaerobe implicated in the bacteremia depends on the portal of entry and the underlying disease, and while other anaerobes are found, Bacteroides fragilis is the isolate most often recovered (36 to 64% of anaerobic blood culture isolates) (44). These infections are more likely to be found in children with predisposing conditions (e.g., malignancies, immunodeficiencies, renal insufficiency, or polymicrobial sepsis).
Infections in newborns.
The involvement of anaerobes in infections in newborns is lower than that in older children. In newborns, anaerobes may be involved in cellulitis, aspiration pneumonia, infant botulism, conjunctivitis, and omphalitis (35). Bacteroides may be involved in cellulitis at the site of fetal monitoring (49). The fact that ETBF-associated diarrhea can be seen in children of 1 to 5 years but not in neonates suggests the possibility of maternal protection (192). In neonatal bacteremia, anaerobes are recovered in 2 to 12% of cultures, and close to half of those are Bacteroides species (47). The overall mortality noted was 26%, and mortality was highest with the B. fragilis group (34%). Inappropriate antimicrobial therapy was often a contributory factor for the high mortality (47). As mentioned above, ETBF is rarely found earlier than the age of 1 year; however, an ETBF strain was the sole organism isolated from the cerebrospinal fluid of a newborn with a complex congenital medullary-colonic fistula (4). No antibodies to the enterotoxin were found in the patient's serum.
Bacteroides as a Reservoir of Resistance Determinants
Clearly, human intestinal bacteria can have neutral or beneficial effects on human health and are, in fact, essential for the proper functioning of the digestive system. The other side of the coin is, in the words of Salyers et al., their “sinister role in human health as reservoirs for antibiotic resistance genes” (228). Thus, Bacteroides isolates, dwelling as seemingly innocuous members of the human colon, can serve as reservoirs of resistance determinants which they can pass on to much more virulent bacteria that move through the gut only periodically, even respiratory bacteria that are inhaled, swallowed, and pass through the gut in 24 to 48 h. “Viewed in this way, the human colon is the bacterial equivalent of eBay,” says Abigail Salyers, an expert on Bacteroides resistance and resistance transfer. “Instead of creating a new gene the hard way—through mutation and natural selection—you can just stop by and obtain a resistance gene that has been created by some other bacterium” (224).
This model suggests that human intestinal bacteria carry a variety of resistance genes that they can share among themselves (250). This genetic “elasticity” has permitted an unanticipated degree of transfer of resistance genes between species and suggests that multidrug resistance will continue to increase (108). The studies to support this model are retrospective in nature, comparing the DNA sequences of resistance genes found in different bacterial species of the human colon. Carriage of the tetracycline resistance gene (tetQ) in Bacteroides has increased from about 30% to more than 80%, and alleles of tetQ in different Bacteroides species were 96 to 100% identical at the DNA sequence level, which is what would be expected from horizontal gene transfer. Similarly, carriage of the erm gene rose from <2 to 23%. Furthermore, carriage of the tetQ and erm genes was also found in healthy people. If the genes found in different species are >95% identical, it is assumed that the gene was transferred horizontally from one species to the other (as opposed to two functionally similar proteins that evolved separately—in that case the DNA sequences can differ by more than 90%). Comparison of erm gene sequences found in other species that either do not normally reside in the human colon (Streptococcus pneumoniae) or reside there in low numbers (Clostridium perfringens and Enterococcus faecalis) with those in Bacteroides indicate that some transfer (direct or indirect) occurred between the species (250).
The elements containing resistance genes are remarkably stable, even in the absence of antibiotic pressure (227). One mechanism by which their stability is maintained may be the organization of genes into an integron, where the genes for antibiotic resistance are maintained in the same integron as enzymes that provide a benefit for the bacterium (e.g., the ability to colonize efficiently). Also, the ability to transfer these elements, coupled with the ability of tetracycline to induce transfer of these elements, makes it likely that bacteria exposed to low levels of tetracycline will have a tendency to transfer these elements to other bacteria that may have lost these genes (227).
Thus, aside from the danger posed by increasingly resistant B. fragilis, the possibility exists that even respiratory organisms such as Klebsiella pneumoniae (173) and Acinetobacter baumanii (60, 106, 125) may acquire resistance determinants from their temporary neighbors as they pass through the gut. Equally disturbing is the recent evidence that innocuous intestinal bacteria in cattle may be reservoirs for resistance and that mobile DNA elements (e.g., plasmids) were responsible for the rapid spread of drug resistance on farms whether or not therapeutic antibiotic use was involved (234). The likelihood of a meteoric rise in drug resistance is crystal clear; a strategy to halt or delay this potentially catastrophic development is, unfortunately, less obvious.
THE NITTY-GRITTY
Taxonomy
The genus Bacteroides has undergone major revisions in the past 15 years. The inclusion or exclusion of species within the genus Bacteroides changes frequently, and keeping up with the taxonomic revisions is a major undertaking. However, these changes are of importance both to clinicians and to clinical microbiologists, since taxonomic placement is a useful tool that can be an indicator of virulence potential or antimicrobial resistance. Familiarity with Bacteroides taxonomy can also influence the evaluation of published susceptibility assays and aid in predicting susceptibility of a clinical isolate. B. thetaiotaomicron, for example, is much more resistant to many antimicrobials than is B. fragilis, and omission of the more resistant species in a published antibiogram may give misleading results.
In 1989 to 1990, the species within Bacteroides were restricted to members of the B. fragilis group (241), and most of the clinically relevant species that were not retained in the genus Bacteroides were placed in the genus Porphyromonas (242) or Prevotella (240). Bacteroides gracilis, which is often involved in deep-seated infections (126), was moved to the genus Campylobacter and renamed Campylobacter gracilis (286). More recently, a host of other genera were described for Bacteroides species, including, among others, Dialister, Megamonas, Mitsuokella, Tannerella, Tissierella, and Alistipes (260).
Often by using culture-independent approaches such as 16S rRNA gene sequencing, a variety of new species have added to the total number of Bacteroides species (now >20). In the fall of 2005, several species were added to the genus Bacteroides, including Bacteroides goldsteinii (261), Bacteroides nordii and Bacteroides salyersai (262), Bacteroides plebeius and Bacteroides coprocola from human feces (135), and Bacteroides massiliensis isolated from the blood culture of a newborn (86). Recently, B. goldsteinii, along with Bacteroides distasonis and Bacteroides merdae, were moved to a new genus, Parabacteroides (225).
Isolation and Identification
Laboratories experienced in processing specimens for anaerobic bacteria will be familiar with the principles used in isolating and identifying Bacteroides strains, and the reader is referred to the Wadsworth-KTL Anaerobic Bacteriology Manual (127) and the Manual of Clinical Microbiology (128). A very brief summary of points to be aware of in processing the specimen is as follows: (i) collect the specimen in a manner to avoid contamination with normal flora; (ii) use an oxygen-free transport medium system and avoid drying; (iii) Bacteroides spp. grow relatively rapidly compared to most other anaerobes, and their growth on selective medium (e.g., Bacteroides bile esculin agar) is quite distinctive; and (iv) B. fragilis is resistant to kanamycin, vancomycin, and colistin and is stimulated by 20% bile (which is inhibitory for most other anaerobic organisms [except Bilophila]). Other tests to identify the species of Bacteroides isolates are listed in the Wadsworth-KTL Anaerobic Bacteriology Manual (127).
Several PCR schemes to identify Bacteroides species to the genus and/or species level have been developed. One report developed group-specific primers to the β-isopropylmalate dehydrogenase gene leuB and found that this was a useful tool for rapid diagnosis of Bacteroides infections (159). Our laboratory developed a multiplex PCR system with group- and species-specific primers to rapidly identify species of the B. fragilis group (146). Using the latter technique, 10 species of the B. fragilis group could be identified.
Physiology
Bacteroides species are a pleomorphic group of non-spore-forming gram-negative anaerobic bacteria. The cell envelope of B. fragilis is a particularly complex structure consisting of multiple layers, with subunits of one layer protruding through another. Descriptions of these layers come both from structural and functional studies, but results of these studies have not necessarily provided consistent descriptions either of the makeup, function, or relationship to each other of these layers. We have recently published a detailed review of the cell surface structures of Bacteroides (199), and they will be described briefly here. The B. fragilis capsule has already been discussed at length in a previous section (“The bacterial capsule”).
Cell Surface Structures
Pili, fimbriae, and adhesins.
The terms pili, fimbriae, and adhesins are not very distinctly defined. Although adhesins (for example, pili and fimbriae used in adhesion) are often protein, the term is not restricted to protein adhesins, and other structures may be implicated. B. fragilis may possess peritrichous fimbriae (178). These fimbriae have been implicated in adhesion; in one study, trypsin treatment of clinical isolates of B. fragilis inhibited both hemagglutination and adhesion to a human intestinal cell line, suggesting that the responsible adhesins were proteins (87). In various studies, pili have also been implicated in hemagglutination and adhesion (51, 193). Other studies, however, implicated the polysaccharide capsule rather than the protein appendages (177). Recent functional genomic studies classifying databases of specific molecules note that members of the Bacteroides secrete large numbers of lipoproteins with an N-terminal beta-propeller domain, which may form a specialized adhesion module (7, 20). Discrepancies among the various studies may be due to differences in the cell lines used, differences in the pilus type studied, or differences in the capsule characteristics of the particular strains.
Lectin-like adhesins have been demonstrated in B. fragilis (218); correspondingly, sialic acid and other sugars, as well as macromolecules rich in sialic acid, have been identified as the receptors for these adhesins (77). In some cases, the adhesin will bind to the receptor residue only after neuraminidase treatment (110). Indeed, many B. fragilis strains have neuraminidase activity, and Guzman et al. (110) suggest that the bacterial removal of the terminal sialic acid may serve as a mechanism to expose the adhesion sites in a two-step adhesion process. Others found that adherence to WiDr cells and hemagglutination were not affected by neuraminidase activity (167). Hemagglutination appears to be caused by more than one adhesin, at least one of which is a carbohydrate (probably the capsule), with the pili assuming this role in noncapsulated strains (15). Recently, a surface glycoprotein of B. fragilis was implicated in binding to one of the laminin proteins that make part of the extracellular matrix that underlies epithelial, endothelial, and surrounding connective tissue cells (74).
Fibrils.
Fibrils are a class of bacterial appendage consisting of polysaccharide and associated proteins and are much finer and often much shorter than pili. In fact, they may be impossible to distinguish from long LPS side chains in transmission electron microscopy, since the lengths quoted for these structures overlap. However, peritrichous fibrils were reported in only one out of 19 B. fragilis strains studied (178), and these were distinguished from the capsule, pili, and ruthenium red staining layer (probably composed mostly of LPS side chains). Again, not all cells of a given population exhibited these fibrils. Little is known about the function of these fibrils, and their role in adhesion and biofilm formation remains to be determined.
LPS.
The LPS side chains project from the lipid moiety that is anchored in the outer membrane, forming a visible fringe in transmission electron microscopy (the exact nature of this layer remains to be determined, but most authors assume that the fringe overlying the outer membrane comprises the LPS side chains.) In our own recent studies, we noted significant variation in the height and density of the LPS fringe between individual colonies within the same cell population grown under the same conditions (199).
Outer membrane proteins.
Several membrane proteins have so far been characterized in B. fragilis. We demonstrated that OmpA was the major outer membrane protein in B. fragilis (302), and our studies further suggest that B. fragilis OmpA1 is important in maintaining cell structure (unpublished data). We identified four distinct genes that encode OmpA homologs and found that all four ompA genes are transcribed in B. fragilis. In other bacteria OmpA has been shown to be associated with virulence (292), but we have not yet studied this in B. fragilis.
Two porin proteins, B. distasonis HMP-1 and B. fragilis Omp200 (a two-component porin protein composed of Omp121 and Omp71), were described by our laboratory (297, 301). The Omp121 component of Omp200 of B. fragilis had some homology to B. thetaiotaomicron SusC; genomic analysis has indicated that one group of SusC homologs in this species are positioned upstream of SusD homologs that may be involved in acquisition or utilization of polysaccharides (308). The other group of the SusC homologs (including Omp121) are positioned upstream of homologs of Omp71, a protein whose function is not yet defined. Xu and Gordon speculated that many of the organism's SusC homologs are conserved components of a series of multifunctional outer membrane porins (308). Researchers in other laboratories have described a 45-kDa porin-like protein and 51-, 92-, and 125-kDa porin-like proteins (130, 171). Two large porins of 210 and 135 kDa that include the 45-kDa outer membrane protein were also described (170).
The membrane also contains an iron-regulated outer membrane protein involved in heme uptake (176). Iron starvation triggers the expression of this heme importer. Since iron is limiting in the host, iron uptake mechanisms are undoubtedly an important colonization factor. There is apparently no evidence for the secretion of iron chelators by B. fragilis.
Outer membrane vesicles.
Bacteroides fragilis characteristically produces numerous outer membrane vesicles. These vesicles appear in transmission electron microscopy as surface blebs and detached extracellular vesicles and have been shown to have a hemagglutinin function (185) and to contain sialidase activity that may be correlated to virulence (78). Similar outer membrane vesicles have been observed in Porphyromonas gingivalis, Pseudomonas fragi, and Xenorhabdus nematophilus, where it has been suggested that they may serve as a vehicle for toxins and attachment to host cells (80, 109, 132). Furthermore, similar vesicles in Pseudomonas aeruginosa have been shown to carry quinolone signaling molecules for intercellular signaling (153). Vesicle production varies considerably between strains of B. fragilis; we noted that these vesicles are produced in large amounts by certain clinical isolates but are almost entirely absent in others (199).
Export systems.
The genomic studies of B. fragilis did not show evidence of type III, IV, autotransporter, or two-partner secretion systems. There were genes for Hly type I secretion systems, which are similar to the hemolysin type I secretion system HlyDb from E. coli (288), or the type II general secretion pathway (54). The large quantity of enzyme-containing outer membrane vesicles produced by B. fragilis (185) suggests that this may be an important export pathway (54). The Bacteroides genome also contains large numbers of gene homologs of a variety of efflux pumps systems whose export functions have not yet been defined. Bacteroides fragilis may secrete two toxins into the medium: endotoxin (LPS) and BFT (fragolysin). Intracellular, periplasmic and outer membrane-bound proteases have been identified in B. fragilis (101). Many of the hydrolytic enzymes, which are generally considered to be pathogenic factors, may be membrane bound and/or secreted into the medium. Many histolytic enzymes are associated with the Bacteroides cells during exponential phase but are released in stationary phase, apparently without cell lysis (149). Determining the exact location of hydrolytic enzymes at different phases of growth would aid the understanding of their possible role as invasins.
Bacteriocins.
Bacteriocins are antibacterial chemicals secreted by bacteria to inhibit competitors. Analysis of clinical isolates of B. fragilis revealed that intestinal isolates secreted high levels of a bacteriocin protein, and they themselves were highly resistant to bacteriocins secreted by other B. fragilis strains, while nonintestinal isolates produced only medium levels of bacteriocins and were less resistant (5). A bacteriocin protein has been shown to be produced by a B. fragilis strain that inhibited the RNA polymerases only of other B. fragilis strains in vivo (165). This narrow specificity is typical of bacteriocins that bind to specific receptors and attack species or strains closely related to the producer. Presumably, this is a mechanism to reduce competition between strains occupying the same ecological niche.
Resistance to bile.
In the host, bile functions as a biological detergent with an essential role in fat digestion (i.e., it emulsifies and solubilizes lipids). Deconjugated bile salts are less efficiently reabsorbed than conjugated bile salts; this results in greater excretion of free bile acids in feces, which may result in greater use of cholesterol for de novo synthesis of bile to compensate for this loss. This increased utilization of cholesterol may lower serum cholesterol levels. Alternatively, the lowered ability of free bile salts to solubilize cholesterol may reduce the absorption of cholesterol through the intestinal lumen and into the serum (17). B. fragilis plays a key role in the enterohepatic circulation of bile acids by helping the process of biotransformation between conjugated and deconjugated bile salts. B. fragilis contains many enzymes required for these reactions, including a bile salt hydrolase (66, 267). In turn, these organisms must be able to survive when exposed to the high concentration of bile salts in the intestinal tract. The detergent activity of bile salts permeabilizes bacterial membranes and can eventually lead to membrane collapse and cell damage (16, 34, 70). Factors that enable Bacteroides species to tolerate bile salts are important elements in their ability to survive in the gut. On the other hand, exposure to bile may initiate other survival mechanisms. For example, we have demonstrated that expression of the RND efflux pumps of B. fragilis is increased in response to stress by bile and bile salts (197). Bile salt hydrolase activity may have both beneficial (e.g., in lowering cholesterol levels) and deleterious (e.g., unconjugated bile acids are less efficient than conjugated molecules in digestion of lipids) effects for the host (17).
