Abstract
Cysteine-substitution mutants of yeast DNA topoisomerase II were used to test footprinting of the enzyme by 2-nitro-5-thiocyanobenzoate, which cyanylates exposed cysteines in a native protein for peptide cleavage at the cyanylated sites upon unfolding and incubating the protein at pH 9. For a mutant enzyme containing a single cysteine, the extent of peptide cleavage was found to reflect the accessibility of the residue in the native protein. For proteins with multiple cysteines, however, such a correlation was obscured by the transfer of cyano groups from modified to unmodified cysteines during incubation of the unfolded protein at pH 9; accessibilities of the cysteinyl residues in a native protein could be assessed only if cyano shuffling was prevented by blocking uncyanylated sulfhydryls with a second thiol reagent. The successive use of two reagents in cysteine footprinting was applied in probing the ATP-modulated formation of contacts in yeast DNA topoisomerase II.
Keywords: thiol cyanylation, ligand-induced protein contacts, mutagenesis
The modulation of the chemical and enzymatic reactivities of individual nucleotides in a DNA or RNA by the binding of other molecules, particularly proteins, has been used extensively in mapping the interacting surfaces of nucleic acids (1–4). In a typical experiment, often termed “footprinting” (2), the chemical or enzymatic reagent is one that reacts with a nucleic acid under physiological conditions and directly or indirectly effects cleavage of the polynucleotide at the sites of reaction. When a nucleic acid labeled at one unique end is treated very lightly with such a footprinting reagent, the products can be resolved by electrophoresis in a sequencing gel and the probabilities of cleavage at different positions along the polynucleotide chain are readily deduced from the intensities of the labeled fragments.
The development of methods for the footprinting of proteins has lagged behind that of the nucleic acids. By fusing to one protein terminus a short amino acid stretch that serves as the recognition site of a sequence-specific protein kinase (5) or antibodies (6–8), end-labeling of a polypeptide by radiolabeling or immunochemical staining can be accomplished without resorting to laborious chemical steps (9). However, in contrast to the general susceptibility of nucleic acids to nonspecific nucleases, enzymatic attack of well folded proteins often is restricted to their disordered regions, which, in turn, limit the usefulness of proteolytic probing. In principle, chemical footprinting of proteins should be of broader applicability. There is, however, a scarcity of protein reagents that meet the criteria of reacting under mild conditions and effecting peptide cleavage at the site of modification. Only redox-active metal complexes, which have been used extensively in nucleic acid footprinting, recently have been gaining popularity in protein footprinting (see for examples, refs. 10–13, and references therein). In several studies, reagents that act at particular amino acid residues also have been used. For example, a reversible lysine reagent, citraconic anhydride, was used to footprint lysines (14). After partial citraconylation of a tagged protein in its native state, the protein is unfolded and irreversibly acetylated at the unreacted lysines; reversal of citraconylation then avails the liberated lysines for peptide cleavage by the lysine-specific endoproteinase Lys-C (14). In a methionine-footprinting procedure, a reagent that irreversibly modifies methionyl residues was used as the footprinting reagent. This modification prevents subsequent cleavage at the residue by cyanogen bromide, and, hence, the positions of methionyl residues inaccessible to the footprinting reagent could be deduced (15).
A well known cysteine reagent, 2-nitro-5-thiocyanobenzoate (NTCB), appeared to meet the criteria for a convenient protein footprinting agent. Cyanylation by NTCB occurs specifically at cysteinyl residues under physiological conditions, and cleavage of the peptide bond at an S-cyanocysteine is nearly quantitative upon unfolding and incubating the polypeptide at pH 9 (16, 17). Cysteine footprinting also is attractive because the small size and unique chemical reactivity of a cysteinyl residue already has made it one of the most useful residues in structural and biochemical studies of proteins and their complexes, and cysteine substitution or cysteine-scanning mutagenesis has been widely applied in such studies. Despite the apparent potential of NTCB as a protein footprinting agent, however, its use so far has been limited. For example, in a study of the protection of the cysteinyl residues of Escherichia coli RecA protein by the binding of ATP or ADP, a maleimide was used to modify the cysteines and the positions of the unmodified cysteines in the polypeptide chain then were mapped by using NTCB as a cysteine-specific polypeptide cleavage reagent (18). More recently, Yamashita et al. (19) footprinted the positions of six disulfides in a protein by partial disulfide reduction and alkylation of the sulfhydryls formed; after the isolation of intermediates with different numbers of disulfides, the positions of the remaining disulfides in each were mapped by cleaving the protein with a cyanylation reagent after disulfide reduction. In both examples, NTCB-type compounds were used as a cysteine-specific peptide cleavage agent, but not as footprinting agents that probe the relative reactivities of cysteinyl residues in a native protein.
