Abstract
Mutation rate and cooperation have important ecological and evolutionary consequences and, moreover, can affect pathogen virulence. While hypermutability accelerates adaptation to novel environments, hypermutable lineages (‘mutators’) are selected against in well-adapted populations. Using the model organism Pseudomonas aeruginosa, we previously demonstrated a further potential disadvantage to hypermutability, namely, that it can accelerate the breakdown of cooperation. We now investigate how this property of mutators can affect their persistence in metapopulations. Mutator and wild-type bacteria were competed for 250 generations in globally competing metapopulations, imposing conditions of high or low intra-deme relatedness. High relatedness favours cooperating groups, so we predicted that mutators should achieve lower equilibrium frequencies under high relatedness than under low relatedness. This was observed in our study. Consistent with our hypothesis, there was a positive correlation between mean mutator and cheat frequencies. We conclude that when dense population growth requires cooperation, and when cooperation is favoured (high relatedness), demes containing high frequencies of mutators are likely to be selected against because they also contain high frequencies of non-cooperating cheats. We have also identified conditions where mutator lineages are likely to dominate metapopulations; namely, when low relatedness reduces kin selection for cooperation. These results may help to explain clinical distributions of mutator bacteria.
Keywords: cooperation, mutation rate, Hamilton's rule, metapopulations, Pseudomonas aeruginosa, microbial ecology
1. Introduction
The importance of mutation rate in evolution has been the subject of much theoretical and empirical research (Rainey 1999; Sniegowski et al. 2000; de Visser 2002). Hypermutability is also thought to be a significant risk factor in bacterial infections of animals, including humans (Oliver et al. 2000; Giraud et al. 2002; Ciofu et al. 2005). Experimental studies with hypermutable lineages of bacteria (‘mutators’) have successfully been used to expand our understanding of how and when alterations in mutation rate affect the evolution of a population. When adaptation is limited by the beneficial mutation rate, as is the case in novel or changeable environments, mutator alleles can hitch-hike with beneficial mutations to reach high frequencies (Ishii et al. 1989; Sniegowski et al. 1997; Taddei et al. 1997; de Visser et al. 1999; Tanabe et al. 1999; Giraud et al. 2001; Schaaff et al. 2002). However, when a population is well adapted to its environment, the increased rate of deleterious mutations produced by mutators selects against hypermutability (Trobner & Piechocki 1984; Funchain et al. 2000; Giraud et al. 2001).
A further disadvantage may accrue to hypermutable lineages when population growth depends upon cooperative behaviours. Cooperative behaviours are common in nature, and often underpin virulence-related traits in pathogenic microbes (West & Buckling 2003; Harrison et al. 2006; West et al. 2006). Two key variables which affect the evolution of cooperation are the relatedness of interacting individuals and the scale of competition. The ‘relatedness’ of two individuals can be defined as the probability of them sharing a common allele at a locus governing social interaction or cooperative action (Frank 1998). High relatedness favours cooperation via kin selection (Hamilton 1964), whereas competition between less related individuals favours the evolution of non-cooperating social cheats, which pay none of the costs of cooperation but enjoy the benefits of their neighbours' efforts. Understanding the evolution and ecology of cooperative traits is important when trying to predict the virulence of mixed (i.e. low relatedness) infections (Brown et al. 2002). Mutators are predicted to reduce relatedness and hence generate cheating genotypes more readily. Mutators are also predicted to have access to a greater area of the adaptive landscape, allowing them to produce ‘fitter’ cheats which can persist at higher relative densities (Harrison & Buckling 2005; Harrison & Buckling in preparation). While it is possible that mutators could also have access to fitter cooperating genotypes (e.g. with reduced costs of production), the reduction in relatedness generated by elevated mutation rates leads to an asymmetry in the strength of selection in favour of cheating over cooperation. We would thus expect mutators to be selected against under conditions that favour cooperation, namely, where ecological conditions result in high relatedness within demes.
The effect of relatedness on cooperation is itself mediated by the scale of competition (Hamilton 1964; West & Buckling 2003; Griffin et al. 2004). When competition is entirely local (i.e. due to within-patch or soft selection), selection will favour those individuals who are most successful within their group: the overall success of the group is unimportant. This creates a significant selective advantage for cheats, and cooperation breaks down under such conditions (West & Buckling 2003; Griffin et al. 2004). Under global competition, on the other hand, groups compete with groups and cooperating demes grow to higher densities than do cheating demes. This leads to cooperators persisting in the metapopulation and visible differences in equilibrium levels of cooperation under high versus low relatedness (West & Buckling 2003; Griffin et al. 2004). Selection for cheating under local competition can counteract kin selection due to high relatedness. This is because neighbours compete intensely with one another regardless of kinship. Under local competition, therefore, relatedness affects only the speed with which cooperation declines. Further, entirely local competition is biologically unrealistic as patches are highly unlikely to experience no migration (e.g. Saccheri & Hanski 2006). Any experiment designed to investigate the effect of relatedness on mutator dynamics should, therefore, incorporate a degree of global competition.