Oxidative stress response.
The ability of B. fragilis to grow in the presence of nanomolar concentrations of O2 has important implications for the interaction of this opportunistic pathogen with its oxygenated hosts. During the establishment of an infection, the ability to grow in the presence of nanomolar concentrations of O2 would enable proliferation in host tissues even before the anaerobic abscess is formed (13). Many studies of the oxidative stress response in B. fragilis have focused on the ability of enzymes to detoxify or otherwise protect the organism from oxygen radicals; these proteins include catalase, superoxide dismutase, alkyl hydroperoxide reductase, and nonspecific DNA binding protein (32, 217, 271). These genes are, in part, under the control of the redox-sensitive regulator OxyR (253). Recently, six new genes were identified upon exposure of B. fragilis to O2, including an aspartate decarboxylase gene, a putative outer membrane protein gene, a cation efflux pump gene, a heat shock protein gene, and two ribonucleotide reductase genes which may have a role in maintaining deoxyribonucleotide pools for DNA repair and growth recovery (253).
Susceptibility Patterns of Bacteroides
Bacteroides species are most commonly found in mixed infections, and the commonly prescribed treatments are designed accordingly. Frequently prescribed antibiotics include β-lactams (with or without β-lactamase inhibitors), carbapenems, clindamycin, and metronidazole. Fluoroquinolones are also used in combination with clindamycin and metronidazole (41). Bacteroides species are potentially resistant to a broad range of antibiotics, and resistance to a given antimicrobial can vary greatly between geographical locations and institutions.
Resistance rates can also vary widely among the different species of the Bacteroides fragilis group. For example, reported resistance rates vary widely for clindamycin (15 to 44%), cefotetan (13 to 94%), and cefoxitin (3.5 to 41.5%) (63). Resistance to even the most active drugs, such as imipenem, piperacillin-tazobactam, ampicillin-sulbactam, and metronidazole, is found in occasional strains (201, 236, 290). Frequently, the other species in the B. fragilis group are more resistant than B. fragilis to many antibiotics. Current antibiograms of anaerobic bacteria have been summarized and reviewed recently (114, 115, 256, 258) and results of several studies have been collated in Table 3.
TABLE 3.
Susceptibility of Bacteroides species to antimicrobial agents
| Organism | Antimicrobial | MIC (μg/ml)
|
% of isolates
|
Reference | ||||
|---|---|---|---|---|---|---|---|---|
| 90% | Geometric mean | Breakpoint | Susceptible | Intermediate | Resistant | |||
| B. fragilis | Amoxicillin + clavulanic acid | >64 | ≥16/8 | 4.8 | 19 | |||
| Ampicillin-sulbactam | 16 | 1.7 | 256 | |||||
| Cefotetan | 64 | ≥64 | 19 | 19 | ||||
| Cefoxitin | 32 | ≥64 | 3.4 | 255 | ||||
| Cefoxitin | 32 | 5.2 | 256 | |||||
| Ceftriaxone | >32 | 25.7 | 15 | 48 | 37 | 296 | ||
| Chloramphenicol | 8 | 100 | 0 | 0 | 24 | |||
| Clinafloxacin | 2 | 257 | ||||||
| Clindamycin | >128 | ≥8 | 24.9 | 255 | ||||
| Clindamycin | 256 | 19.3 | 256 | |||||
| Doripenem | 1 | 0.5 | 96 | 3 | 1 | 296 | ||
| DX-619 | 0.5 | 275 | ||||||
| Ertapenem | 0.5 | 0.3 | 100 | 0 | 0 | 296 | ||
| Garenoxacin | 4 | ≥4 | 14.8 | 255 | ||||
| Garenoxacin | 0.5 | 81 | ||||||
| Imipenem | 0.5 | 0.2 | 100 | 0 | 0 | 296 | ||
| Levofloxacin | 2 | 1.5 | 91 | 5 | 4 | 296 | ||
| Linezolid | 8 | ≥4 | 94.3 | 255 | ||||
| Meropenem | 0.5 | 0.2 | 97 | 3 | 0 | 296 | ||
| Metronidazole | 4 | ≥32 | 0.5 | 19 | ||||
| Metronidazole | 2 | 100 | 0 | 0 | 24 | |||
| Moxifloxacin | 8 | ≥4 | 36.4 | 255 | ||||
| Moxifloxacin | 8 | 27.3 | 256 | |||||
| Moxifloxacin | 1 | 81 | ||||||
| Moxifloxacin | 4 | 25 | ||||||
| Moxifloxacin | 8 | 257 | ||||||
| NVP-LMB415 | 0.5 | ≥4 | 0 | 255 | ||||
| Piperacillin-tazobactam | 4 | 0.4 | 256 | |||||
| Sitafloxacin | 1 | 257 | ||||||
| Ticarcillin | >256 | ≥128 | 28.6 | 19 | ||||
| Ticarcillin + clavulanic acid | 4 | ≥128/2 | 1.6 | 19 | ||||
| Tigecycline | 8 | ≥4 | 14.8 | 255 | ||||
| Tigecycline | 8 | 5.1 | 256 | |||||
| Tigecycline | 8 | 24 | ||||||
| B. ovatus | Cefoxitin | 32 | ≥64 | 6.2 | 255 | |||
| Ceftriaxone | >32 | 25 | 20 | 50 | 30 | 296 | ||
| Chloramphenicol | 8 | 100 | 0 | 0 | 24 | |||
| Clinafloxacin | 4 | 257 | ||||||
| Clindamycin | >128 | ≥8 | 40.6 | 255 | ||||
| Doripenem | 1 | 0.4 | 100 | 0 | 0 | 296 | ||
| DX-619 | 1.125 | 275 | ||||||
| Ertapenem | 1 | 0.5 | 100 | 0 | 0 | 296 | ||
| Garenoxacin | 8 | ≥4 | 31.3 | 255 | ||||
| Garenoxacin | 1 | 81 | ||||||
| Imipenem | 0.5 | 0.2 | 100 | 0 | 0 | 296 | ||
| Levofloxacin | 16 | 7.5 | 0 | 25 | 75 | 296 | ||
| Linezolid | 8 | ≥4 | 78.1 | 255 | ||||
| Meropenem | 0.5 | 0.2 | 100 | 0 | 0 | 296 | ||
| Metronidazole | 4 | 100 | 0 | 0 | 24 | |||
| Moxifloxacin | 64 | ≥4 | 81.3 | 255 | ||||
| Moxifloxacin | 2 | 81 | ||||||
| Moxifloxacin | 32 | 257 | ||||||
| Moxifloxacin | 16 | 25 | ||||||
| NVP-LMB415 | 0.25 | ≥4 | 0 | 255 | ||||
| Sitafloxacin | 4 | 257 | ||||||
| Tigecycline | 8 | ≥4 | 18.8 | 255 | ||||
| Tigecycline | 16 | 24 | ||||||
| B. thetaiotaomicron | Cefoxitin | 64 | ≥64 | 10.5 | 255 | |||
| Ceftriaxone | >32 | 31.1 | 2 | 58 | 40 | 296 | ||
| Ceftriaxone | >32 | 30.3 | 4 | 53 | 43 | 296 | ||
| Chloramphenicol | 8 | 100 | 0 | 0 | 24 | |||
| Clinafloxacin | 4 | 257 | ||||||
| Clindamycin | >128 | ≥8 | 42.1 | 255 | ||||
| Doripenem | 2 | 0.6 | 98 | 0 | 2 | 296 | ||
| Doripenem | 1 | 0.6 | 100 | 0 | 0 | 296 | ||
| DX-619 | 0.5 | 275 | ||||||
| Ertapenem | 2 | 0.6 | 100 | 0 | 0 | 296 | ||
| Ertapenem | 2 | 0.6 | 100 | 0 | 0 | 296 | ||
| Garenoxacin | 4 | ≥4 | 18.4 | 255 | ||||
| Garenoxacin | 2 | 81 | ||||||
| Imipenem | 1 | 0.4 | 100 | 0 | 0 | 296 | ||
| Levofloxacin | 8 | 6.3 | 2 | 2 | 69 | 296 | ||
| Levofloxacin | 16 | 3.9 | 52 | 13 | 35 | 296 | ||
| Linezolid | 8 | ≥4 | 93.4 | 255 | ||||
| Meropenem | 0.5 | 0.3 | 100 | 0 | 0 | 296 | ||
| Meropenem | 0.5 | 0.3 | 100 | 0 | 0 | 296 | ||
| Metronidazole | 2 | 100 | 0 | 0 | 24 | |||
| Moxifloxacin | 32 | ≥4 | 47.4 | 255 | ||||
| Moxifloxacin | 2 | 81 | ||||||
| Moxifloxacin | 8 | 25 | ||||||
| Moxifloxacin | 32 | 257 | ||||||
| NVP-LMB415 | 0.025 | ≥4 | 0 | 255 | ||||
| Sitafloxacin | 4 | 257 | ||||||
| Tigecycline | 8 | ≥4 | 27.6 | 255 | ||||
| Tigecycline | 8 | 24 | ||||||
| B. distasonis | Cefoxitin | 128 | ≥64 | 39.3 | 255 | |||
| Clinafloxacin | 8 | 257 | ||||||
| Clindamycin | >128 | ≥8 | 21.4 | 255 | ||||
| DX-619 | 1 | 275 | ||||||
| Ertapenem | 4 | 1.77 | 1.5 | 256 | ||||
| Garenoxacin | 8 | ≥4 | 35.7 | 255 | ||||
| Garenoxacin | 1 | 81 | ||||||
| Imipenem | 2 | ≥16 | 0 | 255 | ||||
| Imipenem | 2 | 0.95 | 0.4 | 256 | ||||
| Linezolid | 4 | ≥4 | 92.9 | 255 | ||||
| Meropenem | 2 | 0.58 | 1.1 | 256 | ||||
| Moxifloxacin | 32 | ≥4 | 50.0 | 255 | ||||
| Moxifloxacin | 2 | 81 | ||||||
| Moxifloxacin | 16 | 25 | ||||||
| Moxifloxacin | 32 | 257 | ||||||
| NVP-LMB415 | 0.5 | ≥4 | 0 | 255 | ||||
| Sitafloxacin | 4 | 257 | ||||||
| Tigecycline | 8 | ≥4 | 39.3 | 255 | ||||
| B. uniformis | Cefoxitin | 32 | ≥64 | 4.5 | 255 | |||
| Chloramphenicol | 8 | 100 | 0 | 0 | 24 | |||
| Clinafloxacin | 4 | 257 | ||||||
| Clindamycin | >128 | ≥8 | 18.2 | 255 | ||||
| Ertapenem | 2 | 0.88 | 0.5 | 256 | ||||
| Garenoxacin | 8 | ≥4 | 27.3 | 255 | ||||
| Garenoxacin | >8.0 | 81 | ||||||
| Imipenem | 0.5 | ≥16 | 0 | 255 | ||||
| Imipenem | 1 | 0.43 | 0.5 | 256 | ||||
| Linezolid | 8 | ≥4 | 68.2 | 255 | ||||
| Meropenem | 1 | 0.41 | 0.5 | 256 | ||||
| Metronidazole | 4 | 100 | 0 | 0 | 24 | |||
| Moxifloxacin | 64 | ≥4 | 59.1 | 255 | ||||
| Moxifloxacin | 32 | 25 | ||||||
| Moxifloxacin | >8.0 | 81 | ||||||
| Moxifloxacin | 32 | 257 | ||||||
| NVP-LMB415 | 0.25 | ≥4 | 0 | 255 | ||||
| Sitafloxacin | 4 | 257 | ||||||
| Tigecycline | 8 | ≥4 | 18.2 | 255 | ||||
| Tigecycline | 8 | 24 | ||||||
| B. vulgatus | Cefoxitin | 32 | ≥64 | 4.5 | 255 | |||
| Chloramphenicol | 8 | 100 | 0 | 0 | 24 | |||
| Clinafloxacin | 16 | 257 | ||||||
| Clindamycin | >128 | ≥8 | 50.0 | 255 | ||||
| Ertapenem | 2 | 0.82 | 0.3 | 256 | ||||
| Garenoxacin | 16 | ≥4 | 50.0 | 255 | ||||
| Garenoxacin | 1 | 81 | ||||||
| Imipenem | 1 | ≥16 | 0 | 255 | ||||
| Linezolid | 4 | ≥4 | 68.2 | 255 | ||||
| Meropenem | 1 | 0.47 | 0.0 | 256 | ||||
| Metronidazole | 4 | 100 | 0 | 0 | 24 | |||
| Moxifloxacin | 64 | ≥4 | 72.7 | 255 | ||||
| Moxifloxacin | 4 | 81 | ||||||
| Moxifloxacin | 4 | 25 | ||||||
| Moxifloxacin | 128 | 257 | ||||||
| NVP-LMB415 | 0.5 | ≥4 | 0 | 255 | ||||
| Sitafloxacin | 16 | 257 | ||||||
| Tigecycline | 4 | ≥4 | 18.2 | 255 | ||||
| Tigecycline | 8 | 24 | ||||||
| B. fragilis group | Amoxicillin + clavulanic acid | 4 | ≥16/8 | 5.6 | 19 | |||
| Cefotetan | >128 | ≥64 | 44 | 19 | ||||
| Cefoxitin | 32 | ≥64 | 7.4 | 255 | ||||
| Chloramphenicol | 8 | 100 | 0 | 0 | 24 | |||
| Clinafloxacin | 4 | 257 | ||||||
| Clindamycin | >128 | ≥8 | 30.9 | 255 | ||||
| Ertapenem | 4 | 1.04 | 1.0 | 256 | ||||
| Garenoxacin | 8 | ≥4 | 21.2 | 255 | ||||
| Imipenem | 1 | ≥16 | 0.07 | 255 | ||||
| Imipenem | 1 | 0.38 | 0.4 | 256 | ||||
| Linezolid | 8 | ≥4 | 90.2 | 255 | ||||
| Meropenem | 1 | 0.42 | 0.6 | 256 | ||||
| Metronidazole | 2 | ≥32 | 0.3 | 19 | ||||
| Metronidazole | 2 | 100 | 0 | 0 | 24 | |||
| Moxifloxacin | 32 | ≥4 | 48.0 | 255 | ||||
| Moxifloxacin | 4 | 25 | ||||||
| Moxifloxacin | 32 | 257 | ||||||
| NVP-LMB415 | 0.5 | ≥4 | 0 | 255 | ||||
| Sitafloxacin | 2 | 257 | ||||||
| Ticarcillin | >256 | ≥128 | 33.7 | 19 | ||||
| Ticarcillin + clavulanic acid | 16 | ≥128/2 | 2.2 | 19 | ||||
| Tigecycline | 2 | ≥4 | 20.1 | 255 | ||||
| Tigecycline | 8 | 24 | ||||||
| Other Bacteroides spp. | Cefoxitin | 32 | ≥64 | 0 | 255 | |||
| Chloramphenicol | 8 | 100 | 0 | 0 | 24 | |||
| Clinafloxacin | 4 | 257 | ||||||
| Clindamycin | >128 | ≥8 | 31.3 | 255 | ||||
| Ertapenem | 4 | 1.03 | 0.5 | 256 | ||||
| Garenoxacin | 4 | ≥4 | 25.0 | 255 | ||||
| Imipenem | 0.05 | ≥16 | 0 | 255 | ||||
| Linezolid | 8 | ≥4 | 100 | 255 | ||||
| Meropenem | 1 | 0.4 | 0.5 | 256 | ||||
| Metronidazole | 4 | 100 | 0 | 0 | 24 | |||
| Moxifloxacin | 32 | ≥4 | 81.3 | 255 | ||||
| Moxifloxacin | 16 | 257 | ||||||
| NVP-LMB415 | 0.5 | ≥4 | 0 | 255 | ||||
| Sitafloxacin | 2 | 257 | ||||||
| Tigecycline | 8 | ≥4 | 25.0 | 255 | ||||
| Tigecycline | 8 | 24 | ||||||
The most recent multicenter study by Snydman and colleagues reviewed results for 11 antimicrobials and more than 5,000 isolates referred by 10 medical centers (256) and presented the results as trends of antimicrobial susceptibility over an 8-year time period. Resistance to carbapenems was rarely seen in this study (<1.5%). The trends in resistance to piperacillin-tazobactam, ampicillin-sulbactam, and cefoxitin were species dependent and did not necessarily correlate with each other. A worrisome development was the increase in resistance of B. distasonis (which is no longer technically in the genus Bacteroides [225]) to ampicillin-sulbactam in 2002 and 2004 (>20%). Resistance of B. fragilis, B. ovatus, and B. thetaiotaomicron to clindamycin increased significantly, as did the geometric mean MICs of these agents; similar results were seen for moxifloxacin. Resistance rates for tigecycline were low and stable during the 5-year period during which this agent was studied.
