Abstract
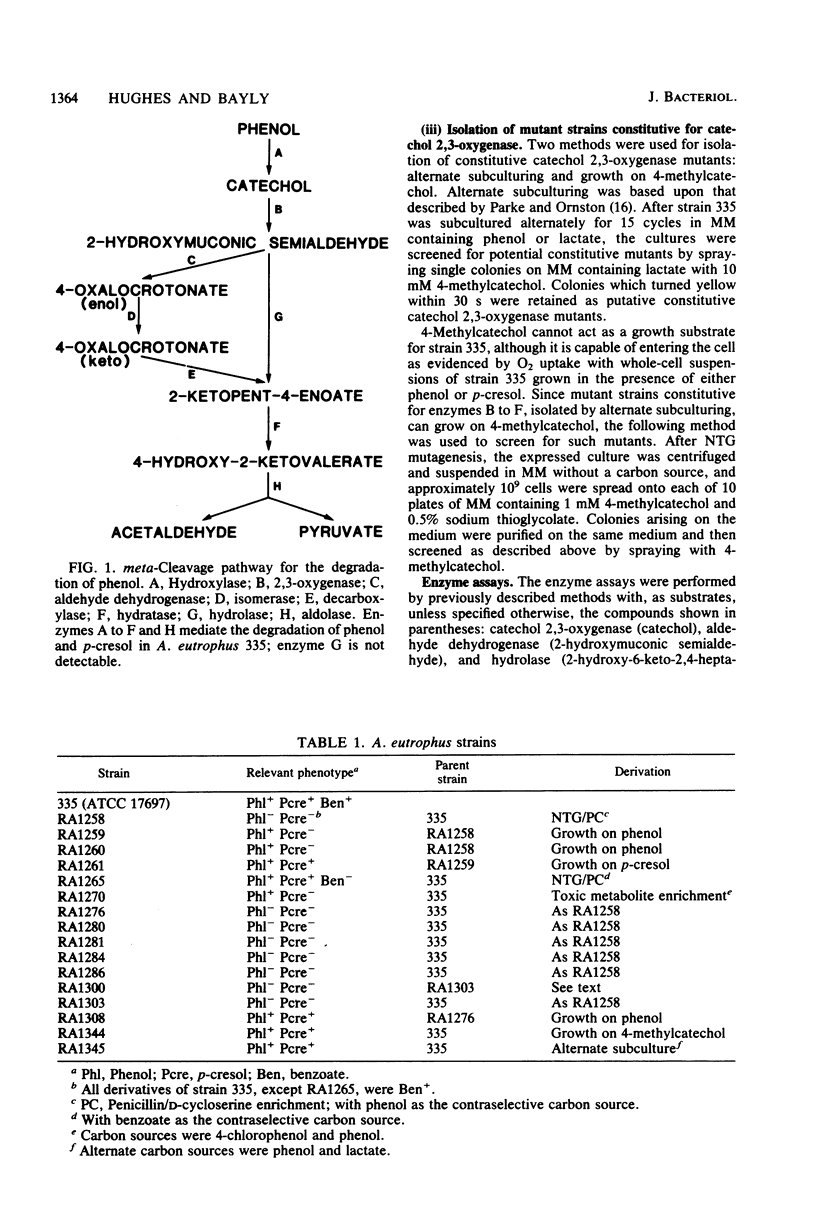
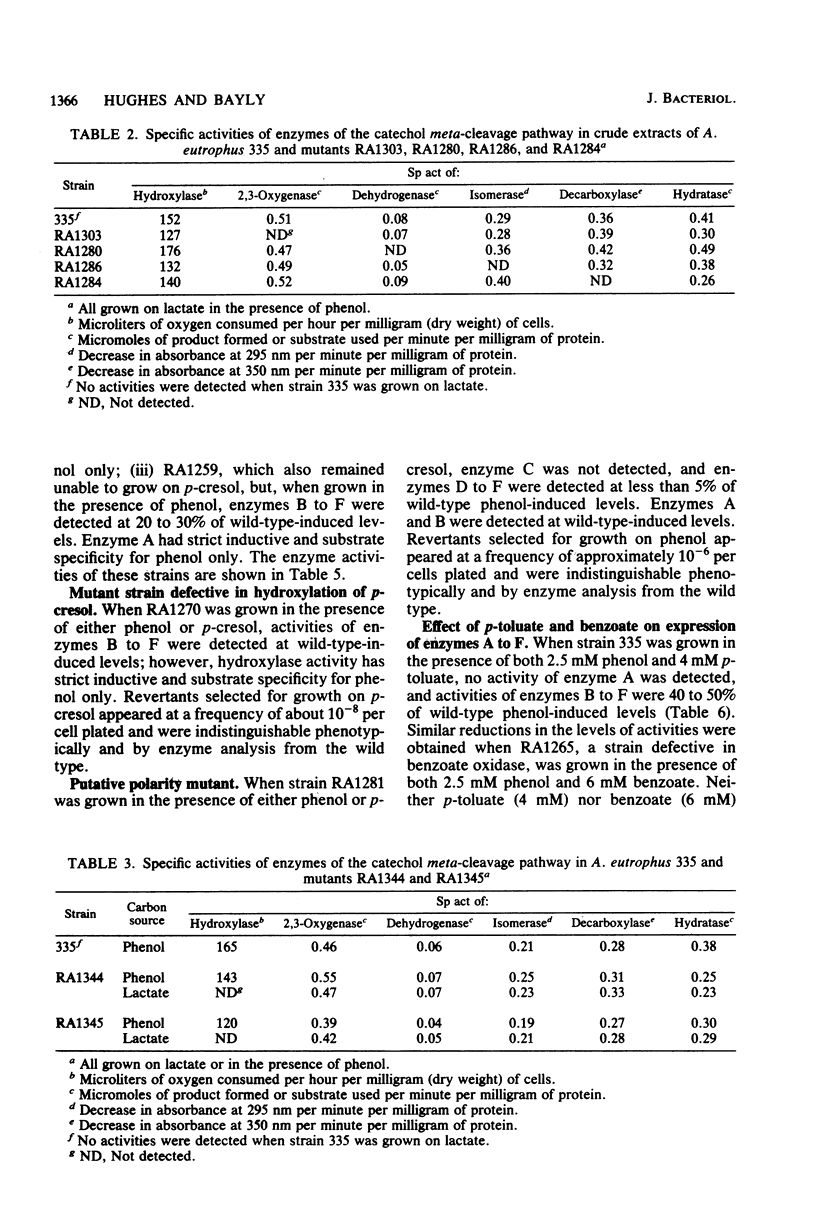
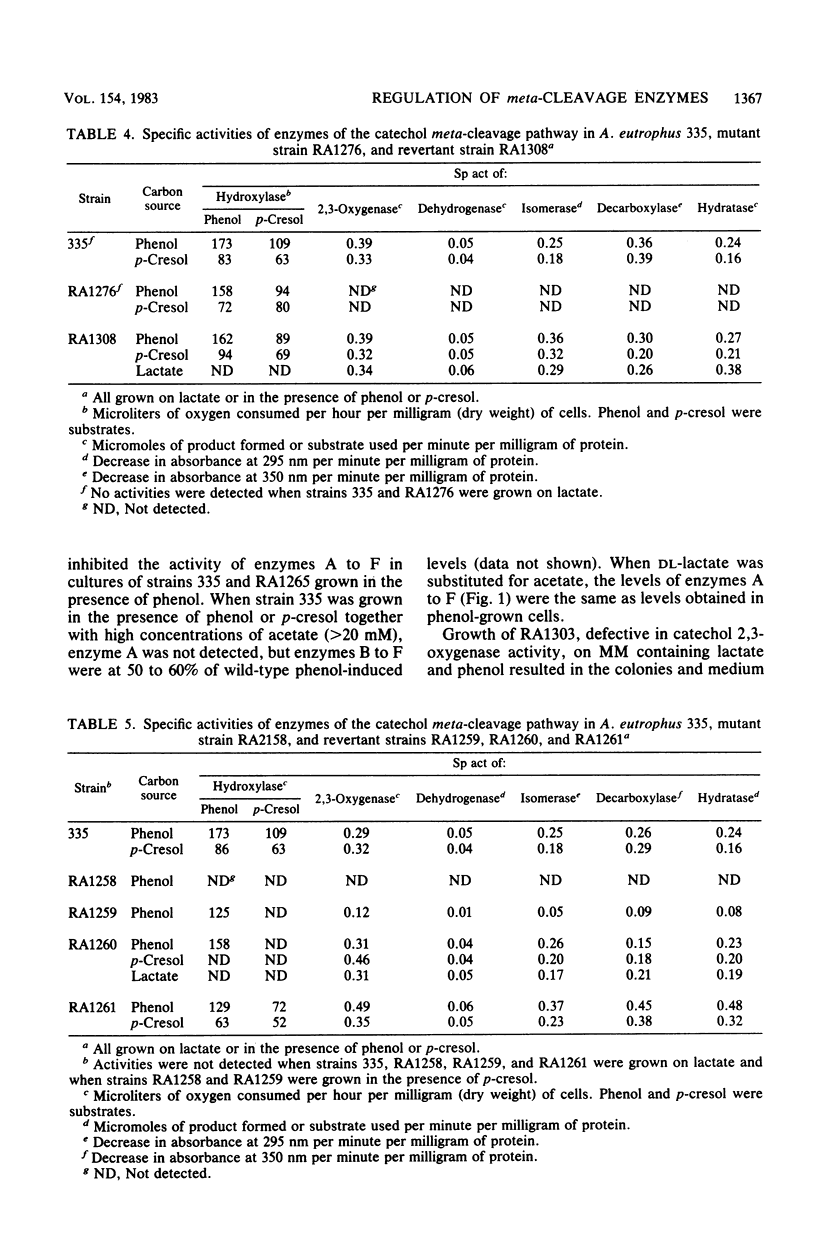
Alcaligenes eutrophus 335 (ATCC 17697) metabolizes phenol and p-cresol via a catechol meta-cleavage pathway. Studies with mutant strains, each defective in an enzyme of the pathway, showed that the six enzymes assayed are induced by the primary substrate. Studies with a putative polarity mutant defective in the expression of aldehyde dehydrogenase suggested that the structural genes encoding this and subsequent enzymes of the pathway exist in the same operon. From studies with mutant strains that constitutively synthesize catechol 2,3-oxygenase and subsequent enzymes and from the coordination of repression of these enzymes by p-toluate, benzoate, and acetate, it is proposed the catechol 2,3-oxygenase structural gene is situated in this operon (2,3-oxygenase operon). Studies with regulatory mutant strains suggest that the 2,3-oxygenase operon is under negative control.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbour M. G., Bayly R. C. Control of meta-cleavage degradation of 4-hydroxyphenylacetate in Pseudomonas putida. J Bacteriol. 1981 Sep;147(3):844–850. doi: 10.1128/jb.147.3.844-850.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour M. G., Bayly R. C. Mutants defective in isomerase and decarboxylase activities of the 4-hydroxyphenylacetic acid meta-cleavage pathway in Pseudomonas putida. J Bacteriol. 1980 May;142(2):480–485. doi: 10.1128/jb.142.2.480-485.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly R. C., McKenzie D. I. Catechol oxygenases of Pseudomonas putida mutant strains. J Bacteriol. 