The production of iron-scavenging siderophores by Pseudomonas aeruginosa is a classic example of cooperation via production of a ‘public good’ (West & Buckling 2003; Griffin et al. 2004) and a useful model system for studying cooperation (Griffin et al. 2004; Harrison & Buckling 2005). Siderophores enhance population growth under iron-limited conditions and are necessary for virulence in acute P. aeruginosa infections (Meyer et al. 1996; Harrison et al. 2006), meaning that useful inferences regarding cooperation, mutation and pathogen virulence can be made. We have previously used this system to show that hypermutability accelerates the breakdown of cooperation when competition is entirely local (Harrison & Buckling 2005). We now use the same system to examine the fate of hypermutable lineages in globally competing metapopulations (within- and between-patch competition) under conditions of high or low intra-deme relatedness.
Our experimental design was based on that of Griffin et al. (2004) and comprised two treatments in which relatedness was either high or low. Metapopulations comprising six demes (iron-limited broth microcosms) were initially inoculated with wild-type and mutator bacteria in a 1 : 1 ratio. For high relatedness, three demes were inoculated with a single wild-type clone and three with a single mutator clone. For low relatedness, each deme was inoculated with one wild-type clone and one mutator clone. Colonies from each deme were transferred to fresh growth medium every 24 h. Global competition was imposed via hard selection: all six microcosms within a population were mixed before transferring single (high-relatedness treatment) or pairs (low-relatedness treatment) of colonies to fresh microcosms as appropriate. Mixing demes represents global competition as the probability of colonies from any given deme being selected for transfer depends upon the total productivity of that deme relative to the others in the metapopulation (Saccheri & Hanski 2006). It should be noted that the treatments do not differ in relatedness at time zero, as this is defined with respect only to loci governing siderophore production. After a few transfers, most demes in the low-relatedness treatment will be inoculated with one cooperator and one cheat clone and the relatedness will be low. Iron-limited CAA represents a novel environment for the bacteria, and as such we expect mutator lineages to adapt more rapidly to the growth in these microcosms.
2. Experimental procedures
(a) Bacterial strains
The tetracycline-resistant strain PAO985 (University of Washington) was used as the wild type, and the strain PAOΔmutS (Oliver et al. 2004), which has a deletion of the mismatch repair gene mutS, was used as the mutator. This strain has a spontaneous mutation rate more than two orders of magnitude higher (Oliver et al. 2004) than that of strain ATC 15692 (PAO1), from which both PAOΔmutS and PAO985 are derived.
(b) Growth conditions
Our study comprised 16 metapopulations of bacteria, divided into two treatment groups of eight metapopulations each; the experiment was carried out in two equal blocks. Each metapopulation comprised six demes (glass microcosms). Microcosms contained 6 ml casamino acids medium (CAA: 5 g casamino acids, 1.18 g K2HPO4·3H2O, 0.25 g MgSO4·7H2O, per litre), made iron limited by the addition of 70 μg ml−1 human apotransferrin (a natural iron chelator) and 20 mM sodium bicarbonate (necessary for iron chelator activity; Meyer et al. 1996).
High-relatedness metapopulations initially comprised three microcosms inoculated with a single colony of PAO985, and three microcosms inoculated with a single colony of PAOΔmutS. In low-relatedness metapopulations, each of the six microcosms was inoculated with a single colony of both strains.
Microcosms were incubated for 24 h at 37°C on an orbital shaker. Individual microcosms were then homogenized using a vortex mixer. Aliquots of culture from each of the six microcosms within a metapopulation were mixed and the mixture plated onto King's B agar (KB: 10 g glycerol, 20 g proteose peptone no. 3, 1.5 g K2HPO4·3H2O, 1.5 g MgSO4·7H2O, per litre). Single colonies (high relatedness) or pairs of colonies (low relatedness) were picked at random and used to inoculate six fresh microcosms. While our design meant that low-relatedness demes were always inoculated with twice as many cells as were high-relatedness treatments (approx. 4.6×107 versus 2.3×107 cells), the total number of cell divisions occurring within demes should not differ between the two treatments as stationary phase will be reached long before transfers take place.
This evolution was continued for 25 transfers (approx. 250 bacterial generations). Every fifth day, aliquots from all six microcosms within a metapopulation were mixed together in equal proportions, and the relative densities of PAO985 and PAOΔmutS, the frequency of siderophore-negative cheats and the total siderophore production were measured in each metapopulation as described below.