Mechanisms of Antimicrobial Resistance
Species of the genus Bacteroides have the most antibiotic resistance mechanisms and the highest resistance rates of all anaerobic pathogens (259). B. fragilis is intrinsically resistant to several classes of structurally unrelated antibiotics (114), and the mechanisms are often poorly understood. Logically, resistance to any antimicrobial agent should be due to one of three mechanisms: altered target binding affinity, decreased penetration for the antibiotic due to permeability or efflux changes, or the presence of an inactivating enzyme. Clinically, Bacteroides species have exhibited increasing resistance to many antibiotics, including cefoxitin, clindamycin, metronidazole, carbapenems, and fluoroquinolones (e.g., gatifloxacin, levofloxacin, and moxifloxacin). The newer fluoroquinolones, including sitafloxacin, clinafloxacin, and garenoxacin, are generally more active against Bacteroides species (257). Bacteroides species are adept at antimicrobial evasion and may use any or all of the above-mentioned mechanisms to thwart effective clinical therapy.
β-Lactam agents.
The most common mechanism of resistance to β-lactam agents is β-lactamase (product of the cepA gene), a chromosomally encoded class 2e cephalosporinase (113), which is found in nearly all Bacteroides species (219). This enzyme is inhibited by the most commonly used β-lactamase inhibitors (sulbactam, clavulanic acid, and tazobactam), and agents containing these inhibitors are active against strains that produce the β-lactamase enzyme. The cefoxitin resistance gene, cfxA (182), has been shown to be distantly related to the B. fragilis endogenous cepA (219).
Carbapenems.
Resistance to carbapenems remains rare. Genes for two enzymes that can degrade these agents have been identified: cfiA and ccrA, both encoding class B metallo-β-lactamases (147). These enzymes confer resistance to carbapenems, β-lactams, and β-lactamase inhibitor combination agents. Some strains of Bacteroides contain “silent” carbapenemase genes, and expression levels are dependent on promoter-containing insertion sequences (IS) inserted upstream of the cfiA/ccrA gene sequence. Changes in penicillin binding proteins (PBPs) or changes in porins that allow permeation of the carbapenems are other theoretically possible carbapenem resistance mechanisms but have only rarely been described in clinical isolates (e.g., PBP2Bfr, a homolog of E. coli PBP3, was implicated in imipenem resistance in one strain [6]). On the other hand, we have recently studied multidrug-resistant strains with increased efflux pump activity, and we have found clinical isolates that are resistant to carbapenems because of increased efflux activity (194, 201).
PBPs.
Penicillin and other β-lactams are cell wall-active drugs that interfere with the final transpeptidation step in the synthesis of the peptidoglycan. The structure of the β-lactam antibiotics facilitates their binding to the active site of PBPs; this irreversible inhibition of the PBPs prevents the final cross-linking (transpeptidation) of the nascent peptidoglycan layer. The correlation of β-lactam activity and PBP affinity in B. fragilis has not been straightforward, partly because there are conflicting results from investigations into the numbering and molecular masses of PBPs from this bacterium. Most studies were done by labeling cells or cell extracts with a labeled antimicrobial and analyzing the results by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (82, 83, 298). The completion of the genome sequence of B. fragilis allowed a more complete analysis, and seven putative PBP genes were identified. The gene sequences for the closest homologs to the E. coli PBPs in the B. fragilis genome (pbp1abBfr, pbp1cBfr, pbpABfr, pbpBBfr, pbp4Bfr, and pbp7Bfr) were deduced as the orthologs for the E. coli genes ponBEco (PBP1bEco), pbpCEco (PBP1cEco), pbpAEco (PBP2Eco), pbpBEco (PBP3Eco), dacBEco (PBP4Eco), and pbpGEco (PBP7Eco), respectively (188). In any case, as the authors finally conclude, it is very difficult to correlate the 50% inhibitory concentration affinity of a particular PBP for an antibiotic with the MIC, since this is not generally the only mechanism responsible for the resistance.
In spite of this difficulty, several workers have reported an association between the reduced affinity of β-lactam compounds for the PBPs of Bacteroides species and resistance to these agents. Reduced affinity of an 80-kDa PBP for imipenem, piperacillin, cefoperazone, cefotaxime, and ceftazidime was observed in a resistant strain of B. fragilis, although no precise 50% inhibitory concentration calculations were performed (99). Alterations in PBPs, mainly changes in PBP1 and/or PBP2, have been implicated in non-β-lactamase-mediated cefoxitin resistance in cefoxitin-resistant mutants of B. fragilis group species (187, 298). The affinities of the PBP1 complex (86 kDa) of B. thetaiotaomicron 238m for cefoxitin and piperacillin were >100-fold and ∼70-fold reduced, respectively, compared with those of its parent strain (85). The ortholog of the E. coli PBP3 gene (pbpBBfr, which encodes the protein PBP2Bfr) was implicated in binding to imipenem (6, 188).
Outer membrane proteins.
There are some data correlating changes in outer membrane porin proteins and antimicrobial resistance in Bacteroides. A 45-kDa porin-like protein from B. fragilis was potentially correlated with antibiotic resistance in B. fragilis (171), and a putative porin of ∼70 kDa of B. thetaiotaomicron was potentially associated with resistance to β-lactam agents (18). To date, however, the relationships between outer membrane (porin) proteins and antimicrobial resistance have been associative, and no definitive correlation has been shown.
Aminoglycosides.
Bacteroides species are inherently resistant to aminoglycosides (protein synthesis inhibitors that bind the 30S subunit of the ribosome), as uptake of this drug requires an oxygen- or nitrate-dependent electron transport chain which is lacking in these anaerobes (52).
Macrolides, lincosamides, and chloramphenicol.
Macrolides (e.g., erythromycin) inhibit protein synthesis by binding to the 23S rRNA molecule (in the 50S subunit) of the bacterial ribosome, blocking the exit of the growing peptide chain. Lincosamides (e.g., clindamycin) also bind to the 50S ribosomal subunit. Resistance to clindamycin, a commonly used antianaerobic drug during the last few decades, has increased steadily. Homologs of the macrolide-lincosamide-streptogramin B resistance genes confer both clindamycin and erythromycin resistance in Bacteroides (89, 215). These genes are similar to the macrolide-lincosamide-streptogramin B resistance genes in gram-positive organisms that cause resistance by methylation of the ribosome target.
Chloramphenicol binds to the ribosomal peptidyl transferase in bacteria and thus prevents protein biosynthesis; the enzyme chloramphenicol acetyltransferase transfers an acetyl group from acetyl coenzyme A to the primary hydroxyl on C-6 of chloramphenicol, preventing the modified antibiotic from binding to the ribosome and exerting its effect (33). In the most recent national survey of multiple hospitals within the United States, no resistance to chloramphenicol (MIC, >16 μg/ml) was seen (256). However, the MICs of chloramphenicol seem to cluster just around breakpoint levels (in our own studies, we have seen many strains of B. fragilis with chloramphenicol MICs of 8 μg/ml [300]), so a slight shift of MICs could change this picture, since many strains would then be considered resistant.
Tetracycline.
Tetracyclines inhibit bacterial protein synthesis by blocking the attachment of the tRNA-amino acid to the ribosome. This agent was once the first-line antibiotic for treatment of anaerobic infections, and all strains of Bacteroides isolated in the 1950s were susceptible (103). This antibiotic remained a useful antianaerobic agent until the early 1970s (134); today, nearly all clinical isolates of Bacteroides are resistant (80 to 90%). tetQ is one of several tetracycline resistance genes that codes for a cytoplasmic protein that interacts with the ribosome, making it insensitive to tetracycline blocking (232). TetQ is the most common type of tetracycline resistance in Bacteroides (140). Efflux of tetracycline has also long been described as a resistance mechanism in B. fragilis (203).
Nitroimidazoles.
Metronidazole is a commonly used drug for anaerobic infections and resistance to this drug is rare, but there is evidence both of increasing resistance and of artificially low resistance rates, in some cases, due to the screening method used for detecting anaerobes. The true incidence of metronidazole resistance of B. fragilis may be much higher than normally assumed, at least in the United Kingdom, where most laboratories use resistance to metronidazole as an indication that the organisms are facultative anaerobes (as opposed to obligate anaerobes with reduced susceptibilities to metronidazole) (30, 290); thus, a metronidazole-resistant B. fragilis strain would be mistakenly assumed to be a facultative aerobe and not tested further.
Resistance is generally attributed to changes in the nim genes or associated genes (e.g., IS) (30, 56, 75, 259). The nim gene can be “silent” unless activated by IS elements (111). Four nim genes (nimA to -D) can occur in all Bacteroides species; each of these genes is associated with a distinct mobile genetic element, and each also has a specific activating IS element (111, 148, 211, 278). The presence of any one of these genes can be detected by PCR (279). Three additional nim genes (nimE, nimF, and nimG) have also been described (148), but less is known about their mobility and association with IS elements. However, the presence of a nim gene does not necessarily confer clinical levels of metronidazole resistance; in some cases, the gene may not be expressed or may be expressed only at very low levels (96).
Quinolones.
Quinolones inhibit two specific enzymes, DNA gyrase and DNA topoisomerase IV, which aid in bacterial DNA replication, and mutations in these enzymes are the most common causes of quinolone resistance. Mutations in GyrA causing fluoroquinolone resistance in B. fragilis have been identified at hot-spot positions 82 and 86 (equivalent to positions 83 and 87 in E. coli) (174, 213). Substitutions in GyrB, ParC, and ParE have so far proven uncommon and are not well established in B. fragilis. Bacteria may also become resistant by increased export of quinolones out of the cell, and Miyamae et al. found active efflux of norfloxacin by B. fragilis and suggested that the efflux is catalyzed by a pump similar to that of NorA/Bmr (160). Also, B. thetaiotaomicron possesses a MATE (multidrug and toxic compound extrusion) family multidrug efflux exporter (BexA) that pumps out fluoroquinolones (161). We have also demonstrated quinolone resistance in clinical isolates and in laboratory mutants that is due to increased efflux activity of pumps of the RND family (194, 196, 200, 201).
IS Elements
IS typically contain DNA coding for a transposase that allow this element to generate mutations and genome rearrangements, as well as sometimes providing efficient promoters, either carried entirely by the IS element or created as a result of the hybrid sequence between the IS and the target sequence. This is the case for the IS-borne promoters that can be found upstream of the cfiA or ccrA gene (189), macrolide resistance genes (205), and metronidazole resistance nim genes (259, 278).
Mobile Genetic Elements in Bacteroides
Bacteroides species, like other bacteria, have a variety of mechanisms for exchanging genetic information. The frequent presence of mobile genetic elements in Bacteroides is not merely an interesting topic for molecular biologists but critically relevant to both the clinical microbiologist and the infectious disease practitioner. These elements are vitally important in the spread of antibiotic resistance genes. Mobile elements found in Bacteroides include plasmids, transposons, conjugative transposons, and bacteriophages (254), and all but bacteriophages have been implicated in transfer of antimicrobial resistance genes.
Plasmids.
Plasmids are very common in Bacteroides species and are found in ∼20 to 50% of strains (254). Plasmids typically replicate as separate elements in the host cell, although there are some plasmids in other genera that can either integrate into the chromosome or replicate as plasmids, depending on the host (231). Many plasmids possess oriT and a trans-acting mobilization gene, which allow them to be transferred by conjugation (note that trans-acting factors may act on entities that are not in close proximity.) There are also plasmids that do not self-transfer but may be transferred via mobilization by other chromosomal elements (e.g., pBFTM10) (254).
Genes conferring resistance to many different classes of antibiotics have been found on plasmids in Bacteroides. Resistance genes nimA to -F, which have been implicated in sporadic cases of metronidazole resistance in several hospitals worldwide, have been identified on transferable plasmids (56, 148, 279). Transferable plasmid-linked chloramphenicol acetyltransferase conferring high-level resistance was observed in a clinical isolate of Bacteroides uniformis (152). The cfiA gene, conferring resistance to carbapenems, has also been found on a 6.4-kb plasmid in clinical isolates (166). Clindamycin and erythromycin resistance can be transferable between Bacteroides species, either via a chromosomal element (273) or in association with a conjugative plasmid (249, 294).
Conjugative transposons.
Transposons, both mobilizable and conjugative, do not replicate independently; rather, they excise from and integrate into chromosomal DNA and are copied along with the chromosomal DNA. Conjugative transposons have a mechanism of excision and integration that resemble some features of both plasmids and bacteriophage (231). Conjugative transposons are practically ubiquitous among the Bacteroides: over 80% of Bacteroides strains contain at least one conjugative transposon (250).
The particular attributes of conjugative transposons have been elegantly summarized by Salyers et al. (231). Conjugative transposons have certain characteristics of plasmids, of transposons, and of phages. Often, but not always, they are designated by CTn (conjugative transposon) as opposed to Tn (transposon). The conjugative transposons of Bacteroides belong to at least two families; CTnDot is the best described (231, 254). Often, the name of the strain in which they are found is added to the designation (e.g., CTnDot, found in the DOT strain of B. thetaiotaomicron). In addition to being able to insert into the chromosome, Bacteroides conjugative transposons can insert into coresident plasmids and mobilize them in cis (i.e., they can act on entities that are physically adjacent) by integrating themselves into the plasmid and facilitating transfer of the plasmid-conjugative transposon hybrid into another cell (230). They can also mobilize coresident plasmids “in trans” by supplying factors needed to facilitate transfer of the plasmid, while remaining physically separate from the plasmid.
The conjugative transposons do not exclude each other as do plasmids, so a strain can accumulate more than one conjugative transposon. Furthermore, there is some evidence that the presence of more than one copy of the conjugative transposon in the strain results in a stimulation of transposition (transactivation) (231). Theoretically, this suggests that as more conjugative transposons with antibiotic resistance genes accumulate in the environment, the transfer of these genes to other bacteria will also increase, and there will be a significant upward spiraling of antibiotic resistance.
Conjugative transposons have been largely responsible for the spread of tetracycline and erythromycin resistance in clinical isolates of Bacteroides (250). Fifty years ago, tetracycline resistance was almost nonexistent in this genus (103). By the 1980s a majority of strains were resistant, and today almost all strains are resistant. Given the potentially rapid increase in resistance rates, the importance of this mode of antibiotic gene transfer can hardly be overstated.
Many of the Bacteroides transposons carry the tetQ gene and thus confer tetracycline resistance. Further, self-transfer and other activities are significantly stimulated by low levels of tetracycline, regulated by the tetQ-rteA-rteB operon (230). Tetracycline increases transcription of rteA and -B, which code for the sensor and activator components of a two-component regulatory system (discussed above). In turn, RteB activates expression of rteC, which is necessary for self-transfer (231).
Mobilizable transposons.
Mobilizable transposons, like mobilizable plasmids, cannot self-transfer but can transfer between species in the presence of the TcR helper element (254). The most commonly discussed Bacteroides transposons of this class include Tn4399, Tn4555, and the nonreplicating Bacteroides units. The mobilizable transposon Tn4555, for example, was first detected during studies of transmissible cefoxitin resistance in a clinical isolate of Bacteroides vulgatus (182).
Multidrug Resistance
Data from other gram-negative bacteria, including P. aeruginosa, have shown that RND efflux systems can be a major cause of clinically relevant multidrug resistance (191). In contrast, very little is known about efflux pumps in anaerobic bacteria. We described 16 homologs of RND pumps in Bacteroides fragilis (284), and there are two reports indirectly implicating efflux pumps in B. fragilis in antimicrobial resistance, including resistance to norfloxacin (160, 213); also, a MATE-type efflux system has been characterized in B. thetaiotaomicron (161).
RND efflux systems in B. fragilis.
We identified 16 RND family efflux pumps in B. fragilis; we named these bmeABC1 to -16 (B. fragilis multidrug efflux) (284). This system differs from the RND pump system of P. aeruginosa in several respects: (i) each operon has all three components (which was not seen in the Pseudomonas RND pumps [also known as the Mex system pumps]); (ii) the bmeC10 outer membrane component is part of a contiguous gene with the bmeB10 pump gene; (iii) there are two functional pump genes (bmeB11 and bmeB11′) which are transcribed separately in bme11; and (iv) at least 15 of the 16 genes are transcribed (200), whereas in Pseudomonas a few of the pumps are transcribed, one or two more may be induced, and the others are normally silent.
Characterization of bmeB gene function.
