Abstract
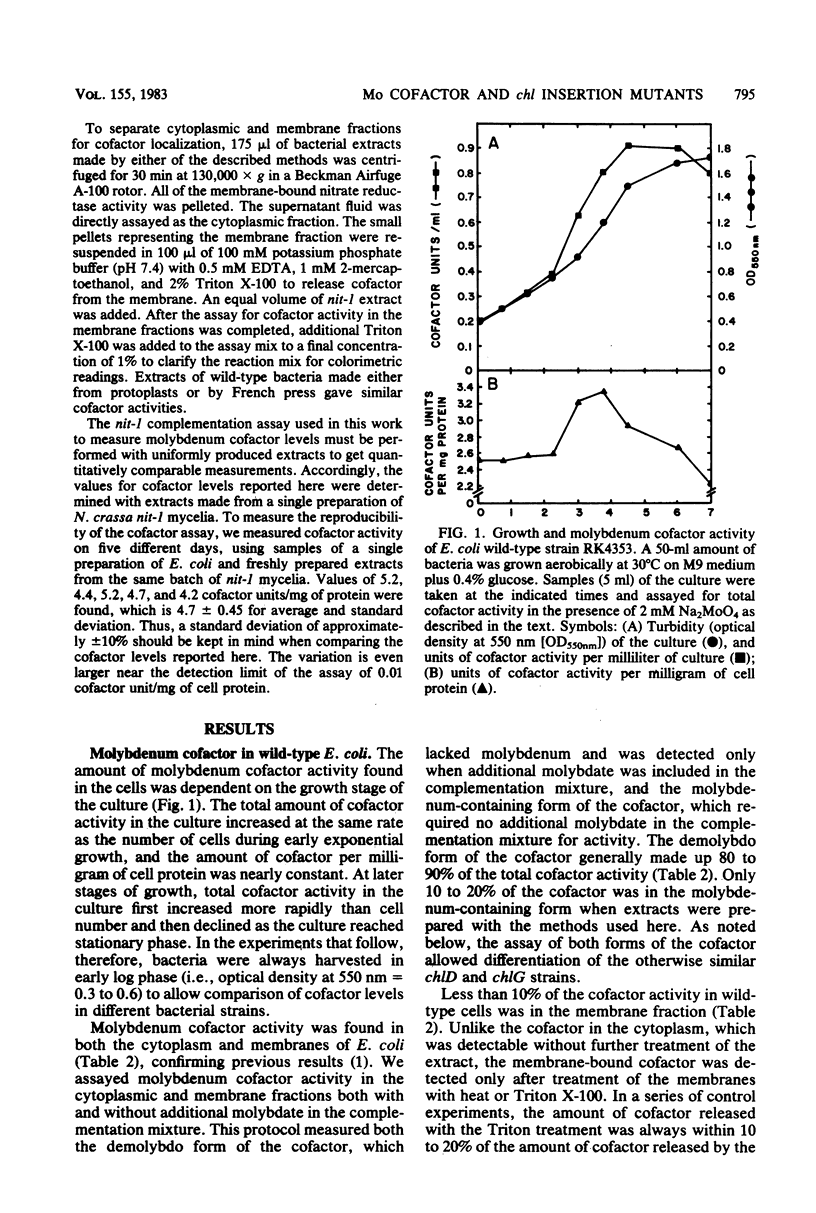
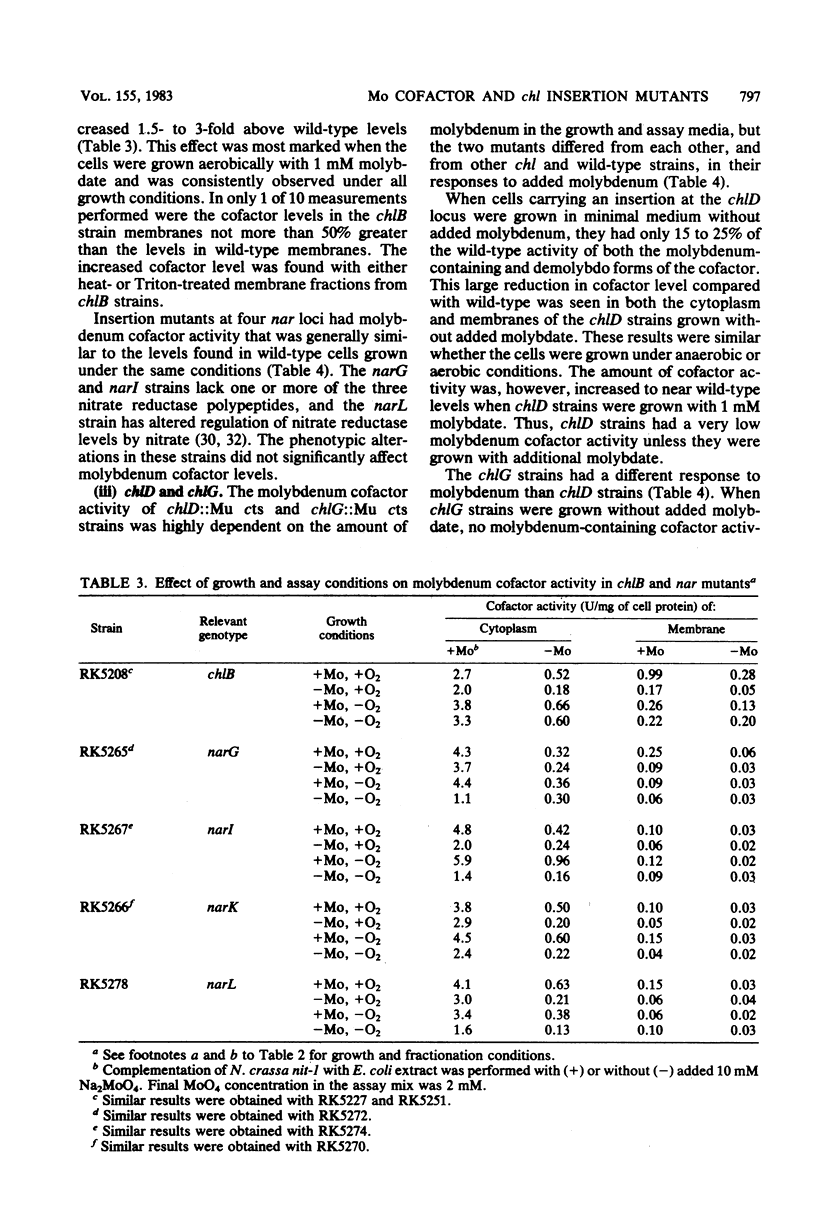
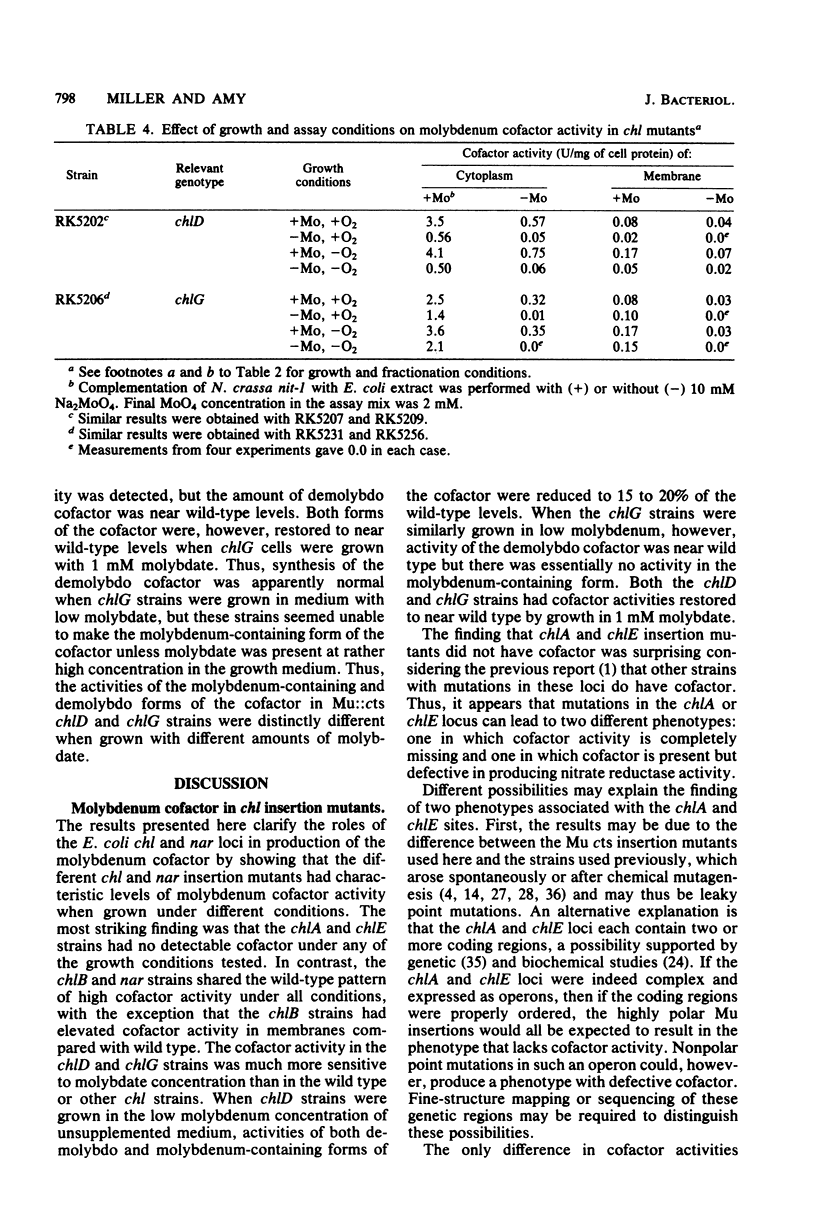
We examined molybdenum cofactor activity in chlorate-resistant (chl) and nitrate reductase-deficient (nar) insertion mutants and wild-type strains of Escherichia coli K-12. The bacterial molybdenum cofactor was assayed by its ability to restore activity to the cofactor-deficient nitrate reductase found in the nit-1 strain of Neurospora crassa. In the wild-type E. coli strains, molybdenum cofactor was synthesized constitutively and found in both cytoplasmic and membrane fractions. Cofactor was found in two forms: the demolybdo form required additional molybdate in the assay mix for detection, whereas the molybdenum-containing form was active without additional molybdate. The chlA and chlE mutants had no detectable cofactor. The chlB and the narG, narI, narK, and narL (previously designated chlC) strains had cofactor levels similar to those of the wild-type strains, except the chlB strains had two to threefold more membrane-bound cofactor. Cofactor levels in the chlD and chlG strains were sensitive to molybdate. When grown in 1 microM molybdate, the chlD strains had only 15 to 20% of the wild-type levels of the demolybdo and molybdenum-containing forms of the cofactor. In contrast, the chlG strains had near wild-type levels of demolybdo cofactor when grown in 1 microM molybdate, but none of the molybdenum-containing form of the cofactor. Near wild-type levels of both forms of the cofactor were restored to the chlD and chlG strains by growth in 1 mM molybdate.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amy N. K. Identification of the molybdenum cofactor in chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1981 Oct;148(1):274–282. doi: 10.1128/jb.148.1.274-282.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amy N. K., Rajagopalan K. V. Characterization of molybdenum cofactor from Escherichia coli. J Bacteriol. 1979 Oct;140(1):114–124. doi: 10.1128/jb.140.1.114-124.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casse F. Mapping of the gene chl-B controlling membran bound nitrate reductase and formic hydrogen-lyase activities in Escherichia coli K 12. Biochem Biophys Res Commun. 1970 May 11;39(3):429–436. doi: 10.1016/0006-291x(70)90596-6. [DOI] [PubMed] [Google Scholar]