In this study, we explored the utility of NTCB as a protein footprinting reagent by applying it to the identification of interdomainal contacts that are formed in yeast DNA topoisomerase II upon binding a nonhydrolyzable ATP analog 5′-adenylyl-β,γ-imidodiphosphate (AMPPNP). By studying a number of cysteine-substitution mutants of the enzyme, we found that footprinting by NTCB was straightforward for polypeptides with a single cysteine. For a polypeptide containing multiple cysteines, however, during the rather slow peptide cleavage step a cyano group apparently could transfer from a cyanylated cysteine to a free sulfhydryl. Thus, the peptide cleavage pattern would no longer reflect the relative reactivities of the individual cysteinyl residues during the footprinting step. We found that cyano shuffling could be prevented by adding an excess of a second thiol reagent during peptide cleavage to block cysteines that were not cyanylated in the footprinting step. Using such a strategy, we were able to affirm the involvement of a particular region of the homodimeric yeast DNA topoisomerase II in the formation of a dimer interface upon the binding of AMPPNP. We believe that a combination of cysteine-substitution (or -scanning) mutagenesis and cysteine footprinting by NTCB should add a useful tool in the mapping of protein–protein contacts or contacts between proteins and other molecules.
MATERIALS AND METHODS
Site-Directed Cysteine Substitution.
Site-directed mutagenesis within the yeast TOP2 gene in an expression plasmid YEpTOP2PGAL1 was carried out as described (20), and the mutated regions of the resulting plasmids were sequenced to confirm the introduction of the desired mutations. The ability of the plasmid-borne top2 mutants to complement the top2–4 temperature-sensitive defect of a yeast strain (21) then was carried out to test the functionality of the mutant enzyme; mutants that could not complement the top2–4 defect were not used in further studies. For N-terminal tagging of unsubstituted and mutant polypeptides, a DNA segment containing codons for an influenza virus hemagglutinin epitope and a heart muscle kinase phosphorylation site was inserted into the expression clone, as described (22). The addition of these tags to the N terminus of the yeast enzyme has no major effect on its catalytic activities (22). Overexpression and purification of the proteins were done as described (20, 22), but 2-mercaptoethanol was omitted in the final elution buffer. This omission does not affect appreciably the activity of the yeast enzyme (23). Purified proteins were tested further for their formation of contacts between the N-terminal ATPase domains upon binding of AMPPNP (closed-clamp formation), using the filter-binding assay described (24).
Cysteine Cyanylation and Peptide Cleavage at S-Cyanocysteines.