In one of our initial experiments exploring bmeB gene function, bme3 was expressed in a hypersusceptible strain of E. coli and resulted in higher MICs of several antimicrobial agents (284). Analysis of mutants with deletions of several of the pump genes (MIC patterns coupled with quantitative analysis of their transcription levels) demonstrated that these efflux systems transport a variety of antimicrobial agents (antibiotics, detergents, dyes, and biocides), have overlapping substrate profiles, and confer intrinsic resistance. Deleting three or more efflux pump genes resulted in overexpression of others pumps, possibly as a compensatory response by the bacterium (200). This increased overexpression of some of these efflux pumps caused an increase in MICs of different antimicrobial agents; these increases could be reversed by the addition of broad-spectrum efflux pump inhibitor compounds such as carbonyl cyanide m-chlorophenylhydrazone, reserpine, MC-207110, and verapamil. The data strongly suggest a complex regulatory feedback system simultaneously involving multiple pumps.
Selection of multidrug-resistant mutants.
We used a variety of antimicrobial agents, including therapeutic choice drugs, to select for multidrug-resistant mutants of B. fragilis (196). We selected mutants from both from a laboratory strain and a series of single, double, triple, and quadruple efflux pump deletants. We used a total of 21 agents (antimicrobials, detergents, and dyes) and were able to obtain mutants using cefoxitin, doripenem, imipenem, metronidazole, levofloxacin, and SDS. The mutants selected by these agents exhibited multidrug resistance to unrelated classes of antimicrobial agents. Ten of the 16 bme efflux pump genes were overexpressed in one or more mutants, strongly suggesting that bmeB overexpression is a major mechanism of multidrug resistance in B. fragilis. The implications of this study corroborate the alarms raised about the overuse of antibiotics and even biocides and demonstrate that exposure to a wide variety of agents can result in multidrug-resistant strains overexpressing multiple efflux pumps. We are currently testing a wide variety of antimicrobial agents, commonly used biocides, and homeopathic “antibacterial” treatments to determine their effect on inducing multidrug resistance (unpublished data).
Multidrug resistance in clinical isolates.
Wareham et al. (290) reported the isolation of a B. fragilis strain from a patient with anaerobic sepsis. The strain was simultaneously resistant to metronidazole, β-lactams, β-lactam/β-lactamase inhibitor combinations, carbapenems, macrolides, and tetracyclines. Although microbiological cure was apparently achieved with linezolid (an oxazolidinone antibiotic), the patient ultimately succumbed to ischemic bowel disease and died.
We studied this isolate and tested it for the presence and expression of genes associated with a variety of resistance mechanisms (201). In addition to the standard 5.31-Mbp chromosome, WI1 possessed a 16-kb low copy-number native plasmid, pBHag, which was absent in the B. fragilis type strain; sequencing of this plasmid revealed that this plasmid contained the gene for tetQ as well as a number of genes that normally reside on the conjugative transposon CTn341. Failed curing attempts suggested that it was stable without obvious selection. We found that the cfiA, ermF, tetQ2, and tetQ3 genes were expressed in the total cellular RNA; neither tetX nor nim genes were detected. Sequencing of the gyrA quinolone resistance-determining region revealed a mutation causing a Ser83→Ile substitution. The efflux pump genes bmeB9 and bmeB15 were significantly overexpressed, and addition of efflux pump inhibitors significantly increased susceptibility of the isolate to several unrelated antibiotics. These data suggest that this isolate was highly multidrug resistant due to additive effects of chromosomally and plasmid-encoded resistance determinants.
We also studied recent multidrug-resistant clinical isolates and found that they overexpressed several bmeB efflux pumps. The data suggest that bmeB efflux pump overexpression can cause low- to intermediate-level clinically relevant fluoroquinolone resistance and can contribute to high-level clinically relevant resistance to β-lactams; moreover, it can be coupled with GyrA substitutions to cause high-level fluoroquinolone resistance (194).
We demonstrated that bmeB5 was overexpressed in metronidazole-resistant laboratory mutants of Bacteroides fragilis, and we identified an upstream putative TetR family regulator gene (bmeR5). Deletant strains lacking bmeB5 or bmeR5 were constructed and characterized. The MICs of ampicillin, cefoperazone, metronidazole, and SDS were reduced by approximately twofold in ADB77 ΔbmeB5. Deletion of bmeR5 (ADB77 ΔbmeR5) resulted in a significant (P < 0.05) increase in expression of bmeA5, bmeB5, and bmeC5 and a >2-fold increase in MICs of ampicillin, cefoxitin, cefoperazone, ciprofloxacin, imipenem, metronidazole, ethidium bromide, and SDS. We found that BmeR5-His6 bound specifically to the bmeR5-bmeA5 intergenic region (IT1). A multidrug (metronidazole)-resistant, nim-negative B. fragilis clinical isolate overexpressed bmeABC5 genes, had a G-T point mutation in IT1, and significantly reduced binding to BmeR5-His6 (195). A schematic description of bmeRABC5 is depicted in Fig. 2.
FIG. 2.
Gene arrangement in the B. fragilis bme5 efflux pump operon and analysis of the B. fragilis bme5 gene sequences. The bme operon codes for the three components of the RND efflux pumps: bmeA codes for the linker protein which connects the pump to the outer membrane barrel, bmeB codes for the efflux pump, and bmeC codes for the outer membrane barrel through which the substance is exported. Most of the B. fragilis bme pump operons are in the order (bmeA-bmeB-bmeC) but some (e.g., bmeABC5, shown here) are different. Another distinctive feature of bmeABC5 is that it has two promoter regions (indicated by the gold stars). Genomic analysis revealed a putative regulator sequence upstream of bmeABC5 (bmeR5). We demonstrated that the BmeR5 protein binds to the first intergenic region (IT1) but not to the second or third intergenic region (IT2 or IT3). A closer analysis of IT1 revealed that it had two inverted repeats (IR) (GGGAAT******ATTCCC) separated by six nucleotides. We analyzed a clinical metronidazole-resistant isolate of B. fragilis and found a G→ T mutation in the IT1 region. Gel shift assays demonstrated that BmeR5 was no longer able to bind to this region.
IMPORTANT FUTURE RESEARCH AVENUES
With the adaptability of a chameleon, the Bacteroides species can function as an integral partner in the human metabolic system and yet can be the cause of serious, life-threatening disease (Table 4). A cartoon description of the some of the most interesting features of a theoretical composite strain of B. fragilis and B. distasonis is depicted in Fig. 3. Despite its predominance as the most numerous organism in the human host, progress in understanding Bacteroides has consistently lagged behind that for aerobic bacteria. However, with the publication of the genome sequences of B. fragilis and B. thetaiotaomicron, several tantalizing potential avenues of research are now within reach. Constructing microarrays for Bacteroides to measure expression of various genes would be a major step toward unraveling some of the complex regulatory relationships found in this organism. For example, the regulation of efflux pump expression appears to be an important factor in multidrug resistance. The multiplicity of the pumps, coupled with their overlapping substrate profiles, suggests that inhibiting their action may need to happen at a regulatory level. These efflux pumps may also be involved in the ability of the organism to thrive in the GI tract. Maintaining the proper balance of GI microbes has been long recognized as an important mechanism to avoid colonization by dangerous pathogens; it is now also recognized as a potentially important factor in obesity. A recent report suggested that understanding the manner by which bacteria adapt to specific niches may aid in designing site-directed bacterial vehicles for delivering beneficial molecules to the human GI tract (90). Happily, many of these long-range research goals appear to be intertwined: efflux pumps that affect antimicrobial resistance may be involved in virulence and adhesion, sugar-acquiring pathways and metabolism that allow adaptation to factors in the GI tract may be important in obesity control, and understanding the regulatory factors responsible for controlling the complex face of the Bacteroides outer membrane may ultimately allow manipulation to reduce resistance and pathogenicity and to exploit its particular characteristics for beneficial purposes.
TABLE 4.
Bacteroides: the good and the bad
| Characteristic |
|---|
| Beneficial effects associated with Bacteroides |
| Starch utilizations systems to break down complex polysaccharides |
| Zwitterionic polysaccharide mediates T-cell activation |
| Aids in development of GALT and a mature immune system |
| Directs Paneth cells to produce antibacterial molecules, including defensins and lectins |
| Deters colonization of the GI tract by pathogens |
| May aid in properly development of immune tolerance and avoidance of allergy and asthma |
| May have a role in preventing obesity |
| Bacteroides as pathogens |
| Can be involved in: |
| Infection after colonic contamination of the abdominal cavity and tissues |
| Bacteremia |
| Skin and soft tissue infections |
| Osteomyelitis |
| May produce enterotoxin and cause GI illness |
| Produces hemolysins, histolytic enzymes that cause tissue destruction |
| May be implicated in Crohn's disease and other IBDs |
| Causes abscess formation |
FIG. 3.
Cartoon depiction of the salient features of a theoretical composite of Bacteroides fragilis and Bacteroides thetaiotaomicron that underscores the extent of its interaction with its environment. (A) Tetracycline has a stimulatory effect on expression of the RteA-RteB two-component regulatory system and induces expression and transfer of tetracycline resistance. The RteA-RteB two-component system controls the expression of a third regulatory gene, rteC, which, in turn controls excision and transfer of CTnDot, a conjugative transposon carrying the tetracycline resistance gene (tetQ). CTnDot can then transfer tetQ to other bacteria. (B and C) ECF-type σ-factor and its membrane-tethered cognate anti-σ-factor. These factors are frequently associated with operons for starch utilization and may be involved in the adaptive ability of B. thetaiotomicron to express different enzymes according to specific nutrient availability. (D) ETBF can excrete an endotoxin implicated in GI illness and possibly IBD. (E) B. fragilis may resist antimicrobial action in a number of ways. Two of these mechanisms are periplasmic β-lactamase enzymes that digest β-lactamases (and, depending on the enzyme, carbapenems). Another mechanism is the RND-type efflux pumps, which can expel the antibiotic and may contribute to multiple antimicrobial resistance. (F) A hybrid two-component signal system interprets the nutrient environment and may also be involved in sensing the neighboring polysaccharide landscape and modulating “mimicry” so that the surface polysaccharide structure of the bacterium can be altered to match the surrounding landscape. This may allow the bacterium to avoid eliciting a host immune response. (G) The capsule of B. fragilis can induce abscesses in the host. Somewhat ironically, the zwitterionic nature of the capsule allows it to “dock” on a groove on the APC and be presented to the CD4+ T cell, which then produces a number of cytokines, including IL-10, which can inhibit abscess formation in the host. (H) Expression of the bmeB RND pumps in B. fragilis can be induced by a variety of agents, including bile, antimicrobial agents, cleansers, and autoinducers important in quorum sensing among bacteria.
Acknowledgments
H.M.W. is supported by Merit Review Funds from the Department of Veterans Affairs.
REFERENCES
- 1.Abi, K. G., H. Awada, and R. Nasnas. 2001. Isolated septic arthritis of a lumbar facet joint. J. Med. Liban. 49:228-230. [PubMed] [Google Scholar]
- 2.Anderson, K. L., and A. A. Salyers. 1989. Genetic evidence that outer membrane binding of starch is required for starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 171:3199-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, K. L., and A. A. Salyers. 1989. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J. Bacteriol. 171:3192-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aucher, P., J. P. Saunier, G. Grollier, M. Sebald, and J. L. Fauchere. 1996. Meningitis due to enterotoxigenic Bacteroides fragilis. Eur. J. Clin. Microbiol. Infect. Dis. 15:820-823. [DOI] [PubMed] [Google Scholar]
- 5.Avelar, K. E., L. J. Pinto, L. C. Antunes, L. A. Lobo, M. C. Bastos, R. M. Domingues, and M. C. Ferreira. 1999. Production of bacteriocin by Bacteriodes fragilis and partial characterization. Lett. Appl. Microbiol. 29:264-268. [DOI] [PubMed] [Google Scholar]
- 6.Ayala, J., A. Quesada, S. Vadillo, J. Criado, and S. Piriz. 2005. Penicillin-binding proteins of Bacteroides fragilis and their role in the resistance to imipenem of clinical isolates. J. Med. Microbiol. 54:1055-1064. [DOI] [PubMed] [Google Scholar]
- 7.Babu, M. M., M. L. Priya, A. T. Selvan, M. Madera, J. Gough, L. Aravind, and K. Sankaran. 2006. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J. Bacteriol. 188:2761-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 9.Bajzer, M., and R. J. Seeley. 2006. Physiology: obesity and gut flora. Nature 444:1009-1010. [DOI] [PubMed] [Google Scholar]
- 10.Bamba, T., H. Matsuda, M. Endo, and Y. Fujiyama. 1995. The pathogenic role of Bacteroides vulgatus in patients with ulcerative colitis. J. Gastroenterol. 30(Suppl. 8):45-47. [PubMed] [Google Scholar]
- 11.Bamias, G., A. Okazawa, J. Rivera-Nieves, K. O. Arseneau, S. A. De La Rue, T. T. Pizarro, and F. Cominelli. 2007. Commensal bacteria exacerbate intestinal inflammation but are not essential for the development of murine ileitis. J. Immunol. 178:1809-1818. [DOI] [PubMed] [Google Scholar]
- 12.Basset, C., J. Holton, A. Bazeos, D. Vaira, and S. Bloom. 2004. Are Helicobacter species and enterotoxigenic Bacteroides fragilis involved in inflammatory bowel disease? Dig. Dis. Sci. 49:1425-1432. [DOI] [PubMed] [Google Scholar]
- 13.Baughn, A. D., and M. H. Malamy. 2004. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427:441-444. [DOI] [PubMed] [Google Scholar]
- 14.Baumann, H., A. O. Tzianabos, J. R. Brisson, D. L. Kasper, and H. J. Jennings. 1992. Structural elucidation of two capsular polysaccharides from one strain of Bacteroides fragilis using high-resolution NMR spectroscopy. Biochemistry 31:4081-4089. [DOI] [PubMed] [Google Scholar]
- 15.Beena, V. K., and P. G. Shivananda. 1997. In vitro adhesiveness of Bacteroides fragilis group in relation to encapsulation. Indian J. Med. Res. 105:258-261. [PubMed] [Google Scholar]
- 16.Begley, M., C. G. Gahan, and C. Hill. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29:625-651. [DOI] [PubMed] [Google Scholar]
- 17.Begley, M., C. Hill, and C. G. Gahan. 2006. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 72:1729-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Behra-Miellet, J., L. Calvet, and L. Dubreuil. 2004. A Bacteroides thetaiotamicron porin that could take part in resistance to beta-lactams. Int. J. Antimicrob. Agents 24:135-143. [DOI] [PubMed] [Google Scholar]
- 19.Behra-Miellet, J., L. Calvet, F. Mory, C. Muller, M. Chomarat, M. C. Bezian, S. Bland, M. E. Juvenin, T. Fosse, F. Goldstein, B. Jaulhac, and L. Dubreuil. 2003. Antibiotic resistance among anaerobic Gram-negative bacilli: lessons from a French multicentric survey. Anaerobe 9:105-111. [DOI] [PubMed] [Google Scholar]
- 20.Bendtsen, J. D., T. T. Binnewies, P. F. Hallin, T. Sicheritz-Ponten, and D. W. Ussery. 2005. Genome update: prediction of secreted proteins in 225 bacterial proteomes. Microbiology 151:1725-1727. [DOI] [PubMed] [Google Scholar]
- 21.Bennion, R. S., E. J. Baron, J. E. Thompson, Jr., J. Downes, P. Summanen, D. A. Talan, and S. M. Finegold. 1990. The bacteriology of gangrenous and perforated appendicitis—revisited. Ann. Surg. 211:165-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berg, J. O., L. Lindqvist, G. Andersson, and C. E. Nord. 1983. Neuraminidase in Bacteroides fragilis. Appl. Environ. Microbiol. 46:75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergan, T. 1983. Anaerobic bacteria as cause of infections in female genital organs. Scand. J. Gastroenterol. Suppl. 85:37-47. [PubMed] [Google Scholar]
- 24.Betriu, C., E. Culebras, M. Gomez, I. Rodriguez-Avial, and J. J. Picazo. 2005. In vitro activity of tigecycline against Bacteroides species. J. Antimicrob. Chemother. 56:349-352. [DOI] [PubMed] [Google Scholar]
- 25.Betriu, C., I. Rodriguez-Avial, M. Gomez, E. Culebras, and J. J. Picazo. 2005. Changing patterns of fluoroquinolone resistance among Bacteroides fragilis group organisms over a 6-year period (1997-2002). Diagn. Microbiol. Infect. Dis. 53:221-223. [DOI] [PubMed] [Google Scholar]
- 26.Bisharat, N., L. Goldstein, R. Raz, and M. Elias. 2001. Gram-negative anaerobic endocarditis: two case reports and review of the literature. Eur. J. Clin. Microbiol. Infect. Dis. 20:651-654. [DOI] [PubMed] [Google Scholar]
- 27.Bjursell, M. K., E. C. Martens, and J. I. Gordon. 2006. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J. Biol. Chem. 281:36269-36279. [DOI] [PubMed] [Google Scholar]
- 28.Boquet, P., P. Munro, C. Fiorentini, and I. Just. 1998. Toxins from anaerobic bacteria: specificity and molecular mechanisms of action. Curr. Opin. Microbiol. 1:66-74. [DOI] [PubMed] [Google Scholar]
- 29.Borer, A., G. Weber, K. Riesenberg, F. Schlaeffer, and J. Horowitz. 1997. Septic arthritis due to Bacteroides fragilis after pilonidal sinus resection in a patient with rheumatoid arthritis. Clin. Rheumatol. 16:632-634. [DOI] [PubMed] [Google Scholar]
- 30.Brazier, J. S., S. L. Stubbs, and B. I. Duerden. 1999. Metronidazole resistance among clinical isolates belonging to the Bacteroides fragilis group: time to be concerned? J. Antimicrob. Chemother. 44:580-581. (Letter.) [DOI] [PubMed] [Google Scholar]
- 31.Brigham, C. J., and M. H. Malamy. 2005. Characterization of the RokA and HexA broad-substrate-specificity hexokinases from Bacteroides fragilis and their role in hexose and N-acetylglucosamine utilization. J. Bacteriol. 187:890-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brioukhanov, A. L., and A. I. Netrusov. 2004. Catalase and superoxide dismutase: distribution, properties, and physiological role in cells of strict anaerobes. Biochemistry (Moscow) 69:949-962. [DOI] [PubMed] [Google Scholar]
- 33.Britz, M. L., and R. G. Wilkinson. 1978. Chloramphenicol acetyl-transferase of Bacteroides fragilis. Antimicrob. Agents Chemother. 14:105-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bron, P. A., M. Marco, S. M. Hoffer, M. E. Van, W. M. de Vos, and M. Kleerebezem. 2004. Genetic characterization of the bile salt response in Lactobacillus plantarum and analysis of responsive promoters in vitro and in situ in the gastrointestinal tract. J. Bacteriol. 186:7829-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brook, I. 2002. Anaerobic infections in children. Microbes. Infect. 4:1271-1280. [DOI] [PubMed] [Google Scholar]
- 36.Brook, I. 2002. Joint and bone infections due to anaerobic bacteria in children. Pediatr. Rehabil. 5:11-19. [DOI] [PubMed] [Google Scholar]
- 37.Brook, I. 2002. Microbiology and management of post-surgical wounds infection in children. Pediatr. Rehabil. 5:171-176. [DOI] [PubMed] [Google Scholar]
- 38.Brook, I. 2003. Microbiology and management of intra-abdominal infections in children. Pediatr. Int. 45:123-129. [DOI] [PubMed] [Google Scholar]
- 39.Brook, I. 2004. Intra-abdominal, retroperitoneal, and visceral abscesses in children. Eur. J. Pediatr. Surg. 14:265-273. [DOI] [PubMed] [Google Scholar]
- 40.Brook, I. 2002. Pericarditis due to anaerobic bacteria. Cardiology 97:55-58. [DOI] [PubMed] [Google Scholar]
- 41.Brook, I. 2002. Microbiology of polymicrobial abscesses and implications for therapy. J. Antimicrob. Chemother. 50:805-810. [DOI] [PubMed] [Google Scholar]
- 42.Brook, I. 2002. Endocarditis due to anaerobic bacteria. Cardiology 98:1-5. [DOI] [PubMed] [Google Scholar]
- 43.Reference deleted.