(c) Assays
Aliquots of diluted culture were replica plated on King's B (KB) agar (for a measure of total density), KB agar+60 μg ml−1 tetracycline (to select for growth of PAO985 only) and CAA agar (to score siderophore producers and non-producers visually).
- An aliquot of the whole-population mix was centrifuged to pellet the cells, and the supernatant (containing siderophores) was stored at −20°C. The total siderophore content of these supernatants was later determined using the chrome azurol S (CAS) method described by Schwyn & Neilands (1987), with the modification that we diluted Schwyn & Neilands' CAS recipe 1 : 1 with ddH2O. The relative absorbency at 630 nm of a mixture of 50 μl supernatant, and 100 μl CAS solution (all chemicals from Sigma) decreases linearly as siderophore concentration rises. Thus, a measure of mean siderophore production per colony-forming unit (CFU) in the ith microcosm is given by
where Ai is the absorbency of the ith sample; Aref is the absorbency of a reference solution comprising 50 μl sterile growth medium plus 100 μl CAS; and density is the CFU in 50 μl of the population sample.
(d) Statistical analyses
All data were analysed using Minicab v. 13. All frequency data were arcsine square root transformed prior to analysis, except in the case of the analysis of mean mutator versus mean cheat frequencies (figure 3), where a log transformation was used. These transformations provided the best fit to the assumptions of general linear modelling in each case. For the calculation of mean cheat frequencies, only data from timepoints 5–25 were used to avoid the initial frequency of zero influencing the results.
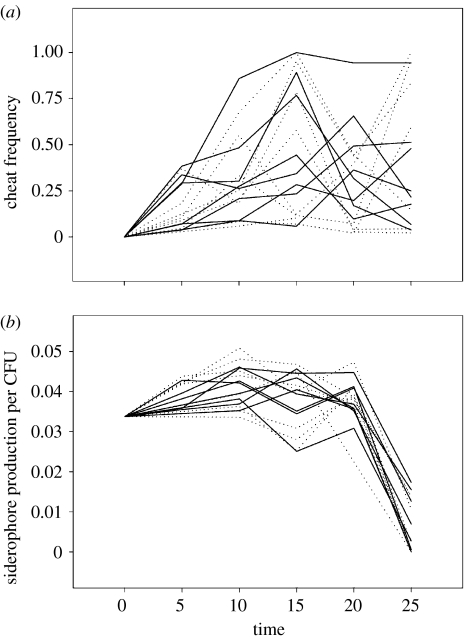
Figure 3.
Siderophore cheats appeared in all metapopulations and their frequency oscillated over time. (a) Pyoverdin-negative cheat frequencies; (b) total siderophore production per CFU. Dotted lines show metapopulations with high intra-deme relatedness, solid lines show metapopulations with low intra-deme relatedness.
3. Results
(a) Mutators are more able to dominate metapopulations when intra-deme relatedness is low
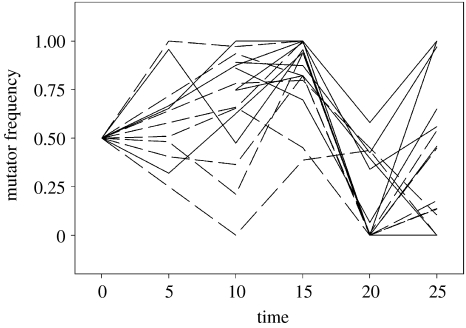
Mutator frequencies oscillated over time in all populations (figure 1). This was unsurprising, given the stochasticity expected to result from bottlenecking at each transfer, sampling effects and the nature of mutators themselves. We therefore calculated the mean mutator frequency over time for each population and used these data for our analyses, and we stress that our results should be viewed as demonstrating how relatedness affects the range of possible outcomes of mutator/wild-type competition.
Figure 1.
Mutator frequencies oscillated over time in all metapopulations. Dotted lines show metapopulations with high intra-deme relatedness, solid lines show metapopulations with low intra-deme relatedness. NB, the data for timepoint 5 are missing for one of the blocks; lines have been drawn connecting timepoints 0 and 10 in these cases.
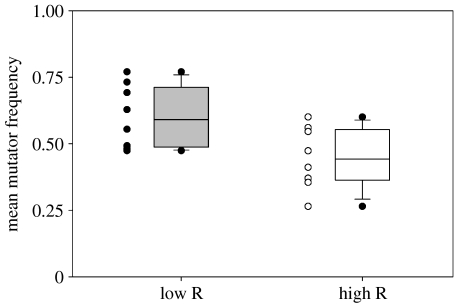
As shown in figure 2, the mean equilibrium mutator frequency was significantly higher in low-relatedness metapopulations (ANOVA with block and treatment fitted as factors: F1,13=6.46; p=0.025). Among high-relatedness metapopulations, the mean equilibrium mutator frequency was not significantly different from 0.5 (T7=1.26, p=0.247: range 0.26–0.60). Among low-relatedness metapopulations, however, mutators were more often able to rise to dominance (range 0.47–0.77). The mean equilibrium mutator frequency for this treatment was significantly greater than 0.5 (T7=2.45, p=0.021).