Approximately 20 μg of a tagged protein (approximately 0.12 nmol protein or 1.1 nmol cysteinyl residues), in 200 μl of buffer containing 50 mM Tris⋅HCl, pH 7.7/1 mM EDTA/1 mM EGTA/5% glycerol/150 mM KCl/1 μM each of leupeptin and pepstatin A, was used in each footprinting reaction. The reaction mixtures were supplemented with MgCl2 to 10 mM and AMPPNP to 1 mM if present. After 15 min at 30° to allow the formation of AMPPNP-modulated dimer contacts, approximately 1 μl of 0.5 mM NTCB (Aldrich; 50 mM stock in EtOH, diluted with H2O) was added. This corresponded to a molar ratio of NTCB/protein of about 4:1 or NTCB/cysteinyl residue of about 0.4:1. After 10 min at 30°, cyanylation was terminated by the addition of acetic acid to 20% (vol/vol). The reaction mixtures were dialyzed against two changes of 15% (vol/vol) acetic acid over a period of 4.5 h to remove unreacted NTCB (all dialysis steps were carried out at 4°C). Each dialyzed mixture was dried under reduced pressure (in a Savant Speed Vac) to ≤5μl, thoroughly mixed with 40 μl of distilled H2O, and then dried to completion. Each dried protein was resuspended in ∼110 μl of an alkaline denaturing solution containing 8 M urea, 0.2 M N-[2-hydroxyethyl]piperazine-N′-[3-propanesulfonic acid] (EPPS) titrated with NaOH to pH 9, and 10 mM N-ethylmaleimide (NEM; from freshly prepared 0.2 M stock in H2O), and incubated at 37° for 12–16 h for polypeptide cleavage at the cyanylated cysteines. The reaction mixtures then were dialyzed against a buffer containing 10 mM Tris⋅HCl, pH 7.7/0.01% SDS for 3 h and then against 10 mM Tris⋅HCl, pH 7.7/0.001% SDS for another 3 h. SDS was added to the dialysis buffers to prevent formation of proteinaceous precipitates. The heart muscle kinase phosphorylation site (HMK) tag of the polypeptide fragments was then radiolabeled by using [γ-32P]ATP and the protein kinase A catalytic subunit (New England Biolabs), as described (22). Radiolabeling was performed in a buffer containing 20 mM Tris⋅HCl, pH 7.7/10 mM MgCl2/50 mM NaCl/5 mM 2-mercaptoethanol. The low level of SDS in the protein samples was tolerated by the kinase. The polypeptide fragments were separated by SDS/PAGE, using 15% gel in most of the experiments. Before loading, samples were heated at 65° rather than 100° for 3 min to minimize degradation of the polypeptide. Size markers formed by cleavage of HMK-tagged yeast DNA topoisomerase II at the cysteines were prepared as described (14, 22).
RESULTS
Construction of Cysteine-Substitution Mutants of Yeast DNA Topoisomerase II and Cysteine Footprinting by Cyanylation.
Various experiments have indicated that the binding of a nonhydrolyzable ATP analog and, by implication, ATP itself leads to dimerization of the pair of ATPase domains of a type II DNA topoisomerase (24–27). In the case of E. coli DNA gyrase (DNA topoisomerase II), the solution of the crystal structure of a 43-kDa B-subunit fragment with a bound AMPPNP showed that in the crystal the AMPPNP-GyrB complex exists as a dimer, with the dimer interface formed on one side by residues near the N terminus and on the other side by a cluster of residues including Ile-94, His-99, and Leu-115 (25).
The GyrB crystal structure and biochemical studies of the ATP-modulated dimerization of GyrB suggest that the dimer interface observed in the crystal is also formed in solution upon binding of ATP or its analogs (25–27). Furthermore, because of the high degree of sequence homology between the ATPase domains of bacterial gyrase and other type IIA DNA topoisomerases including yeast DNA topoisomerase II (28), a very similar ligand-induced dimer interface presumably is formed in all these enzymes. We therefore decided to explore the utility of cysteine footprinting by applying it to a test of the AMPPNP-dependent formation of this dimer interface.
Based on the E. coli GyrB fragment crystal structure (25) and alignment of the amino acid sequences of type IIA DNA topoisomerases (28), a set of cysteine-substitution mutants I116C, I120C, L125C, Y130C, and R141C of the yeast DNA topoisomerase II was constructed. In each mutant, the specified amino acid residue was replaced by a cysteine. Three of the mutants, I120C, L125C, and R141C, overexpressed well, and the expression of each mutant protein from a plasmid-borne gene was found to complement the thermal sensitivity of a yeast strain carrying a mutation top2–4 (21) in the chromosomal DNA topoisomerase II gene. Thus, the three mutant enzymes appeared to be functional in vivo, and these mutations were moved into a plasmid originally constructed for overexpression of DNA topoisomerase II with an HMK added to its N terminus (22). Each mutant protein was purified to near homogeneity and tested for its conversion to the closed-clamp form upon binding of AMPPNP, which signifies dimerization of the pair of ATPase domains of the homodimeric enzyme (24). All three were found to undergo the AMPPNP-mediated formation of dimer contacts (results not shown). Surprisingly, when the three HMK-tagged I120C, L125C, and R141C proteins were lightly treated with NTCB and then unfolded at pH 9 to effect peptide cleavage at the cyanocysteines, none showed significant AMPPNP-dependence of polypeptide cleavage.