- 44.Brook, I. 2002. Clinical review: bacteremia caused by anaerobic bacteria in children. Crit. Care 6:205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brook, I. 1989. Aerobic and anaerobic microbiology of Bartholin's abscess. Surg. Gynecol. Obstet. 169:32-34. [PubMed] [Google Scholar]
- 46.Brook, I. 1986. Anaerobic osteomyelitis in children. Pediatr. Infect. Dis. 5:550-556. [DOI] [PubMed] [Google Scholar]
- 47.Brook, I. 1990. Bacteremia due to anaerobic bacteria in newborns. J. Perinatol. 10:351-356. [PubMed] [Google Scholar]
- 48.Brook, I., J. C. Coolbaugh, and R. I. Walker. 1984. Pathogenicity of piliated and encapsulated Bacteroides fragilis. Eur. J. Clin. Microbiol. 3:207-209. [DOI] [PubMed] [Google Scholar]
- 49.Brook, I., and E. H. Frazier. 1992. Microbiology of scalp abscess in newborns. Pediatr. Infect. Dis. J. 11:766-768. [PubMed] [Google Scholar]
- 50.Brook, I., N. Johnson, G. D. Overturf, and J. Wilkins. 1977. Mixed bacterial meningitis: a complication of ventriculo- and lumboperitoneal shunts. J. Neurosurg. 47:961-964. [DOI] [PubMed] [Google Scholar]
- 51.Brook, I., and M. L. Myhal. 1991. Adherence of Bacteroides fragilis group species. Infect. Immun. 59:742-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bryan, L. E., S. K. Kowand, and H. M. Van Den Elzen. 1979. Mechanism of aminoglycoside antibiotic resistance in anaerobic bacteria: Clostridium perfringens and Bacteroides fragilis. Antimicrob. Agents Chemother. 15:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cash, H. L., C. V. Whitham, C. L. Behrendt, and L. V. Hooper. 2006. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313:1126-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cerdeno-Tarraga, A. M., S. Patrick, L. C. Crossman, G. Blakely, V. Abratt, N. Lennard, I. Poxton, B. Duerden, B. Harris, M. A. Quail, A. Barron, L. Clark, C. Corton, J. Doggett, M. T. Holden, N. Larke, A. Line, A. Lord, H. Norbertczak, D. Ormond, C. Price, E. Rabbinowitsch, J. Woodward, B. Barrell, and J. Parkhill. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307:1463-1465. [DOI] [PubMed] [Google Scholar]
- 55.Reference deleted.
- 56.Chaudhry, R., P. Mathur, B. Dhawan, and L. Kumar. 2001. Emergence of metronidazole-resistant Bacteroides fragilis, India. Emerg. Infect. Dis. 7:485-486. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen, Y., T. Kinouchi, K. Kataoka, S. Akimoto, and Y. Ohnishi. 1995. Purification and characterization of a fibrinogen-degrading protease in Bacteroides fragilis strain YCH46. Microbiol. Immunol. 39:967-977. [DOI] [PubMed] [Google Scholar]
- 58.Cho, K. H., D. Cho, G. R. Wang, and A. A. Salyers. 2001. New regulatory gene that contributes to control of Bacteroides thetaiotaomicron starch utilization genes. J. Bacteriol. 183:7198-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho, K. H., and A. A. Salyers. 2001. Biochemical analysis of interactions between outer membrane proteins that contribute to starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 183:7224-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cisneros, J. M., J. Rodriguez-Bano, F. Fernandez-Cuenca, A. Ribera, J. Vila, A. Pascual, L. Martinez-Martinez, G. Bou, and J. Pachon. 2005. Risk-factors for the acquisition of imipenem-resistant Acinetobacter baumannii in Spain: a nationwide study. Clin. Microbiol. Infect. 11:874-879. [DOI] [PubMed] [Google Scholar]
- 61.Claros, M. C., Z. C. Claros, D. W. Hecht, D. M. Citron, E. J. Goldstein, J. Silva, Jr., Y. Tang-Feldman, and A. C. Rodloff. 2006. Characterization of the Bacteroides fragilis pathogenicity island in human blood culture isolates. Anaerobe 12:17-22. [DOI] [PubMed] [Google Scholar]
- 62.Claros, M. C., Z. C. Claros, Y. J. Tang, S. H. Cohen, J. Silva, Jr., E. J. Goldstein, and A. C. Rodloff. 2000. Occurrence of Bacteroides fragilis enterotoxin gene-carrying strains in Germany and the United States. J. Clin. Microbiol. 38:1996-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clinical and Laboratory Standards Institute. 2006. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard, 7th ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 64.Cohavy, O., D. Bruckner, L. K. Gordon, R. Misra, B. Wei, M. E. Eggena, S. R. Targan, and J. Braun. 2000. Colonic bacteria express an ulcerative colitis pANCA-related protein epitope. Infect. Immun. 68:1542-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Comstock, L. E., and M. J. Coyne. 2003. Bacteroides thetaiotaomicron: a dynamic, niche-adapted human symbiont. Bioessays 25:926-929. [DOI] [PubMed] [Google Scholar]
- 66.Corzo, G., and S. E. Gilliland. 1999. Measurement of bile salt hydrolase activity from Lactobacillus acidophilus based on disappearance of conjugated bile salts. J. Dairy Sci. 82:466-471. [DOI] [PubMed] [Google Scholar]
- 67.Coyne, M. J., W. Kalka-Moll, A. O. Tzianabos, D. L. Kasper, and L. E. Comstock. 2000. Bacteroides fragilis NCTC9343 produces at least three distinct capsular polysaccharides: cloning, characterization, and reassignment of polysaccharide B and C biosynthesis loci. Infect. Immun. 68:6176-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coyne, M. J., B. Reinap, M. M. Lee, and L. E. Comstock. 2005. Human symbionts use a host-like pathway for surface fucosylation. Science 307:1778-1781. [DOI] [PubMed] [Google Scholar]
- 69.Coyne, M. J., K. G. Weinacht, C. M. Krinos, and L. E. Comstock. 2003. Mpi recombinase globally modulates the surface architecture of a human commensal bacterium. Proc. Natl. Acad. Sci. USA 100:10446-10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Boever, P., and W. Verstraete. 1999. Bile salt deconjugation by Lactobacillus plantarum 80 and its implication for bacterial toxicity. J. Appl. Microbiol. 87:345-352. [DOI] [PubMed] [Google Scholar]
- 71.Delahooke, D. M., G. R. Barclay, and I. R. Poxton. 1995. A re-appraisal of the biological activity of bacteroides [sic] LPS. J. Med. Microbiol. 42:102-112. [DOI] [PubMed] [Google Scholar]
- 72.D'Elia, J. N., and A. A. Salyers. 1996. Contribution of a neopullulanase, a pullulanase, and an alpha-glucosidase to growth of Bacteroides thetaiotaomicron on starch. J. Bacteriol. 178:7173-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D'Elia, J. N., and A. A. Salyers. 1996. Effect of regulatory protein levels on utilization of starch by Bacteroides thetaiotaomicron. J. Bacteriol. 178:7180-7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de O. Ferreira, E., L. Araujo Lobo, D. Barreiros Petropolis, K. dos S. Avelar, M. C. Ferreira, F. Costa e Silva Filho, and R. M. Domingues. 2006. A Bacteroides fragilis surface glycoprotein mediates the interaction between the bacterium and the extracellular matrix component laminin-1. Res. Microbiol. 157:960-966. [DOI] [PubMed] [Google Scholar]
- 75.Diniz, C. G., L. M. Farias, M. A. Carvalho, E. R. Rocha, and C. J. Smith. 2004. Differential gene expression in a Bacteroides fragilis metronidazole-resistant mutant. J. Antimicrob. Chemother. 54:100-108. [DOI] [PubMed] [Google Scholar]
- 76.Dodd, M. J., I. D. Griffiths, and R. Freeman. 1982. Pyogenic arthritis due to bacteroides [sic] complicating rheumatoid arthritis. Ann. Rheum. Dis. 41:248-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Domingues, R. M., S. M. Cavalcanti, A. F. Andrade, and M. C. Ferreira. 1992. Sialic acid as receptor of Bacteroides fragilis lectin-like adhesin. Zentbl. Bakteriol. 277:340-344. [DOI] [PubMed] [Google Scholar]
- 78.Domingues, R. M., W. das G. Silva e Souza, S. R. Moraes, K. E. Avelar, J. R. Hirata, M. E. Fonseca, and M. C. Ferreira. 1997. Surface vesicles: a possible function in commensal relations of Bacteroides fragilis. Zentbl. Bakteriol. 285:509-517. [DOI] [PubMed] [Google Scholar]
- 79.Duncan, S. H., A. Belenguer, G. Holtrop, A. M. Johnstone, H. J. Flint, and G. E. Lobley. 2007. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 73:1073-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dutson, T. R., A. M. Pearson, J. F. Price, G. C. Spink, and P. J. Tarrant. 1971. Observations by electron microscopy on pig muscle inoculated and incubated with Pseudomonas fragi. Appl. Microbiol. 22:1152-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edmiston, C. E., Jr., C. J. Krepel, K. S. Kehl, G. R. Seabrook, L. B. Somberg, G. H. Almassi, T. L. Smith, T. A. Loehrl, K. R. Brown, B. D. Lewis, and J. B. Towne. 2005. Comparative in vitro antimicrobial activity of a novel quinolone, garenoxacin, against aerobic and anaerobic microbial isolates recovered from general, vascular, cardiothoracic and otolaryngologic surgical patients. J. Antimicrob. Chemother. 56:872-878. [DOI] [PubMed] [Google Scholar]
- 82.Edwards, R. 1997. Resistance to beta-lactam antibiotics in Bacteroides spp. J. Med. Microbiol. 46:979-986. [DOI] [PubMed] [Google Scholar]
- 83.Edwards, R., and D. Greenwood. 1996. Mechanisms responsible for reduced susceptibility to imipenem in Bacteroides fragilis. J. Antimicrob. Chemother. 38:941-951. [DOI] [PubMed] [Google Scholar]
- 84.Elliott, D., J. A. Kufera, and R. A. Myers. 2000. The microbiology of necrotizing soft tissue infections. Am. J. Surg. 179:361-366. [DOI] [PubMed] [Google Scholar]
- 85.Fang, H., C. Edlund, C. E. Nord, and M. Hedberg. 2002. Selection of cefoxitin-resistant Bacteroides thetaiotaomicron mutants and mechanisms involved in β-lactam resistance. Clin. Infect. Dis. 35:S47-S53. [DOI] [PubMed] [Google Scholar]
- 86.Fenner, L., V. Roux, M. N. Mallet, and D. Raoult. 2005. Bacteroides massiliensis sp. nov., isolated from blood culture of a newborn. Int. J. Syst. Evol. Microbiol. 55:1335-1337. [DOI] [PubMed] [Google Scholar]
- 87.Ferreira, R., M. C. Alexandre, E. N. Antunes, A. T. Pinhao, S. R. Moraes, M. C. Ferreira, and R. M. Domingues. 1999. Expression of Bacteroides fragilis virulence markers in vitro. J. Med. Microbiol. 48:999-1004. [DOI] [PubMed] [Google Scholar]
- 88.Feuillet, L., J. Carvajal, I. Sudre, J. Pelletier, J. M. Thomassin, M. Drancourt, and A. A. Cherif. 2005. First isolation of Bacteroides thetaiotaomicron from a patient with a cholesteatoma and experiencing meningitis. J. Clin. Microbiol. 43:1467-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fletcher, H. M., and F. L. Macrina. 1991. Molecular survey of clindamycin and tetracycline resistance determinants in Bacteroides species. Antimicrob. Agents Chemother. 35:2415-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Forum on Microbial Threats. 2006. Manipulating host-microbe interactions: probiotic research and regulations, p. 207. In Ending the war metaphor: the changing agenda for unraveling the host-microbe relationship. The National Academies Press, Washington, DC. [PubMed]
- 91.Franco, A. A. 2004. The Bacteroides fragilis pathogenicity island is contained in a putative novel conjugative transposon. J. Bacteriol. 186:6077-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Franco, A. A., R. K. Cheng, G. T. Chung, S. Wu, H. B. Oh, and C. L. Sears. 1999. Molecular evolution of the pathogenicity island of enterotoxigenic Bacteroides fragilis strains. J. Bacteriol. 181:6623-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Frank, G. R., and L. G. Rubin. 1989. Mixed meningitis with Bacteroides ovatus caused by an occult congenital dermal sinus. Pediatr. Infect. Dis. J. 8:401-403. [DOI] [PubMed] [Google Scholar]
- 94.Freitas, M., E. Tavan, C. Cayuela, L. Diop, C. Sapin, and G. Trugnan. 2003. Host-pathogens cross-talk. Indigenous bacteria and probiotics also play the game. Biol. Cell 95:503-506. [DOI] [PubMed] [Google Scholar]
- 95.Fujita, H., Y. Eishi, I. Ishige, K. Saitoh, T. Takizawa, T. Arima, and M. Koike. 2002. Quantitative analysis of bacterial DNA from Mycobacteria spp., Bacteroides vulgatus, and Escherichia coli in tissue samples from patients with inflammatory bowel diseases. J. Gastroenterol. 37:509-516. [DOI] [PubMed] [Google Scholar]
- 96.Gal, M., and J. S. Brazier. 2004. Metronidazole resistance in Bacteroides spp. carrying nim genes and the selection of slow-growing metronidazole-resistant mutants. J. Antimicrob. Chemother. 54:109-116. [DOI] [PubMed] [Google Scholar]
- 97.Ganz, T. 2000. Paneth cells-guardians of the gut cell hatchery. Nat. Immunol. 1:99-100. [DOI] [PubMed] [Google Scholar]
- 98.Gemmell, C. G., P. K. Peterson, D. Schmeling, J. Mathews, and P. G. Quie. 1983. Antibiotic-induced modification of Bacteroides fragilis and its susceptibility to phagocytosis by human polymorphonuclear leukocytes. Eur. J. Clin. Microbiol. 2:327-334. [DOI] [PubMed] [Google Scholar]
- 99.Georgopapadakou, N. H., S. A. Smith, and R. B. Sykes. 1983. Penicillin-binding proteins in Bacteroides fragilis. J. Antibiot. (Tokyo) 36:907-910. [DOI] [PubMed] [Google Scholar]
- 100.Germain, M., M. A. Krohn, S. L. Hillier, and D. A. Eschenbach. 1994. Genital flora in pregnancy and its association with intrauterine growth retardation. J. Clin. Microbiol. 32:2162-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gibson, S. A., and G. T. Macfarlane. 1988. Characterization of proteases formed by Bacteroides fragilis. J. Gen. Microbiol. 134:2231-2240. [DOI] [PubMed] [Google Scholar]
- 102.Gill, S. R., M. Pop, R. T. DeBoy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gillespie, W. A., and J. Guy. 1956. Bacteroides in intra-abdominal sepsis. Lancet i:1039-1041. [DOI] [PubMed] [Google Scholar]
- 104.Gilmore, M. S., and J. J. Ferretti. 2003. The thin line between gut commensal and pathogen. Science 299:1999-2002. [DOI] [PubMed] [Google Scholar]