1976 Sep;127(3):1098–1107. doi: 10.1128/jb.127.3.1098-1107.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly R. C., Wigmore G. J. Metabolism of phenol and cresols by mutants of Pseudomonas putida. J Bacteriol. 1973 Mar;113(3):1112–1120. doi: 10.1128/jb.113.3.1112-1120.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M., Friello D. A., Bopp L. H. Transposition of plasmid DNA segments specifying hydrocarbon degradation and their expression in various microorganisms. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3109–3112. doi: 10.1073/pnas.75.7.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinsworth W. L., Chapman P. J., Dagley S. Stereospecific enzymes in the degradation of aromatic compounds by pseudomonas putida. J Bacteriol. 1973 Feb;113(2):922–931. doi: 10.1128/jb.113.2.922-931.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farin F., Clarke P. H. Positive regulation of amidase synthesis in Pseudomonas aeruginosa. J Bacteriol. 1978 Aug;135(2):379–392. doi: 10.1128/jb.135.2.379-392.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Williams P. A. Construction of a partial diploid for the degradative pathway encoded by the TOL plasmid (pWWO) from Pseudomonas putida mt-2: evidence for the positive nature of the regulation by the xyIR gene. Mol Gen Genet. 1980 Jan;177(2):321–328. doi: 10.1007/BF00267445. [DOI] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Molecular cloning of TOL genes xylB and xylE in Escherichia coli. J Bacteriol. 1981 Mar;145(3):1137–1143. doi: 10.1128/jb.145.3.1137-1143.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. F., Stanier R. Y. Dissimilation of aromatic compounds by Alcaligenes eutrophus. J Bacteriol. 