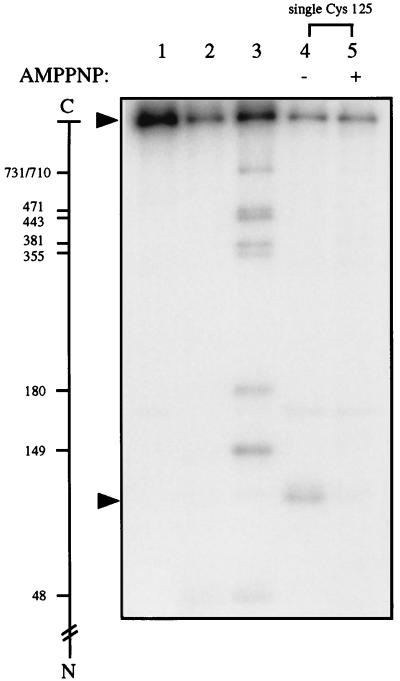
To test whether this failure reflected methodological complications or a misidentification of the ATP-modulated dimer interface, the L125C mutation was introduced into a catalytically active mutant yeast DNA topoisomerase II in which all nine cysteines of the wild-type enzyme had been replaced by alanines (29). The results are displayed in Fig. 1. The first three lanes of Fig. 1 contained control samples in which unsubstituted yeast DNA topoisomerase II with an HMK tag was used. The lane 1 sample was not subjected to footprinting treatments, the lane 2 sample was taken through all the footprinting steps but without the addition of NTCB, and the lane 3 sample was obtained by extensive cyanylation and subsequent cleavage of the polypeptide at the S-cyanocysteines to provide a ladder of size markers, as described previously (14, 22). After radiolabeling of the samples at the HMK tag, the proteins were analyzed by electrophoresis in an SDS-polyacrylamide gel. As shown in the autoradiogram of the resulting gel, the lane 3 size marker showed a distinct pattern of bands, which could be assigned readily as the products of cleavage at Cys-731 and 710 (unresolved), Cys-471, Cys-443, Cys-381, Cys-355, Cys-180, Cys-149, and Cys-48 (22). The pattern of the lane 2 sample indicated that nonspecific degradation of the mock-treated sample was acceptable; other than the intense band of the intact protein, only a few faint bands that did not coincide with the positions of the bands generated by cleavage at the cysteines (lane 3) were observed.
Figure 1.
Cysteine cyanylation by NTCB and subsequent polypeptide cleavage at the cyanylated cysteinyl residues upon unfolding and incubating the protein at pH 9. Lanes 1–3 contained yeast DNA topoisomerase II tagged with an HMK at its N terminus, and lanes 4 and 5 contained a similarly tagged mutant enzyme termed “single Cys-125,” in which all nine cysteines of wild-type yeast DNA topoisomerase II had been replaced by alanines and a cysteinyl residue had replaced Leu-125 of the wild-type enzyme. See text for treatments of the samples. The minus and plus signs over lanes 4 and 5 denote the absence and presence of AMPPNP, respectively, during cysteine cyanylation by NTCB. The scale in the left margin marks the cleavage sites of the 32P-labeled protein in the formation of the radiolabeled fragments by partial cleavage at the cysteines. The arrows near the top and bottom of the gel indicate, respectively, the positions of the intact protein and the radiolabeled fragment generated by cleavage at Cys-125 of the mutant enzyme.