- 105.Godoy, V. G., M. M. Dallas, T. A. Russo, and M. H. Malamy. 1993. A role for Bacteroides fragilis neuraminidase in bacterial growth in two model systems. Infect. Immun. 61:4415-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Goh, B. K., G. Alkouder, T. K. Lama, and C. E. Tan. 2005. Multi-drug-resistant Acinetobacter baumannii intra-abdominal abscess. Surg. Infect. 6:345-347. [DOI] [PubMed] [Google Scholar]
- 107.Goldstein, E. J. 1996. Anaerobic bacteremia. Clin. Infect. Dis. 23(Suppl. 1):S97-S101. [DOI] [PubMed] [Google Scholar]
- 108.Gootz, T. D. 2006. The forgotten Gram-negative bacilli: what genetic determinants are telling us about the spread of antibiotic resistance. Biochem. Pharmacol. 71:1073-1084. [DOI] [PubMed] [Google Scholar]
- 109.Grenier, D., and D. Mayrand. 1987. Functional characterization of extracellular vesicles produced by Bacteroides gingivalis. Infect. Immun. 55:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guzman, C. A., M. Plate, and C. Pruzzo. 1990. Role of neuraminidase-dependent adherence in Bacteroides fragilis attachment to human epithelial cells. FEMS Microbiol. Lett. 59:187-192. [DOI] [PubMed] [Google Scholar]
- 111.Haggoud, A., G. Reysset, H. Azeddoug, and M. Sebald. 1994. Nucleotide sequence analysis of two 5-nitroimidazole resistance determinants from Bacteroides strains and of a new insertion sequence upstream of the two genes. Antimicrob. Agents Chemother. 38:1047-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hart, C. A., V. M. Godfrey, J. C. Woodrow, and A. Percival. 1982. Septic arthritis due to Bacteroides fragilis in a wrist affected by rheumatoid arthritis. Ann. Rheum. Dis. 41:623-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hecht, D. W. 2006. Anaerobes: antibiotic resistance, clinical significance, and the role of susceptibility testing. Anaerobe 12:115-121. [DOI] [PubMed] [Google Scholar]
- 114.Hecht, D. W. 2004. Prevalence of antibiotic resistance in anaerobic bacteria: worrisome developments. Clin. Infect. Dis. 39:92-97. [DOI] [PubMed] [Google Scholar]
- 115.Hedberg, M., and C. E. Nord. 2003. Antimicrobial susceptibility of Bacteroides fragilis group isolates in Europe. Clin. Microbiol. Infect. 9:475-488. [DOI] [PubMed] [Google Scholar]
- 116.Heinonen, P. K., and M. Leinonen. 2003. Fecundity and morbidity following acute pelvic inflammatory disease treated with doxycycline and metronidazole. Arch. Gynecol. Obstet. 268:284-288. [DOI] [PubMed] [Google Scholar]
- 117.Hooper, L. V., and J. I. Gordon. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115-1118. [DOI] [PubMed] [Google Scholar]
- 118.Hooper, L. V., T. Midtvedt, and J. I. Gordon. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22:283-307. [DOI] [PubMed] [Google Scholar]
- 119.Hooper, L. V., T. S. Stappenbeck, C. V. Hong, and J. I. Gordon. 2003. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 4:269-273. [DOI] [PubMed] [Google Scholar]
- 120.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881-884. [DOI] [PubMed] [Google Scholar]
- 121.Hooper, L. V., J. Xu, P. G. Falk, T. Midtvedt, and J. I. Gordon. 1999. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. USA 96:9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hopkins, M. J., and G. T. Macfarlane. 2002. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J. Med. Microbiol. 51:448-454. [DOI] [PubMed] [Google Scholar]
- 123.Hopkins, M. J., and G. T. Macfarlane. 2003. Nondigestible oligosaccharides enhance bacterial colonization resistance against Clostridium difficile in vitro. Appl. Environ. Microbiol. 69:1920-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hung, M. N., S. Y. Chen, J. L. Wang, S. C. Chang, P. R. Hsueh, C. H. Liao, and Y. C. Chen. 2005. Community-acquired anaerobic bacteremia in adults: one-year experience in a medical center. J. Microbiol. Immunol. Infect. 38:436-443. [PubMed] [Google Scholar]
- 125.Jain, R., and L. H. Danziger. 2004. Multidrug-resistant Acinetobacter infections: an emerging challenge to clinicians. Ann. Pharmacother. 38:1449-1459. [DOI] [PubMed] [Google Scholar]
- 126.Johnson, C. C., J. F. Reinhardt, M. A. Edelstein, M. E. Mulligan, W. L. George, and S. M. Finegold. 1985. Bacteroides gracilis, an important anaerobic bacterial pathogen. J. Clin. Microbiol. 22:799-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jousimies-Somer, H., P. Summanen, D. M. Citron, E. J. Baron, H. M. Wexler, and S. M. Finegold. 2002. Wadsworth-KTL anaerobic bacteriology manual. Star Publishing Company, Belmont, CA.
- 128.Jousimies-Somer, H., P. Summanen, and S. M. Finegold. 2003. Bacteroides, Porphyromonas, Prevotella, Fusobacterium and other anaerobic gram-negative rods and cocci, p. 690-711. In P. R. Murray, E. J. Baron, J. H. P. M. A. Jorgensen, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 129.Kalka-Moll, W. M., Y. Wang, L. E. Comstock, S. E. Gonzalez, A. O. Tzianabos, and D. L. Kasper. 2001. Immunochemical and biological characterization of three capsular polysaccharides from a single Bacteroides fragilis strain. Infect. Immun. 69:2339-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kanazawa, K., Y. Kobayashi, H. Nakano, M. Sakurai, N. Gotoh, and T. Nishino. 1995. Identification of three porins in the outer membrane of Bacteroides fragilis. FEMS Microbiol. Lett. 127:181-186. [Google Scholar]
- 131.Kazmierczak, M. J., M. Wiedmann, and K. J. Boor. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69:527-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Khandelwal, P., D. Choudhury, A. Birah, M. K. Reddy, G. P. Gupta, and N. Banerjee. 2004. Insecticidal pilin subunit from the insect pathogen Xenorhabdus nematophila. J. Bacteriol. 186:6465-6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kim, J. M., J. Y. Lee, Y. M. Yoon, Y. K. Oh, J. S. Kang, Y. J. Kim, and K. H. Kim. 2006. Bacteroides fragilis enterotoxin induces cyclooxygenase-2 and fluid secretion in intestinal epithelial cells through NF-kappaB activation. Eur. J. Immunol. 36:2446-2456. [DOI] [PubMed] [Google Scholar]
- 134.Kislak, J. W. 1972. The susceptibility of Bacteroides fragilis to 24 antibiotics. J. Infect. Dis. 125:295-298. [DOI] [PubMed] [Google Scholar]
- 135.Kitahara, M., M. Sakamoto, M. Ike, S. Sakata, and Y. Benno. 2005. Bacteroides plebeius sp. nov. and Bacteroides coprocola sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 55:2143-2147. [DOI] [PubMed] [Google Scholar]
- 136.Kling, J. J., R. L. Wright, J. S. Moncrief, and T. D. Wilkins. 1997. Cloning and characterization of the gene for the metalloprotease enterotoxin of Bacteroides fragilis. FEMS Microbiol. Lett. 146:279-284. [DOI] [PubMed] [Google Scholar]
- 137.Krinos, C. M., M. J. Coyne, K. G. Weinacht, A. O. Tzianabos, D. L. Kasper, and L. E. Comstock. 2001. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414:555-558. [DOI] [PubMed] [Google Scholar]
- 138.Kuwahara, T., A. Yamashita, H. Hirakawa, H. Nakayama, H. Toh, N. Okada, S. Kuhara, M. Hattori, T. Hayashi, and Y. Ohnishi. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. USA 101:14919-14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lassmann, B., D. R. Gustafson, C. M. Wood, and J. E. Rosenblatt. 2007. Reemergence of anaerobic bacteremia. Clin. Infect. Dis. 44:895-900. [DOI] [PubMed] [Google Scholar]
- 140.Leng, Z., D. E. Riley, R. E. Berger, J. N. Krieger, and M. C. Roberts. 1997. Distribution and mobility of the tetracycline resistance determinant tetQ. J. Antimicrob. Chemother. 40:551-559. [DOI] [PubMed] [Google Scholar]
- 141.Leszczynski, P., F. Meisel-Mikolajczyk, M. Dworczynska, L. Cwyl-Zembrzuska, and L. Marianowski. 1995. Occurrence of Bacteroides fragilis strains in full term and post term pregnancies. Ginekol. Pol. 66:324-329. [PubMed] [Google Scholar]
- 142.Leszczynski, P., B. A. Van, H. Pituch, H. Verbrugh, and F. Meisel-Mikolajczyk. 1997. Vaginal carriage of enterotoxigenic Bacteroides fragilis in pregnant women. J. Clin. Microbiol. 35:2899-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ley, R. E., F. Backhed, P. Turnbaugh, C. A. Lozupone, R. D. Knight, and J. I. Gordon. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 102:11070-11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ley, R. E., P. J. Turnbaugh, S. Klein, and J. I. Gordon. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022-1023. [DOI] [PubMed] [Google Scholar]
- 145.Lindner, J. G., F. H. Plantema, and J. A. Hoogkamp-Korstanje. 1978. Quantitative studies of the vaginal flora of healthy women and of obstetric and gynaecological patients. J. Med. Microbiol. 11:233-241. [DOI] [PubMed] [Google Scholar]
- 146.Liu, C., Y. Song, M. McTeague, A. W. Vu, H. Wexler, and S. M. Finegold. 2003. Rapid identification of the species of the Bacteroides fragilis group by multiplex PCR assays using group- and species-specific primers. FEMS Microbiol. Lett. 222:9-16. [DOI] [PubMed] [Google Scholar]
- 147.Livermore, D. M., and N. Woodford. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489-495. [DOI] [PubMed] [Google Scholar]
- 148.Lofmark, S., H. Fang, M. Hedberg, and C. Edlund. 2005. Inducible metronidazole resistance and nim genes in clinical Bacteroides fragilis group isolates. Antimicrob. Agents Chemother. 49:1253-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Macfarlane, G. T., S. Macfarlane, and G. R. Gibson. 1992. Synthesis and release of proteases by Bacteroides fragilis. Curr. Microbiol. 24:55-59. [Google Scholar]
- 150.Mackie, R. I., A. Sghir, and H. R. Gaskins. 1999. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 69:1035S-1045S. [DOI] [PubMed] [Google Scholar]
- 151.Marculescu, C. E., E. F. Berbari, F. R. Cockerill III, and D. R. Osmon. 2006. Unusual aerobic and anaerobic bacteria associated with prosthetic joint infections. Clin. Orthop. Relat. Res. 451:55-63. [DOI] [PubMed] [Google Scholar]
- 152.Martinez-Suarez, J. V., F. Baquero, M. Reig, and J. C. Perez-Diaz. 1985. Transferable plasmid-linked chloramphenicol acetyltransferase conferring high-level resistance in Bacteroides uniformis. Antimicrob. Agents Chemother. 28:113-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mashburn, L. M., and M. Whiteley. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422-425. [DOI] [PubMed] [Google Scholar]
- 154.Mazmanian, S. K., and D. L. Kasper. 2006. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat. Rev. Immunol. 6:849-858. [DOI] [PubMed] [Google Scholar]
- 155.Mazmanian, S. K., C. H. Liu, A. O. Tzianabos, and D. L. Kasper. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107-118. (Comment.). [DOI] [PubMed] [Google Scholar]
- 156.McCarthy, R. E., M. Pajeau, and A. A. Salyers. 1988. Role of starch as a substrate for Bacteroides vulgatus growing in the human colon. Appl. Environ. Microbiol. 54:1911-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Merle-Melet, M., D. Mainard, D. Regent, C. Dopff, J. N. Tamisier, P. Ross, J. P. Delagoutte, and A. Gerard. 1994. An unusual case of hip septic arthritis due to Bacteroides fragilis in an alcoholic patient. Infection 22:353-355. [DOI] [PubMed] [Google Scholar]
- 158.Merriam, C. V., H. T. Fernandez, D. M. Citron, K. L. Tyrrell, Y. A. Warren, and E. J. Goldstein. 2003. Bacteriology of human bite wound infections. Anaerobe 9:83-86. [DOI] [PubMed] [Google Scholar]
- 159.Miki, T., T. Kuwahara, H. Nakayama, N. Okada, K. Kataoka, H. Arimochi, and Y. Ohnishi. 2005. Simultaneous detection of Bacteroides fragilis group species by leuB-directed PCR. J. Med. Investig. 52:101-108. [DOI] [PubMed] [Google Scholar]
- 160.Miyamae, S., H. Nikaido, Y. Tanaka, and F. Yoshimura. 1998. Active efflux of norfloxacin by Bacteroides fragilis. Antimicrob. Agents Chemother. 42:2119-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Miyamae, S., O. Ueda, F. Yoshimura, J. Hwang, Y. Tanaka, and H. Nikaido. 2001. A MATE family multidrug efflux transporter pumps out fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob. Agents Chemother. 45:3341-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Moncrief, J. S., R. Obiso, Jr., L. A. Barroso, J. J. Kling, R. L. Wright, R. L. Van Tassell, D. M. Lyerly, and T. D. Wilkins. 1995. The enterotoxin of Bacteroides fragilis is a metalloprotease. Infect. Immun. 63:175-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Montejo, B. M., A. P. Gonzalez de Zarate, F. J. Barron, and E. C. Aguirre. 1984. Meningitis caused by Bacteroides fragilis. Can. Med. Assoc. J. 131:184-186. [PMC free article] [PubMed] [Google Scholar]
- 164.Moon, K., N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2005. Regulation of excision genes of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 187:5732-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mossie, K. G., F. T. Robb, D. T. Jones, and D. R. Woods. 1981. Inhibition of ribonucleic acid polymerase by a bacteriocin from Bacteroides fragilis. Antimicrob. Agents Chemother. 20:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nakano, V., G. Padilla, M. M. do Valle, and M. J. Vila-Campos. 2004. Plasmid-related beta-lactamase production in Bacteroides fragilis strains. Res. Microbiol. 155:843-846. [DOI] [PubMed] [Google Scholar]
- 167.Namavar, F., M. W. Van der Bijl, B. J. Appelmelk, J. de Graaff, and D. M. Maclaren. 1994. The role of neuraminidase in haemagglutination and adherence to colon WiDr cells by Bacteroides fragilis. J. Med. Microbiol. 40:393-396. [DOI] [PubMed] [Google Scholar]
- 168.Ngan, C. C., and A. L. Tan. 1994. Bacteroides fragilis meningitis. Singapore Med. J. 35:283-285. [PubMed] [Google Scholar]
- 169.Nguyen, M. H., V. L. Yu, A. J. Morris, L. McDermott, M. W. Wagener, L. Harrell, and D. R. Snydman. 2000. Antimicrobial resistance and clinical outcome of Bacteroides bacteremia: findings of a multicenter prospective observational trial. Clin. Infect. Dis. 30:870-876. [DOI] [PubMed] [Google Scholar]