Figure 2.
Box plots and individual data points showing the distributions of mean mutator frequencies. Open symbols correspond to metapopulations with high intra-deme relatedness, closed symbols to metapopulations with low intra-deme relatedness.
(b) Mutators have an increased propensity to generate siderophore cheats
Pyoverdin-negative cheats arose in all metapopulations (figure 3a). Pyoverdin is not the only siderophore produced by this species, so we also used a chemical assay to measure total siderophore production in our microcosms. Consistent with the above, total siderophore production per CFU decreased over time in all metapopulations (figure 3b). As cheat frequency and total siderophore production were subject to severe oscillations and thus showed nonlinear dynamics over time (presumably as a result of the severe bottlenecking imposed), we calculated the mean cheat frequencies over time for all metapopulations and used these data in subsequent analyses.
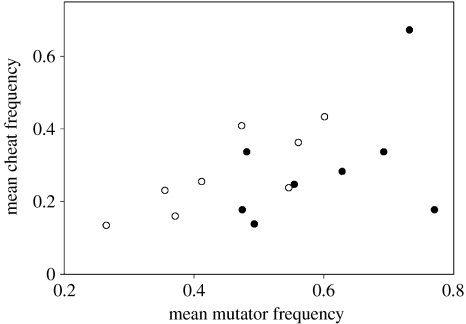
Mutators may affect a variety of traits, including numerous social behaviours. We have hypothesized, however, that mutators are selected against in this system because they break down siderophore-mediated cooperation. We therefore tested for a positive correlation between the mean mutator and mean cheat frequencies in each metapopulation. Consistent with our hypothesis and with our previous work (Harrison & Buckling 2005), a positive correlation was found (general linear model with block and treatment fitted as factors: F1,11 for mean mutator frequency=5.30, p=0.042). The strength of this relationship did not differ between high- and low-relatedness treatments (F1,11 for interaction term=0.04, p=0.842). These results are shown in figure 4 (note the right shift of low-relatedness data relative to high-relatedness data, showing higher mutator and cheat frequencies in these metapopulations).
Figure 4.
Mean mutator frequency and mean cheat frequency are positively correlated (p=0.042). Open circles show high intra-deme relatedness data, closed circles show low intra-deme relatedness data.
4. Discussion
(a) High relatedness selects against mutators in bacterial metapopulations
Here, we tested the hypothesis that kin selection for cooperation can affect the success of mutator alleles in bacterial metapopulations. Global competition favoured groups which contained higher total densities, and hence higher proportions of cooperators. Selection for cooperation was augmented by imposing high intra-deme relatedness or attenuated by imposing low intra-deme relatedness.
We conclude that the increased ability of mutators to respond to individual-level selection for cheating means that demes containing high mutator frequencies are more likely to selected against under high, as opposed to low, intra-deme relatedness. Under such conditions, mutators were unlikely to dominate metapopulations. We have also identified conditions where mutators can readily dominate metapopulations; namely, when relatedness is low. These results are consistent with our previous predictions (Harrison & Buckling 2005) and with work by other authors (Giraud et al. 2001). Previously published observations that migration in a metapopulation can select against mutator lineages (Giraud et al. 2001; Le Chat et al. 2006) may be partially explicable by the effect of hypermutability on cooperation if migration leads to global competition. It should be noted, however, that increases in diversity (i.e. decreases in relatedness) can sometimes reduce the success of cheats if diversification promotes character displacement among cooperators (Brockhurst et al. 2006; see also Smith et al. 2005). It would be very useful to synthesize models of diversification enhancing cooperation with kin selection models in order to obtain a more realistic picture of how the effects of population structure on selective forces interact to influence cooperation.
It is interesting that mutators were able to persist in all metapopulations, despite global competition favouring cooperation. Even under high relatedness, mutators represented 26–60% of the metapopulation. As our experimental microcosms represented a novel environment for the ancestral bacteria, this may be the result of a trade-off between the increased adaptive potential of mutator bacteria, and their increased propensity to break down cooperation. Further, mutators are expected to decrease relatedness and to be able to access cheating genotypes with increased ability to persist at high relative frequencies (Harrison & Buckling 2005; Harrison & Buckling in preparation).