- Claassen V. P., Oltmann L. F., Bus S., v 't Riet J., Stouthamer A. H. The influence of growth conditions on the synthesis of molybdenum cofactor in Proteins mirabilis. Arch Microbiol. 1981 Sep;130(1):44–49. doi: 10.1007/BF00527070. [DOI] [PubMed] [Google Scholar]
- Cox J. C., Edwards E. S., DeMoss J. A. Resolution of distinct selenium-containing formate dehydrogenases from Escherichia coli. J Bacteriol. 1981 Mar;145(3):1317–1324. doi: 10.1128/jb.145.3.1317-1324.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourdieu M., Andrade E., Puig J. Molybdenum and chlorate resistant mutants in Escherichia coli K12. Biochem Biophys Res Commun. 1976 Jun 7;70(3):766–773. doi: 10.1016/0006-291x(76)90658-6. [DOI] [PubMed] [Google Scholar]
- Garrett R. H., Nason A. Further purification and properties of Neurospora nitrate reductase. J Biol Chem. 1969 Jun 10;244(11):2870–2882. [PubMed] [Google Scholar]
- Garrett R. H. The induction of nitrite reductase in Neurospora crassa. Biochim Biophys Acta. 1972 May 16;264(3):481–489. doi: 10.1016/0304-4165(72)90011-6. [DOI] [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Comparison of nitrate reductase mutants of Escherichia coli selected by alternative procedures. Mol Gen Genet. 1972;116(1):1–10. doi: 10.1007/BF00334254. [DOI] [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Phenotypic restoration by molybdate of nitrate reductase activity in chlD mutants of Escherichia coli. J Bacteriol. 1971 Nov;108(2):854–860. doi: 10.1128/jb.108.2.854-860.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Hainline B. E., Rajagopalan K. V. Characterization of the molybdenum cofactor of sulfite oxidase, xanthine, oxidase, and nitrate reductase. Identification of a pteridine as a structural component. J Biol Chem. 1980 Mar 10;255(5):1783–1786. [PubMed] [Google Scholar]