- 170.Odou, M. F., E. Singer, and L. Dubreuil. 2001. Description of complex forms of a porin in Bacteroides fragilis and possible implication of this protein in antibiotic resistance. Anaerobe 7:219-225. [Google Scholar]
- 171.Odou, M. F., E. Singer, M. B. Romond, and L. Dubreuil. 1998. Isolation and characterization of a porin-like protein of 45 kilodaltons from Bacteroides fragilis. FEMS Microbiol. Lett. 166:347-354. [DOI] [PubMed] [Google Scholar]
- 172.Odugbemi, T., S. A. Jatto, and K. Afolabi. 1985. Bacteroides fragilis meningitis. J. Clin. Microbiol. 21:282-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ogawa, W., D. W. Li, P. Yu, A. Begum, T. Mizushima, T. Kuroda, and T. Tsuchiya. 2005. Multidrug resistance in Klebsiella pneumoniae MGH78578 and cloning of genes responsible for the resistance. Biol. Pharm. Bull. 28:1505-1508. [DOI] [PubMed] [Google Scholar]
- 174.Oh, H., N. El Amin, T. Davies, P. C. Appelbaum, and C. Edlund. 2001. gyrA mutations associated with quinolone resistance in Bacteroides fragilis group strains. Antimicrob. Agents Chemother. 45:1977-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Osborne, N. G. 2006. The role of Bacteroides fragilis in pelvic infections. J. Gynecol. Surg. 22:81-82. [Google Scholar]
- 176.Otto, B. R., M. Sparrius, A. M. Verweij-van Vught, and D. M. Maclaren. 1990. Iron-regulated outer membrane protein of Bacteroides fragilis involved in heme uptake. Infect. Immun. 58:3954-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Oyston, P. C., and P. S. Handley. 1990. Surface structures, haemagglutination and cell surface hydrophobicity of Bacteroides fragilis strains. J. Gen. Microbiol. 136:941-948. [DOI] [PubMed] [Google Scholar]
- 178.Oyston, P. C., and P. S. Handley. 1991. Surface components of Bacteroides fragilis involved in adhesion and haemagglutination. J. Med. Microbiol. 34:51-55. [DOI] [PubMed] [Google Scholar]
- 179.Pantosti, A., A. O. Tzianabos, A. B. Onderdonk, and D. L. Kasper. 1991. Immunochemical characterization of two surface polysaccharides of Bacteroides fragilis. Infect. Immun. 59:2075-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pantosti, A., A. O. Tzianabos, B. G. Reinap, A. B. Onderdonk, and D. L. Kasper. 1993. Bacteroides fragilis strains express multiple capsular polysaccharides. J. Clin. Microbiol. 31:1850-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Park, J. T. 2001. Identification of a dedicated recycling pathway for anhydro-N-acetylmuramic acid and N-acetylglucosamine derived from Escherichia coli cell wall murein. J. Bacteriol. 183:3842-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Parker, A. C., and C. J. Smith. 1993. Genetic and biochemical analysis of a novel Ambler class A beta-lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrob. Agents Chemother. 37:1028-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Patey, O., J. Breuil, A. Fisch, C. Burnat, F. Vincent-Ballereau, and A. Dublanchet. 1990. Bacteroides fragilis meningitis: report of two cases. Rev. Infect. Dis. 12:364-365. [DOI] [PubMed] [Google Scholar]
- 184.Patrick, S., D. Gilpin, and L. Stevenson. 1999. Detection of intrastrain antigenic variation of Bacteroides fragilis surface polysaccharides by monoclonal antibody labelling. Infect. Immun. 67:4346-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Patrick, S., J. P. McKenna, S. O'Hagan, and E. Dermott. 1996. A comparison of the haemagglutinating and enzymic activities of Bacteroides fragilis whole cells and outer membrane vesicles. Microb. Pathog. 20:191-202. [DOI] [PubMed] [Google Scholar]
- 186.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 187.Piddock, L. J. V., and R. Wise. 1987. Cefoxitin resistance in Bacteroides species: evidence indicating two mechanisms causing decreased susceptibility. J. Antimicrob. Chemother. 19:161-170. [DOI] [PubMed] [Google Scholar]
- 188.Piriz, S., S. Vadillo, A. Quesada, J. Criado, R. Cerrato, and J. Ayala. 2004. Relationship between penicillin-binding protein patterns and beta-lactamases in clinical isolates of Bacteroides fragilis with different susceptibility to beta-lactam antibiotics. J. Med. Microbiol. 53:213-221. [DOI] [PubMed] [Google Scholar]
- 189.Podglajen, I., J. Breuil, and E. Collatz. 1994. Insertion of a novel DNA sequence, IS1186, upstream of the silent carbapenemase gene cfiA, promotes expression of carbapenem resistance in clinical isolates of Bacteroides fragilis. Mol. Microbiol. 12:105-114. [DOI] [PubMed] [Google Scholar]
- 190.Polk, B. F., and D. L. Kasper. 1977. Bacteroides fragilis subspecies in clinical isolates. Ann. Intern. Med. 86:569-571. [DOI] [PubMed] [Google Scholar]
- 191.Poole, K., and R. Srikumar. 2001. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top. Med. Chem. 1:59-71. [DOI] [PubMed] [Google Scholar]
- 192.Prindiville, T. P., R. A. Sheikh, S. H. Cohen, Y. J. Tang, M. C. Cantrell, and J. Silva, Jr. 2000. Bacteroides fragilis enterotoxin gene sequences in patients with inflammatory bowel disease. Emerg. Infect. Dis. 6:171-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pruzzo, C., B. Dainelli, and M. Ricchetti. 1984. Piliated Bacteroides fragilis strains adhere to epithelial cells and are more sensitive to phagocytosis by human neutrophils than nonpiliated strains. Infect. Immun. 43:189-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pumbwe, L., A. Chang, R. L. Smith, and H. M. Wexler. 2006. Clinical significance of overexpression of multiple RND-family efflux pumps in Bacteroides fragilis isolates. J. Antimicrob. Chemother. 58:543-548. [DOI] [PubMed] [Google Scholar]
- 195.Pumbwe, L., A. Chang, and H. M. Wexler. 2007. The RND-family transport system BmeRABC-5 is a metronidazole efflux pump in Bacteroides fragilis. Microb. Drug Resist. 13:96-101. [DOI] [PubMed] [Google Scholar]
- 196.Pumbwe, L., D. Glass, and H. M. Wexler. 2006. Efflux pump overexpression in multiple antibiotic resistant mutants of Bacteroides fragilis. Antimicrob. Agents Chemother. 50:3150-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Pumbwe, L., C. Skilbeck, A. Oren, and H. M. Wexler. 2007. Bile salts enhance bacterial co-aggregation, bacterial-intestinal epithelial cell adhesion, biofilm formation and antimicrobial resistance of Bacteroides fragilis. Microb. Pathog. 43:78-87. [DOI] [PubMed] [Google Scholar]
- 198.Pumbwe, L., C. Skilbeck, and H. M. Wexler. Impact of anatomic site on growth, efflux pump expression, cell structure and stress responsiveness of Bacteroides fragilis. Curr. Microbiol., in press. [DOI] [PubMed]
- 199.Pumbwe, L., C. A. Skilbeck, and H. M. Wexler. 2006. The Bacteroides fragilis cell envelope: quarterback, linebacker, coach—or all three? Anaerobe 12:211-220. [DOI] [PubMed] [Google Scholar]
- 200.Pumbwe, L., O. Ueda, F. Yoshimura, A. Chang, R. Smith, and H. M. Wexler. 2006. Bacteroides fragilis BmeABC efflux systems additively confer intrinsic antimicrobial resistance. J. Antimicrob. Chemother. 58:37-46. [DOI] [PubMed] [Google Scholar]
- 201.Pumbwe, L., D. W. Wareham, J. Aduse-Opoku, J. S. Brazier, and H. M. Wexler. 2007. Genetic analysis of mechanisms of multidrug resistance in a clinical isolate of Bacteroides fragilis. Clin. Microbiol. Infect. 13:183-189. [DOI] [PubMed] [Google Scholar]
- 202.Ramsay, A. G., K. P. Scott, J. C. Martin, M. T. Rincon, and H. J. Flint. 2006. Cell-associated alpha-amylases of butyrate-producing Firmicute bacteria from the human colon. Microbiology 152:3281-3290. [DOI] [PubMed] [Google Scholar]
- 203.Rasmussen, B. A., K. Bush, and F. P. Tally. 1993. Antimicrobial resistance in Bacteroides. Clin. Infect. Dis. 16(Suppl. 4):S390-S400. [DOI] [PubMed] [Google Scholar]
- 204.Rasmussen, B. A., and E. Kovacs. 1993. Cloning and identification of a two-component signal-transducing regulatory system from Bacteroides fragilis. Mol. Microbiol. 7:765-776. [DOI] [PubMed] [Google Scholar]
- 205.Rasmussen, J. L., D. A. Odelson, and F. L. Macrina. 1987. Complete nucleotide sequence of insertion element IS4351 from Bacteroides fragilis. J. Bacteriol. 169:3573-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Reeves, A. R., J. N. D'Elia, J. Frias, and A. A. Salyers. 1996. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J. Bacteriol. 178:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Reeves, A. R., G. R. Wang, and A. A. Salyers. 1997. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J. Bacteriol. 179:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Reid, G. 2004. When microbe meets human. Clin. Infect. Dis. 39:827-830. [DOI] [PubMed] [Google Scholar]
- 209.Reid, J. H., and S. Patrick. 1984. Phagocytic and serum killing of capsulate and non-capsulate Bacteroides fragilis. J. Med. Microbiol. 17:247-257. [DOI] [PubMed] [Google Scholar]
- 210.Renz, H., N. Blumer, S. Virna, S. Sel, and H. Garn. 2006. The immunological basis of the hygiene hypothesis. Chem. Immunol. Allergy 91:30-48. [DOI] [PubMed] [Google Scholar]
- 211.Reysset, G. 1996. Genetics of 5-nitroimidazole resistance in Bacteroides species. Anaerobe 2:59-69. [DOI] [PubMed] [Google Scholar]
- 212.Rhee, K. J., P. Sethupathi, A. Driks, D. K. Lanning, and K. L. Knight. 2004. Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. J. Immunol. 172:1118-1124. [DOI] [PubMed] [Google Scholar]
- 213.Ricci, V., M. L. Peterson, J. C. Rotschafer, H. Wexler, and L. J. Piddock. 2004. Role of topoisomerase mutations and efflux in fluoroquinolone resistance of Bacteroides fragilis clinical isolates and laboratory mutants. Antimicrob. Agents Chemother. 48:1344-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Riepe, S. P., J. Goldstein, and D. H. Alpers. 1980. Effect of secreted Bacteroides proteases on human intestinal brush border hydrolases. J. Clin. Investig. 66:314-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Roberts, M. C. 2003. Acquired tetracycline and/or macrolide-lincosamides-streptogramin resistance in anaerobes. Anaerobe 9:63-69. [DOI] [PubMed] [Google Scholar]
- 216.Robertson, K. P., C. J. Smith, A. M. Gough, and E. R. Rocha. 2006. Characterization of Bacteroides fragilis hemolysins and regulation and synergistic interactions of HlyA and HlyB. Infect. Immun. 74:2304-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Rocha, E. R., T. Selby, J. P. Coleman, and C. J. Smith. 1996. Oxidative stress response in an anaerobe, Bacteroides fragilis: a role for catalase in protection against hydrogen peroxide. J. Bacteriol. 178:6895-6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Rogemond, V., and R. M. Guinet. 1986. Lectinlike adhesins in the Bacteroides fragilis group. Infect. Immun. 53:99-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Rogers, M. B., A. C. Parker, and C. J. Smith. 1993. Cloning and characterization of the endogenous cephalosporinase gene, cepA, from Bacteroides fragilis reveals a new subgroup of Ambler class A beta-lactamases. Antimicrob. Agents Chemother. 37:2391-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rosenkranz, P., M. M. Lederman, K. V. Gopalakrishna, and J. J. Ellner. 1990. Septic arthritis caused by Bacteroides fragilis. Rev. Infect. Dis. 12:20-30. [DOI] [PubMed] [Google Scholar]
- 221.Rotimi, V. O., T. L. Verghese, N. Al-Sweih, F. B. Khodakhast, and K. Ahmed. 2000. Influence of five antianaerobic antibiotics on endotoxin liberation by gram-negative anaerobes. J. Chemother. 12:40-47. [DOI] [PubMed] [Google Scholar]
- 222.Rudek, W., and R. U. Haque. 1976. Extracellular enzymes of the genus Bacteroides. J. Clin. Microbiol. 4:458-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Russo, T. A., J. S. Thompson, V. G. Godoy, and M. H. Malamy. 1990. Cloning and expression of the Bacteroides fragilis TAL2480 neuraminidase gene, nanH, in Escherichia coli. J. Bacteriol. 172:2594-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sachs, J. 2005. Are antibiotics killing us? Discover 26:36. [Google Scholar]
- 225.Sakamoto, M., and Y. Benno. 2006. Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen.nov, comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int. J. Syst. Bacteriol. 56:1599-1605. [DOI] [PubMed] [Google Scholar]
- 226.Salyers, A. A. 1984. Bacteroides of the human lower intestinal tract. Annu. Rev. Microbiol. 38:293-313. [DOI] [PubMed] [Google Scholar]
- 227.Salyers, A. A., and C. F. Amabile-Cuevas. 1997. Why are antibiotic resistance genes so resistant to elimination? Antimicrob. Agents Chemother. 41:2321-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Salyers, A. A., A. Gupta, and Y. Wang. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12:412-416. [DOI] [PubMed] [Google Scholar]
- 229.Salyers, A. A., and M. Pajeau. 1989. Competitiveness of different polysaccharide utilization mutants of Bacteroides thetaiotaomicron in the intestinal tracts of germfree mice. Appl. Environ. Microbiol. 55:2572-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Salyers, A. A., N. B. Shoemaker, and L. Y. Li. 1995. In the driver's seat: the Bacteroides conjugative transposons and the elements they mobilize. J. Bacteriol. 177:5727-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L. Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Salyers, A. A., B. S. Speer, and N. B. Shoemaker. 1990. New perspectives in tetracycline resistance. Mol. Microbiol. 4:151-156. [DOI] [PubMed] [Google Scholar]
- 233.Salyers, A. A., P. Valentine, and V. Hwa. 1993. Genetics of polysaccharide utilization pathways of colonic Bacteroides species, p. 505-516. In M. Sebald (ed.), Genetics and molecular biology of anaerobic bacteria. Springer-Verlag, New York, NY.