1971 Aug;107(2):468–475. doi: 10.1128/jb.107.2.468-475.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Nakazawa T., Inouye S., Nakazawa A. Physical and functional mapping of RP4-TOL plasmid recombinants: analysis of insertion and deletion mutants. J Bacteriol. 1980 Oct;144(1):222–231. doi: 10.1128/jb.144.1.222-231.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N., Ornston M. K., Chou G. Isolation of spontaneous mutant strains of Pseudomonas putida. Biochem Biophys Res Commun. 1969 Jul 7;36(1):179–184. doi: 10.1016/0006-291x(69)90666-4. [DOI] [PubMed] [Google Scholar]
- Parke D., Ornston L. N. Constitutive synthesis of enzymes of the protocatechuate pathway and of the beta-ketoadipate uptake system in mutant strains of Pseudomonas putida. J Bacteriol. 1976 Apr;126(1):272–281. doi: 10.1128/jb.126.1.272-281.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Evans W. C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971 Jun 11;20(3):400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Sheppard D., Englesberg E. Positive control in the L-arabinose gene-enzyme complex of Escherichia coli B/r exhibited with stable merodiploids. Cold Spring Harb Symp Quant Biol. 1966;31:345–347. doi: 10.1101/sqb.1966.031.01.044. [DOI] [PubMed] [Google Scholar]
- Sparnins V. L., Chapman P. J., Dagley S. Bacterial degradation of 4-hydroxyphenylacetic acid and homoprotocatechuic acid. J Bacteriol. 1974 Oct;120(1):159–167. doi: 10.1128/jb.120.1.159-167.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLSON C., PERRIN D., COHN M., JACOB F., MONOD J. NON-INDUCIBLE MUTANTS OF THE REGULATOR GENE IN THE "LACTOSE" SYSTEM OF ESCHERICHIA COLI. J Mol Biol. 1964 Apr;8:582–592. doi: 10.1016/s0022-2836(64)80013-9. [DOI] [PubMed] [Google Scholar]