(b) Mutators, siderophores and virulence in pathogen metapopulations
The results presented here suggest that further theoretical and empirical work in this area could help to explain environmental and clinical distributions of mutator bacteria, and contribute to our understanding of how mutator variants of pathogenic bacteria affect virulence.
Mutators are much more common in clinical isolates than in conspecific environmental populations (LeClerc et al. 1996). Further, P. aeruginosa mutators are more commonly observed in long-term chronic infections than in acute infections (Oliver et al. 2000). These observations have previously been attributed to the fluctuating environment experienced during long-term in vivo growth as a result of host immune responses and medical intervention (Oliver et al. 2000, 2004; de Visser 2002; Giraud et al. 2002; Schaaff et al. 2002). The appearance of hypermutable variants of pathogenic bacteria is associated with increased antibiotic resistance (Oliver et al. 2000; Ciofu et al. 2005) and as such mutators represent a significant risk factor for chronically colonized patients. Understanding the evolutionary ecology of mutators may thus benefit our understanding of chronic infection progression; indeed, understanding the evolution and ecology of pathogen communities may be vital in the development of new and more effective prophylaxis.
Much of the published work in this area has focused on bacterial populations within the respiratory tracts of cystic fibrosis (CF) patients. Individuals with CF are colonized by bacterial pathogens in early infancy, leading to chronic infection by diverse communities of microbes and, usually, eventual death from respiratory failure (Lyczak et al. 2002). Pseudomonas aeruginosa is one of the commonest CF pathogens, and is associated with a significant increase in patient morbidity (Nixon et al. 2001). Over the course of chronic P. aeruginosa infection, both increases in mutation rate (Oliver et al. 2000; Ciofu et al. 2005) and decreases in siderophore production (De Vos et al. 2001; Smith et al. 2006) are observed. Further, recurrent colonization and genetic diversification of founder clones lead to increases in both inter- and intraspecific diversity within the lung (Bergan & Hoiby 1975; Kresse et al. 2003; Saiman 2004; Moore et al. 2005; Smith et al. 2006). Thus, we expect that relatedness will decrease over the course of chronic infections. The CF lung microflora exists in spatially distinct communities (Armstrong et al. 1996; Gutierrez et al. 2001) which may be expected to undergo periodic mixing as a result of kinesiotherapy. Thus, it is possible that the CF microflora represents a globally competing metapopulation with low relatedness (if inter-deme mixing is common) or numerous distinct locally competing patches with low relatedness (if mixing is less common). Gaining a fuller picture of the structure of this ecosystem could help greatly in ascertaining the role of cooperation, and its effects on virulence, in chronic CF infections (Brown et al. 2002).
This leaves us with several possible explanations for the observed frequencies of P. aeruginosa mutators and siderophore cheats in such communities. Siderophore production is an excellent example of a cooperative trait necessary for virulence in acute infections (Meyer et al. 1996; Harrison et al. 2006), and low relatedness has been shown to lead to decreased virulence in such infections (Harrison et al. 2006). It has been assumed that, as siderophore production declines over time in CF isolates of P. aeruginosa, and as free iron levels can be elevated in the CF respiratory tract (Stites et al. 1998, 1999), siderophores are not necessary in chronic infections. However, this has not been explicitly tested. Declines in siderophore production could also be the result of local competition and/or low relatedness reducing the extent to which cooperative production of siderophores is favoured. This effect could be exacerbated by the appearance of mutator genotypes. As mutators have not been recovered from non-CF patients with acute P. aeruginosa infections (Oliver et al. 2000), it seems that simple environmental novelty is not sufficient to explain the presence of mutators in the CF airways (although the constantly fluctuating conditions experienced during chronic infection may still afford some advantage to mutators). Examining P. aeruginosa populations from different sites within patients could help to narrow down these possibilities and to develop models of ecosystem structure in vivo. Comparing microbial communities from patients with differing treatment histories, or from patients before and after kinesiotherapy, would reveal the effect of these interventions and help to uncover the interplay between adaptation and cooperation in the lung.
Moving on from the distribution of mutators within an infected patient, groups of patients in periodic close contact in CF centres or hospitals may themselves represent a metapopulation, as patient-to-patient transmission of pathogens is known to occur (Cheng et al. 1996; Ledson et al. 1998; Geddes 2001; Salunkhe et al. 2005). Studying groups of cross-infected patients as a metapopulation may help us to understand which strains or clones transmit readily through large numbers of patients. Taking an ecological view of cross-infection could also allow the identification of patients who are likely to be a ‘hub’ of infection. As transmissible clones can have high virulence or be antibiotic resistant (Cheng et al. 1996; Salunkhe et al. 2005), minimizing the contact between such patients and their peers could be a useful addition to preventive treatment.