- 234.Sanchez, S. 2006. Where do antimicrobial resistances reside on the farm?, abstr. A-108. Abstr. 105th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 235.San Joaquin, V., J. C. Griffis, C. Lee, and C. L. Sears. 1995. Association of Bacteroides fragilis with childhood diarrhea. Scand. J. Infect. Dis. 27:211-215. [DOI] [PubMed] [Google Scholar]
- 236.Schapiro, J. M., R. Gupta, E. Stefansson, F. C. Fang, and A. P. Limaye. 2004. Isolation of metronidazole-resistant Bacteroides fragilis carrying the nimA nitroreductase gene from a patient in Washington State. J. Clin. Microbiol. 42:4127-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Schauer, R. 2004. Sialic acids: fascinating sugars in higher animals and man. Zoology (Jena) 107:49-64. [DOI] [PubMed] [Google Scholar]
- 238.Sears, C. L. 2001. The toxins of Bacteroides fragilis. Toxicon 39:1737-1746. [DOI] [PubMed] [Google Scholar]
- 239.Senneville, E., C. Savage, I. Nallet, Y. Yazdanpanah, F. Giraud, H. Migaud, L. Dubreuil, R. Courcol, and Y. Mouton. 2006. Improved aero-anaerobe recovery from infected prosthetic joint samples taken from 72 patients and collected intraoperatively in Rosenow's broth. Acta Orthop. 77:120-124. [DOI] [PubMed] [Google Scholar]
- 240.Shah, H. N., and D. M. Collins. 1990. Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. Int. J. Syst. Bacteriol. 40:205-208. [DOI] [PubMed] [Google Scholar]
- 241.Shah, H. N., and M. D. Collins. 1989. Proposal to restrict the genus Bacteroides (Castellani and Chalmers) to Bacteroides fragilis and closely related species. Int. J. Syst. Bacteriol. 39:85-87. [Google Scholar]
- 242.Shah, H. N., and M. D. Collins. 1988. Proposal for reclassfication of Bacteroides asaccharolyticus, Bacteroides gingivalis, and Bacteroides endodontalis in a new genus, Porphyromonas. Int. J. Syst. Bacteriol. 38:128-131. [Google Scholar]
- 243.Shapiro, M. E., D. L. Kasper, D. F. Zaleznik, S. Spriggs, A. B. Onderdonk, and R. W. Finberg. 1986. Cellular control of abscess formation: role of T cells in the regulation of abscesses formed in response to Bacteroides fragilis. J. Immunol. 137:341-346. [PubMed] [Google Scholar]
- 244.Shapiro, M. E., A. B. Onderdonk, D. L. Kasper, and R. W. Finberg. 1982. Cellular immunity to Bacteroides fragilis capsular polysaccharide. J. Exp. Med. 155:1188-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Shapiro, M. E., A. B. Onderdonk, D. L. Kasper, and R. W. Finberg. 1983. Immune T cells prevent Bacteroides fragilis abscesses. Curr. Surg. 40:123-126. [PubMed] [Google Scholar]
- 246.Shetab, R., S. H. Cohen, T. Prindiville, Y. J. Tang, M. Cantrell, D. Rahmani, and J. Silva, Jr. 1998. Detection of Bacteroides fragilis enterotoxin gene by PCR. J. Clin. Microbiol. 36:1729-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Shipman, J. A., J. E. Berleman, and A. A. Salyers. 2000. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J. Bacteriol. 182:5365-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Shipman, J. A., K. H. Cho, H. A. Siegel, and A. A. Salyers. 1999. Physiological characterization of SusG, an outer membrane protein essential for starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 181:7206-7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Shoemaker, N. B., R. D. Barber, and A. A. Salyers. 1989. Cloning and characterization of a Bacteroides conjugal tetracycline-erythromycin resistance element by using a shuttle cosmid vector. J. Bacteriol. 171:1294-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Simon, G. L., and S. L. Gorbach. 1984. Intestinal flora in health and disease. Gastroenterology 86:174-193. [PubMed] [Google Scholar]
- 252.Simon, G. L., M. S. Klempner, D. L. Kasper, and S. L. Gorbach. 1982. Alterations in opsonophagocytic killing by neutrophils of Bacteroides fragilis associated with animal and laboratory passage: effect of capsular polysaccharide. J. Infect. Dis. 145:72-77. [DOI] [PubMed] [Google Scholar]
- 253.Smalley, D., E. R. Rocha, and C. J. Smith. 2002. Aerobic-type ribonucleotide reductase in the anaerobe Bacteroides fragilis. J. Bacteriol. 184:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Smith, C. J., G. D. Tribble, and D. P. Bayley. 1998. Genetic elements of Bacteroides species: a moving story. Plasmid 40:12-29. [DOI] [PubMed] [Google Scholar]
- 255.Snydman, D. R., N. V. Jacobus, and L. A. McDermott. 2005. Evaluation of the in vitro activity of NVP-LMB415 against clinical anaerobic isolates with emphasis on the Bacteroides fragilis group. J. Antimicrob. Chemother. 55:1024-1028. [DOI] [PubMed] [Google Scholar]
- 256.Snydman, D. R., N. V. Jacobus, L. A. McDermott, R. Ruthazer, Y. Golan, E. J. Goldstein, S. M. Finegold, L. J. Harrell, D. W. Hecht, S. G. Jenkins, C. Pierson, R. Venezia, V. Yu, J. Rihs, and S. L. Gorbach. 2007. National survey on the susceptibility of Bacteroides fragilis group: report and analysis of trends in the United States from 1997 to 2004. Antimicrob. Agents Chemother. 51:1649-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Snydman, D. R., N. V. Jacobus, L. A. McDermott, R. Ruthazer, E. Goldstein, S. Finegold, L. Harrell, D. W. Hecht, S. Jenkins, C. Pierson, R. Venezia, J. Rihs, and S. L. Gorbach. 2002. In vitro activities of newer quinolones against Bacteroides group organisms. Antimicrob. Agents Chemother. 46:3276-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Snydman, D. R., N. V. Jacobus, L. A. McDermott, R. Ruthazer, E. J. Goldstein, S. M. Finegold, L. J. Harrell, D. W. Hecht, S. G. Jenkins, C. Pierson, R. Venezia, J. Rihs, and S. L. Gorbach. 2002. National survey on the susceptibility of Bacteroides fragilis group: report and analysis of trends for 1997-2000. Clin. Infect. Dis. 35:S126-S134. [DOI] [PubMed] [Google Scholar]
- 259.Soki, J., M. Gal, J. S. Brazier, V. O. Rotimi, E. Urban, E. Nagy, and B. I. Duerden. 2006. Molecular investigation of genetic elements contributing to metronidazole resistance in Bacteroides strains. J. Antimicrob. Chemother. 57:212-220. [DOI] [PubMed] [Google Scholar]
- 260.Song, Y., C. Liu, M. Bolanos, J. Lee, M. McTeague, and S. M. Finegold. 2005. Evaluation of 16S rRNA sequencing and reevaluation of a short biochemical scheme for identification of clinically significant Bacteroides species. J. Clin. Microbiol. 43:1531-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Song, Y., C. Liu, J. Lee, M. Bolanos, M. L. Vaisanen, and S. M. Finegold. 2005. “Bacteroides goldsteinii sp. nov.” isolated from clinical specimens of human intestinal origin. J. Clin. Microbiol. 43:4522-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Song, Y. L., C. X. Liu, M. McTeague, and S. M. Finegold. 2004. “Bacteroides nordii” sp. nov. and “Bacteroides salyersae” sp. nov. isolated from clinical specimens of human intestinal origin. J. Clin. Microbiol. 42:5565-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Sonnenburg, E. D., J. L. Sonnenburg, J. K. Manchester, E. E. Hansen, H. C. Chiang, and J. I. Gordon. 2006. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc. Natl. Acad. Sci. USA 103:8834-8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Sonnenburg, J. L., L. T. Angenent, and J. I. Gordon. 2004. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 5:569-573. [DOI] [PubMed] [Google Scholar]
- 265.Sonnenburg, J. L., J. Xu, D. D. Leip, C. H. Chen, B. P. Westover, J. Weatherford, J. D. Buhler, and J. I. Gordon. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955-1959. [DOI] [PubMed] [Google Scholar]
- 266.Stappenbeck, T. S., L. V. Hooper, and J. I. Gordon. 2002. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA 99:15451-15455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Stellwag, E. J., and P. B. Hylemon. 1976. Purification and characterization of bile salt hydrolase from Bacteroides fragilis subsp. fragilis. Biochim. Biophys. Acta 452:165-176. [DOI] [PubMed] [Google Scholar]
- 268.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 269.Strober, W. 2006. Unraveling gut inflammation. Science 313:1052-1054. [DOI] [PubMed] [Google Scholar]
- 270.Strober, W., I. Fuss, and P. Mannon. 2007. The fundamental basis of inflammatory bowel disease. J. Clin. Investig. 117:514-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Takeuchi, T., Y. Nakaya, N. Kato, K. Watanabe, and K. Morimoto. 1999. Induction of oxidative DNA damage in anaerobes. FEBS Lett. 450:178-180. [DOI] [PubMed] [Google Scholar]
- 272.Talan, D. A., D. M. Citron, F. M. Abrahamian, G. J. Moran, E. J. Goldstein, et al. 1999. Bacteriologic analysis of infected dog and cat bites. N. Engl. J. Med. 340:85-92. [DOI] [PubMed] [Google Scholar]
- 273.Tally, F. P., G. J. Cuchural, Jr., and M. H. Malamy. 1984. Mechanisms of resistance and resistance transfer in anaerobic bacteria: factors influencing antimicrobial therapy. Rev. Infect. Dis. 6(Suppl. 1):S260-S269. [DOI] [PubMed] [Google Scholar]
- 274.Tanaka, H., F. Ito, and T. Iwasaki. 1992. Purification and characterization of a sialidase from Bacteroides fragilis SBT3182. Biochem. Biophys. Res. Commun. 189:524-529. [DOI] [PubMed] [Google Scholar]
- 275.Tanaka, K., H. Mikamo, K. Nakao, and K. Watanabe. 2006. In vitro antianaerobic activity of DX-619, a new des-fluoro(6) quinolone. Antimicrob. Agents Chemother. 50:3908-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Tancula, E., M. J. Feldhaus, L. A. Bedzyk, and A. A. Salyers. 1992. Location and characterization of genes involved in binding of starch to the surface of Bacteroides thetaiotaomicron. J. Bacteriol. 174:5609-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Toprak, N. U., A. Yagci, B. M. Gulluoglu, M. L. Akin, P. Demirkalem, T. Celenk, and G. Soyletir. 2006. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin. Microbiol. Infect. 12:782-786. [DOI] [PubMed] [Google Scholar]
- 278.Trinh, S., A. Haggoud, G. Reysset, and M. Sebald. 1995. Plasmids pIP419 and pIP421 from Bacteroides: 5-nitroimidazole resistance genes and their upstream insertion sequence elements. Microbiology 141:927-935. [DOI] [PubMed] [Google Scholar]
- 279.Trinh, S., and G. Reysset. 1996. Detection by PCR of the nim genes encoding 5-nitroimidazole resistance in Bacteroides spp. J. Clin. Microbiol. 34:2078-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Turnbaugh, P. J., R. E. Ley, M. A. Mahowald, V. Magrini, E. R. Mardis, and J. I. Gordon. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027-1131. [DOI] [PubMed] [Google Scholar]
- 281.Tzianabos, A. O., D. L. Kasper, and A. B. Onderdonk. 1995. Structure and function of Bacteroides fragilis capsular polysaccharides: relationship to induction and prevention of abscesses. Clin. Infect. Dis. 20(Suppl. 2):S132-S140. [DOI] [PubMed] [Google Scholar]
- 282.Tzianabos, A. O., A. B. Onderdonk, B. Rosner, R. L. Cisneros, and D. L. Kasper. 1993. Structural features of polysaccharides that induce intra-abdominal abscesses. Science 262:416-419. [DOI] [PubMed] [Google Scholar]
- 283.Tzianabos, A. O., A. Pantosti, H. Baumann, J. R. Brisson, H. J. Jennings, and D. L. Kasper. 1992. The capsular polysaccharide of Bacteroides fragilis comprises two ionically linked polysaccharides. J. Biol. Chem. 267:18230-18235. [PubMed] [Google Scholar]
- 284.Ueda, O., H. M. Wexler, K. Hirai, Y. Shibata, F. Yoshimura, and S. Fujimura. 2005. Sixteen homologs of the Mex-type multidrug resistance efflux pump in Bacteroides fragilis. Antimicrob. Agents Chemother. 49:2807-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.University of Capetown. 11 April 2005, posting date. The enemy inside. In UCT Online-Monday paper. University of Capetown, Capetown, South Africa. http://www.news.uct.ac.za/mondaypaper/archives/?id=5053.
- 286.Vandamme, P., M. I. Daneshvar, F. E. Dewhirst, B. J. Paster, K. Kersters, H. Goossens, and C. W. Moss. 1995. Chemotaxonomic analyses of Bacteroides gracilis and Bacteroides ureolyticus and reclassification of B. gracilis as Campylobacter gracilis comb. nov. Int. J. Syst. Bacteriol. 45:145-152. [DOI] [PubMed] [Google Scholar]
- 287.Vieira, J. M., D. C. Vallim, E. O. Ferreira, S. H. Seabra, R. C. Vommaro, K. E. Avelar, S. W. De, M. C. Ferreira, and R. M. Domingues. 2005. Bacteroides fragilis interferes with iNOS activity and leads to pore formation in macrophage surface. Biochem. Biophys. Res. Commun. 326:607-613. [DOI] [PubMed] [Google Scholar]
- 288.Wang, R. C., S. J. Seror, M. Blight, J. M. Pratt, J. K. Broome-Smith, and I. B. Holland. 1991. Analysis of the membrane organization of an Escherichia coli protein translocator, HlyB, a member of a large family of prokaryote and eukaryote surface transport proteins. J. Mol. Biol. 217:441-454. [DOI] [PubMed] [Google Scholar]
- 289.Wang, Y., W. M. Kalka-Moll, M. H. Roehrl, and D. L. Kasper. 2000. Structural basis of the abscess-modulating polysaccharide A2 from Bacteroides fragilis. Proc. Natl. Acad. Sci. USA 97:13478-13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Wareham, D. W., M. Wilks, D. Ahmed, J. S. Brazier, and M. Millar. 2005. Anaerobic sepsis due to multidrug-resistant Bacteroides fragilis: microbiological cure and clinical response with linezolid therapy. Clin. Infect. Dis. 40:e67-e68. [DOI] [PubMed] [Google Scholar]
- 291.Weintraub, A., B. E. Larsson, and A. A. Lindberg. 1985. Chemical and immunochemical analyses of Bacteroides fragilis lipopolysaccharides. Infect. Immun. 49:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Weiser, J. N., and E. C. Gotschlich. 1991. Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect. Immun. 59:2252-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Weiss, S. T. 2002. Eat dirt: the hygiene hypothesis and allergic diseases. N. Engl. J. Med. 347:930-931. [DOI] [PubMed] [Google Scholar]
- 294.Welch, R. A., K. R. Jones, and F. L. Macrina. 1979. Transferable lincosamide-macrolide resistance in Bacteroides. Plasmid 2:261-268. [DOI] [PubMed] [Google Scholar]
- 295.Wells, C. L., M. A. Maddaus, R. P. Jechorek, and R. L. Simmons. 1988. Role of intestinal anaerobic bacteria in colonization resistance. Eur. J. Clin. Microbiol. Infect. Dis. 7:107-113. [DOI] [PubMed] [Google Scholar]
- 296.Wexler, H. M., A. E. Engel, D. Glass, and C. Li. 2005. In vitro activities of doripenem and comparator agents against 364 anaerobic clinical isolates. Antimicrob. Agents Chemother. 49:4413-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Wexler, H. M., C. Getty, and G. Fisher. 1992. The isolation and characterisation of a major outer-membrane protein from Bacteroides distasonis. J. Med. Microbiol. 37:165-175. [DOI] [PubMed] [Google Scholar]
- 298.Wexler, H. M., and S. Halebian. 1990. Alterations to the penicillin-binding proteins in the Bacteroides fragilis group: a mechanism for non-β-lactamase mediated cefoxitin resistance. J. Antimicrob. Chemother. 26:7-20. [DOI] [PubMed] [Google Scholar]
- 299.Wexler, H. M., E. Molitoris, D. Molitoris, and S. M. Finegold. 1998. In vitro activity of levofloxacin against a selected group of anaerobic bacteria isolated from skin and soft tissue infections. Antimicrob. Agents Chemother. 42:984-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Wexler, H. M., E. Molitoris, D. Reeves, and S. M. Finegold. 1994. In vitro activity of DU-6859a against anaerobic bacteria. Antimicrob. Agents Chemother. 38:2504-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Wexler, H. M., E. K. Read, and T. J. Tomzynski. 2002. Characterization of omp200, a porin gene complex from Bacteroides fragilis: omp121 and omp71, gene sequence, deduced amino acid sequence and predictions of porin structure. Gene 283:95-105. [DOI] [PubMed] [Google Scholar]
- 302.Wexler, H. M., E. K. Read, and T. J. Tomzynski. 2002. Identification of an OmpA protein from Bacteroides fragilis: ompA gene sequence, OmpA amino acid sequence and predictions of protein structure. Anaerobe 8:180-191. [Google Scholar]
- 303.Wu, S., K. C. Lim, J. Huang, R. F. Saidi, and C. L. Sears. 1998. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl. Acad. Sci. USA 95:14979-14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Wu, S., J. Powell, N. Mathioudakis, S. Kane, E. Fernandez, and C. L. Sears. 2004. Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-κB pathway. Infect. Immun. 72:5832-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Wu, S., J. Shin, G. Zhang, M. Cohen, A. Franco, and C. L. Sears. 2006. The Bacteroides fragilis toxin binds to a specific intestinal epithelial cell receptor. Infect. Immun. 74:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
- 307.Xu, J., H. C. Chiang, M. K. Bjursell, and J. I. Gordon. 2004. Message from a human gut symbiont: sensitivity is a prerequisite for sharing. Trends Microbiol. 12:21-28. [DOI] [PubMed] [Google Scholar]
- 308.Xu, J., and J. I. Gordon. 2003. Honor thy symbionts. Proc. Natl. Acad. Sci. USA 100:10452-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Yang, S. H., R. S. Yang, and C. L. Tsai. 2001. Septic arthritis of the hip joint in cervical cancer patients after radiotherapy: three case reports. J. Orthop. Surg. (Hong Kong) 9:41-45. [DOI] [PubMed] [Google Scholar]
- 310.Zaleznik, D. F., R. W. Finberg, M. E. Shapiro, A. B. Onderdonk, and D. L. Kasper. 1985. A soluble suppressor T cell factor protects against experimental intraabdominal abscesses. J. Clin. Investig. 75:1023-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]