- Wigmore G. J., Bayly R. C. A mutant of Pseudomonas putida with altered regulation of the enzymes for degradation of phenol and cresols. Biochem Biophys Res Commun. 1974 Sep 9;60(1):48–55. doi: 10.1016/0006-291x(74)90170-3. [DOI] [PubMed] [Google Scholar]
- Wigmore G. J., Bayly R. C., Di Berardino D. Pseudomonas putida mutants defective in the metabolism of the products of meta fission of catechol and its methyl analogues. J Bacteriol. 1974 Oct;120(1):31–37. doi: 10.1128/jb.120.1.31-37.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigmore G. J., Ribbons D. W. Selective enrichment of Pseudomonas spp. defective in catabolism after exposure to halogenated substrates. J Bacteriol. 1981 Jun;146(3):920–927. doi: 10.1128/jb.146.3.920-927.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigmore G. J., Ribbons D. W. p-Cymene pathway in Pseudomonas putida: selective enrichment of defective mutants by using halogenated substrate analogs. J Bacteriol. 1980 Aug;143(2):816–824. doi: 10.1128/jb.143.2.816-824.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Franklin F. C., Williams P. A. Regulation of the degradative pathway enzymes coded for by the TOL plasmid (pWWO) from Pseudomonas putida mt-2. J Bacteriol. 1978 Jun;134(3):757–764. doi: 10.1128/jb.134.3.757-764.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Characterization of a spontaneously occurring mutant of the TOL20 plasmid in Pseudomonas putida MT20: possible regulatory implications. J Bacteriol. 1977 Jun;130(3):1149–1158. doi: 10.1128/jb.130.3.1149-1158.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]