5. Concluding remarks
In summary, we have shown that high relatedness can select against mutator lineages of bacteria in experimental metapopulations. This is because high relatedness favours cooperation via kin selection. On the other hand, when kin selection is tempered by decreases in relatedness, mutators are more likely to dominate metapopulations. It is clear that simple in vitro studies of microbes can make a valuable contribution to our understanding of pathogen ecology and evolution, and how these affect virulence-related traits. This could aid in targeting the existing and future therapies more effectively.
Acknowledgments
We would like to thank Antonio Oliver for kindly supplying PAOΔmutS and Mike Brockhurst, Andy Gardner and two anonymous referees for commenting on an early version of the manuscript. This work was supported by the Royal Society and NERC (UK). F.H. is supported by the Newton-Abraham Foundation.
References
- Armstrong D.S, Grimwood K, Carlin J.B, Carzino R, Olinsky A, Phelan P.D. Bronchoalveolar lavage or oropharyngeal cultures to identify lower respiratory pathogens in infants with cystic fibrosis. Pediatr. Pulmonol. 1996;21:267–275. doi: 10.1002/(SICI)1099-0496(199605)21:5<267::AID-PPUL1>3.0.CO;2-K. doi:10.1002/(SICI)1099-0496(199605)21:5<267::AID-PPUL1>3.0.CO;2-K [DOI] [PubMed] [Google Scholar]
- Bergan T, Hoiby N. Epidemiological markers for Pseudomonas aeruginosa. 6. Relationship between concomitant non-mucoid and mucoid strains from the respiratory tract in cystic fibrosis. Acta Pathol. Microbiol. Scand. Suppl. 1975;83:553–560. [PubMed] [Google Scholar]
- Brockhurst M.A, Hochberg M.E, Bell T, Buckling A. Character displacement promotes cooperation in bacterial biofilms. Curr. Biol. 2006;16:2030–2034. doi: 10.1016/j.cub.2006.08.068. doi:10.1016/j.cub.2006.08.068 [DOI] [PubMed] [Google Scholar]
- Brown S.P, Hochberg M.E, Grenfell B.T. Does multiple infection select for raised virulence? Trends Microbiol. 2002;10:401–405. doi: 10.1016/s0966-842x(02)02413-7. doi:10.1016/S0966-842X(02)02413-7 [DOI] [PubMed] [Google Scholar]
- Cheng K, Smyth R.L, Govan J.R, Doherty C, Winstanley C, Denning N, Heaf D.P, van Saene H, Hart C.A. Spread of [beta]-lactam-resistant Pseudomonas aeruginosa in a cystic fibrosis clinic. The Lancet. 1996;348:639–642. doi: 10.1016/S0140-6736(96)05169-0. doi:10.1016/S0140-6736(96)05169-0 [DOI] [PubMed] [Google Scholar]
- Ciofu O, Riis B, Pressler T, Poulsen H.E, Hoiby N. Occurrence of hypermutable Pseudomonas aeruginosa in cystic fibrosis patients is associated with the oxidative stress caused by chronic lung inflammation. Antimicrob. Agents Chemother. 2005;49:2276–2282. doi: 10.1128/AAC.49.6.2276-2282.2005. doi:10.1128/AAC.49.6.2276-2282.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser J.A. The fate of microbial mutators. Microbiology. 2002;148:1247–1252. doi: 10.1099/00221287-148-5-1247. [DOI] [PubMed] [Google Scholar]
- de Visser J.A.G.M, Zeyl C.W, Gerrish P.J, Blanchard J.L, Lenski R.E. Diminishing returns from mutation supply rate in asexual populations. Science. 1999;283:404–406. doi: 10.1126/science.283.5400.404. doi:10.1126/science.283.5400.404 [DOI] [PubMed] [Google Scholar]
- De Vos D, De Chial M, Cochez C, Jansen S, Tummler B, Meyer J.M, Cornelis P. Study of pyoverdine type and production by Pseudomonas aeruginosa isolated from cystic fibrosis patients: prevalence of type II pyoverdine isolates and accumulation of pyoverdine-negative mutations. Arch. Microbiol. 2001;175:384–388. doi: 10.1007/s002030100278. doi:10.1007/s002030100278 [DOI] [PubMed] [Google Scholar]
- Frank, S. A. 1998 Foundations of social evolution Monographs in behavior and ecology. Princeton, NJ: Princeton University Press.