- Johnson J. L., Rajagopalan K. V. Structural and metabolic relationship between the molybdenum cofactor and urothione. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6856–6860. doi: 10.1073/pnas.79.22.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchum P. A., Cambier H. Y., Frazier W. A., 3rd, Madansky C. H., Nason A. In vitro assembly of Neurospora assimilatory nitrate reductase from protein subunits of a Neurospora mutant and the xanthine oxidizing or aldehyde oxidase systems of higher animals. Proc Natl Acad Sci U S A. 1970 Jul;66(3):1016–1023. doi: 10.1073/pnas.66.3.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchum P. A., Owens M. S. Production of molybdenum-coordinating compound by Bacillus thuringiensis. J Bacteriol. 1975 May;122(2):412–417. doi: 10.1128/jb.122.2.412-417.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchum P. A., Swarin R. S. In vitro formation of assimilatory nitrate reductase: presence of the constitutive component in bacteria. Biochem Biophys Res Commun. 1973 Jun 19;52(4):1450–1456. doi: 10.1016/0006-291x(73)90663-3. [DOI] [PubMed] [Google Scholar]
- PATEMAN J. A., COVE D. J., REVER B. M., ROBERTS D. B. A COMMON CO-FACTOR FOR NITRATE REDUCTASE AND XANTHINE DEHYDROGENASE WHICH ALSO REGULATES THE SYNTHESIS OF NITRATE REDUCTASE. Nature. 1964 Jan 4;201:58–60. doi: 10.1038/201058a0. [DOI] [PubMed] [Google Scholar]
- Page W. J., von Tigerstrom M. Iron- and molybdenum-repressible outer membrane proteins in competent Azotobacter vinelandii. J Bacteriol. 1982 Jul;151(1):237–242. doi: 10.1128/jb.151.1.237-242.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal M. C., Burini J. F., Ratouchniak J., Chippaux M. Regulation of the nitrate reductase operon: effect of mutations in chlA, B, D and E genes. Mol Gen Genet. 1982;188(1):103–106. doi: 10.1007/BF00333001. [DOI] [PubMed] [Google Scholar]
- Pascal M. C., Chippaux M. Involvement of a gene of the chl E locus in the regulation of the nitrate reductase operon. Mol Gen Genet. 1982;185(2):334–338. doi: 10.1007/BF00330808. [DOI] [PubMed] [Google Scholar]
- Pienkos P. T., Shah V. K., Brill W. J. Molybdenum cofactors from molybdoenzymes and in vitro reconstitution of nitrogenase and nitrate reductase. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5468–5471. doi: 10.1073/pnas.74.12.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig J., Azoulay E. Etude génétique et biochimique des mutants résistant au Clo minus 3 (gènes chl A, chl B, chl C) C R Acad Sci Hebd Seances Acad Sci D. 1967 Apr 10;264(15):1916–1918. [PubMed] [Google Scholar]