- Funchain P, Yeung A, Stewart J.L, Lin R, Slupska M.M, Miller J.H. The consequences of growth of a mutator strain of Escherichia coli as measured by loss of function among multiple gene targets and loss of fitness. Genetics. 2000;154:959–970. doi: 10.1093/genetics/154.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes D.M. Of isolates and isolation: Pseudomonas aeruginosa in adults with cystic fibrosis. Lancet. 2001;358:522–523. doi: 10.1016/S0140-6736(01)05742-7. doi:10.1016/S0140-6736(01)05742-7 [DOI] [PubMed] [Google Scholar]
- Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M, Taddei F. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science. 2001;291:2606–2608. doi: 10.1126/science.1056421. doi:10.1126/science.1056421 [DOI] [PubMed] [Google Scholar]
- Giraud A, Matic I, Radman M, Fons M, Taddei F. Mutator bacteria as a risk factor in treatment of infectious diseases. Antimicrob. Agents Chemother. 2002;46:863–865. doi: 10.1128/AAC.46.3.863-865.2002. doi:10.1128/AAC.46.3.863-865.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin A.S, West S.A, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430:1024–1027. doi: 10.1038/nature02744. doi:10.1038/nature02744 [DOI] [PubMed] [Google Scholar]
- Gutierrez J.P, Grimwood K, Armstrong D.S, Carlin J.B, Carzino R, Olinsky A, Robertson C.F, Phelan P.D. Interlobar differences in bronchoalveolar lavage fluid from children with cystic fibrosis. Eur. Respir. J. 2001;17:281–286. doi: 10.1183/09031936.01.17202810. doi:10.1183/09031936.01.17202810 [DOI] [PubMed] [Google Scholar]
- Hamilton W.D. The genetical evolution of social behaviour I & II. J. Theor. Biol. 1964;7:1–52. doi: 10.1016/0022-5193(64)90038-4. doi:10.1016/0022-5193(64)90038-4 [DOI] [PubMed] [Google Scholar]
- Harrison F, Buckling A. Hypermutability impedes cooperation in pathogenic bacteria. Curr. Biol. 2005;15:1968–1971. doi: 10.1016/j.cub.2005.09.048. doi:10.1016/j.cub.2005.09.048 [DOI] [PubMed] [Google Scholar]
- Harrison, F. & Buckling, A. In preparation. Wider access to the adaptive landscape facilitates social cheating in a bacterial mutator.
- Harrison F, Browning L.E, Vos M, Buckling A. Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol. 2006;4:21. doi: 10.1186/1741-7007-4-21. doi:10.1186/1741-7007-4-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Matsuda H, Iwasa Y, Sasaki A. Evolutionarily stable mutation rate in a periodically changing environment. Genetics. 1989;121:163–174. doi: 10.1093/genetics/121.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresse A.U, Dinesh S.D, Larbig K, Romling U. Impact of large chromosomal inversions on the adaptation and evolution of Pseudomonas aeruginosa chronically colonizing cystic fibrosis lungs. Mol. Microbiol. 2003;47:145–158. doi: 10.1046/j.1365-2958.2003.03261.x. doi:10.1046/j.1365-2958.2003.03261.x [DOI] [PubMed] [Google Scholar]
- Le Chat L, Fons M, Taddei F. Escherichia coli mutators: selection criteria and migration effect. Microbiology. 2006;152:67–73. doi: 10.1099/mic.0.28418-0. doi:10.1099/mic.0.28418-0 [DOI] [PubMed] [Google Scholar]
- LeClerc J.E, Li B, Payne W.L, Cebula T.A. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. doi:10.1126/science.274.5290.1208 [DOI] [PubMed] [Google Scholar]
- Ledson M.J, Gallagher M.J, Corkill J.E, Hart C.A, Walshaw M.J. Cross infection between cystic fibrosis patients colonised with Burkholderia cepacia. Thorax. 1998;53:432–436. doi: 10.1136/thx.53.5.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak J.B, Cannon C.L, Pier G.B. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. doi:10.1128/CMR.15.2.194-222.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J.M, Neely A, Stintzi A, Georges C, Holder I.A. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 1996;64:518–523. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.E, Shaw A, Millar B.C, Downey D.G, Murphy P.G, Elborn J.S. Microbial ecology of the cystic fibrosis lung: does microflora type influence microbial loading? Br. J. Biomed. Sci. 2005;62:175–178. doi: 10.1080/09674845.2005.11732707. [DOI] [PubMed] [Google Scholar]
- Nixon G.M, Armstrong D.S, Carzino R, Carlin J.B, Olinsky A, Robertson C.F, Grimwood K. Clinical outcome after early Pseudomonas aeruginosa infection in cystic fibrosis. J. Pediatr. 2001;138:699–704. doi: 10.1067/mpd.2001.112897. doi:10.1067/mpd.2001.112897 [DOI] [PubMed] [Google Scholar]