- Puig J., Azoulay E., Pichinoty F., Gendre J. Genetic mapping of the chl C gene of the nitrate reductase A system in Escherichia coli K12. Biochem Biophys Res Commun. 1969 Jun 6;35(5):659–662. doi: 10.1016/0006-291x(69)90455-0. [DOI] [PubMed] [Google Scholar]
- Sperl G. T., DeMoss J. A. chlD gene function in molybdate activation of nitrate reductase. J Bacteriol. 1975 Jun;122(3):1230–1238. doi: 10.1128/jb.122.3.1230-1238.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., MacGregor C. H. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982 Aug;151(2):788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J Bacteriol. 1982 Sep;151(3):1320–1325. doi: 10.1128/jb.151.3.1320-1325.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M., Tsuchiya T., Ishimoto M. Proton translocation coupled to trimethylamine N-oxide reduction in anaerobically grown Escherichia coli. J Bacteriol. 1981 Dec;148(3):762–768. doi: 10.1128/jb.148.3.762-768.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomsett A. B., Garrett R. H. The isolation and characterization of mutants defective in nitrate assimilation in Neurospora crassa. Genetics. 1980 Jul;95(3):649–660. doi: 10.1093/genetics/95.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables W. A. Genetic studies with nitrate reductase-less mutants of Escherichia coli. I. Fine structure analysis of the narA, narB and narE loci. Mol Gen Genet. 1972;114(3):223–231. doi: 10.1007/BF01788891. [DOI] [PubMed] [Google Scholar]
- Venables W. A., Guest J. R. Transduction of nitrate reductase loci of Escherichia coli by phages P-1 and lambda. Mol Gen Genet. 1968;103(2):127–140. doi: 10.1007/BF00427140. [DOI] [PubMed] [Google Scholar]
- Weiss R. L. Protoplast formation in Escherichia coli. J Bacteriol. 1976 Nov;128(2):668–670. doi: 10.1128/jb.128.2.668-670.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campillo-Campbell A., Campbell A. Molybdenum cofactor requirement for biotin sulfoxide reduction in Escherichia coli. J Bacteriol. 1982 Feb;149(2):469–478. doi: 10.1128/jb.149.2.469-478.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]