- Oliver A, Canton R, Campo P, Baquero F, Blazquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1254. doi: 10.1126/science.288.5469.1251. doi:10.1126/science.288.5469.1251 [DOI] [PubMed] [Google Scholar]
- Oliver A, Levin B.R, Juan C, Baquero F, Blazquez J. Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: implications for susceptibility testing and treatment of chronic infections. Antimicrob. Agents Chemother. 2004;48:4226–4233. doi: 10.1128/AAC.48.11.4226-4233.2004. doi:10.1128/AAC.48.11.4226-4233.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey P.B. Evolutionary genetics: the economics of mutation. Curr. Biol. 1999;9:R371–R373. doi: 10.1016/s0960-9822(99)80230-9. doi:10.1016/S0960-9822(99)80230-9 [DOI] [PubMed] [Google Scholar]
- Saccheri I, Hanski I. Natural selection and population dynamics. Trends Ecol. Evol. 2006;21:341–347. doi: 10.1016/j.tree.2006.03.018. doi:10.1016/j.tree.2006.03.018 [DOI] [PubMed] [Google Scholar]
- Saiman L. Microbiology of early CF lung disease. Paediatr. Respir. Rev. 2004;5(Suppl. A):S367–S369. doi: 10.1016/s1526-0542(04)90065-6. doi:10.1016/S1526-0542(04)90065-6 [DOI] [PubMed] [Google Scholar]
- Salunkhe P, Smart C.H, Morgan J.A, Panagea S, Walshaw M.J, Hart C.A, Geffers R, Tummler B, Winstanley C. A cystic fibrosis epidemic strain of Pseudomonas aeruginosa displays enhanced virulence and antimicrobial resistance. J. Bacteriol. 2005;187:4908–4920. doi: 10.1128/JB.187.14.4908-4920.2005. doi:10.1128/JB.187.14.4908-4920.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaff F, Reipert A, Bierbaum G. An elevated mutation frequency favors development of vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2002;46:3540–3548. doi: 10.1128/AAC.46.11.3540-3548.2002. doi:10.1128/AAC.46.11.3540-3548.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwyn B, Neilands J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. doi:10.1016/0003-2697(87)90612-9 [DOI] [PubMed] [Google Scholar]
- Smith E.E, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl Acad. Sci. USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. doi:10.1073/pnas.0602138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.E, Sims E.H, Spencer D.H, Kaul R, Olson M.V. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. J. Bacteriol. 2005;187:2138–2147. doi: 10.1128/JB.187.6.2138-2147.2005. doi:10.1128/JB.187.6.2138-2147.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski P.D, Gerrish P.J, Lenski R.E. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. doi:10.1038/42701 [DOI] [PubMed] [Google Scholar]
- Sniegowski P.D, Gerrish P.J, Johnson T, Shaver A. The evolution of mutation rates: separating causes from consequences. Bioessays. 2000;22:1057–1066. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. doi:10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W [DOI] [PubMed] [Google Scholar]
- Stites S.W, Walters B, O'Brien-Ladner A.R, Bailey K, Wesselius L.J. Increased iron and ferritin content of sputum from patients with cystic fibrosis or chronic bronchitis. Chest. 1998;114:814–819. doi: 10.1378/chest.114.3.814. [DOI] [PubMed] [Google Scholar]
- Stites S.W, Plautz M.W, Bailey K, O'Brien-Ladner A.R, Wesselius L.J. Increased concentrations of iron and isoferritins in the lower respiratory tract of patients with stable cystic fibrosis. Am. J. Respir. Crit. Care Med. 1999;160:796–801. doi: 10.1164/ajrccm.160.3.9811018. [DOI] [PubMed] [Google Scholar]
- Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon P.H, Godelle B. Role of mutator alleles in adaptive evolution. Nature. 1997;387:700–702. doi: 10.1038/42696. doi:10.1038/42696 [DOI] [PubMed] [Google Scholar]
- Tanabe K, Kondo T, Onodera Y, Furusawa M. A conspicuous adaptability to antibiotics in the Escherichia coli mutator strain, dnaQ49. FEMS Microbiol. Lett. 1999;176:191–196. doi: 10.1111/j.1574-6968.1999.tb13661.x. doi:10.1111/j.1574-6968.1999.tb13661.x [DOI] [PubMed] [Google Scholar]
- Trobner W, Piechocki R. Selection against hypermutability in Escherichia coli during long term evolution. Mol. Gen. Genet. 1984;198:177–178. doi: 10.1007/BF00328720. doi:10.1007/BF00328720 [DOI] [PubMed] [Google Scholar]
- West S.A, Buckling A. Cooperation, virulence and siderophore production in bacterial parasites. Proc. R. Soc. B. 2003;270:37–44. doi: 10.1098/rspb.2002.2209. doi:10.1098/rspb.2002.2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S.A, Griffin A.S, Gardner A, Diggle S.P. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. doi:10.1038/nrmicro1461 [DOI] [PubMed] [Google Scholar]