Lanes 4 and 5 of Fig. 1 contained, respectively, the single Cys-125 mutant protein first treated with NTCB in the presence and absence of AMPPNP, and then unfolded and incubated at pH 9 to effect peptide cleavage. In the absence of AMPPNP (lane 4), a major cleavage product migrating near the bottom of the gel photograph was observed. The size of this end-labeled fragment was estimated from the lane 3 size markers, and the estimated size agreed with that expected for cleavage at Cys-125 in the mutant protein. A few faint bands also were seen in the gel autoradiogram; these presumably represented degradation products of the radiolabeled polypeptides, but their exact origin was obscure. Significantly, cleavage at Cys-125 was much reduced when cyanylation of the native protein was first carried out in the presence of AMPPNP (lane 5). This reduction suggests that amino acid residue 125 of yeast DNA topoisomerase II becomes less accessible to NTCB when the enzyme is converted to the AMPPNP-bound form, which supports the presence of this residue at the ATP-mediated dimer interface. Why does cyanylation at Cys-125 appear to depend on the binding of AMPPNP in the single-cysteine mutant but not in the L125C mutant with nine other cysteines?
Cyano Shuffling and the Development of a Modified Cysteine Footprinting Approach.
Polypeptide cleavage at an S-cyanocysteinyl residue at pH 9 is a slow reaction, typically carried out over 12 h or longer (16, 17). At the relatively high pH, an unreacted cysteinyl side chain can deprotonate and act as a potent nucleophile. It therefore appeared plausible that the attack of a cyanylated cysteine by a cysteinyl anion could transfer the cyano group from the modified cysteine to the unmodified one, either through a disulfide intermediate (17, 30–33) or by a direct transfer mechanism. Because under footprinting conditions the extent of reaction at a particular site is always kept at a rather low level, in a polypeptide with multiple cysteines there are many more unmodified cysteines than modified ones after footprinting with NTCB. Therefore, intramolecular or intermolecular transfer of cyano groups during chain cleavage would even out the distribution of a cyano group among different cysteines, and the pattern of chain cleavage at the cysteines would no longer reflect the relative reactivities of the cysteines during initial NTCB treatment. For a protein with a single cysteine, intermolecular transfer of a cyano group from a modified protein to an unmodified one would have no net effect on the extent of cleavage at the cysteine.
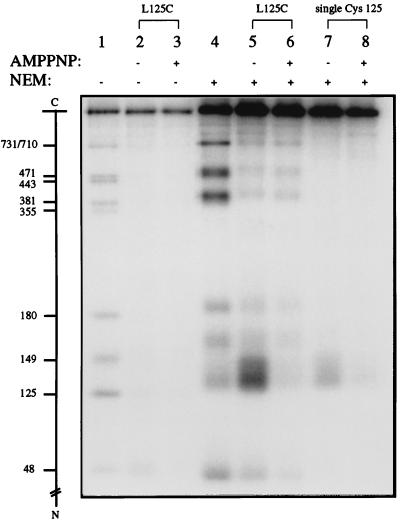
If the shuffling of cyano groups among the cysteines was responsible for the difference in the footprinting of Cys-125 in the L125C mutants with and without other cysteines, then blocking all uncyanylated cysteines by treatment with a second sulfhydryl reagent, such as NEM (see refs. 34 and 35), might alleviate this problem. Fig. 2 illustrates such an experiment. The lane 1 sample contained size markers generated by cleavage at the cysteines of the L125C mutant protein, and lanes 2 and 3 contained samples of the L125C protein footprinted in the absence and presence of AMPPNP. As mentioned in the section above, no difference in cleavage at Cys-125 or any of the other nine cysteines was noticeable between the pair of samples. However, when excess NEM was added to the NTCB-treated samples at the time of unfolding the proteins for peptide cleavage (lanes 5 and 6 of Fig. 2), Cys-125 was found to be more accessible than the other nine cysteines in the absence of AMPPNP, and the accessibility of Cys-125 to cyanylation by NTCB was much reduced in the presence of AMPPNP (compare the intensities of the Cys-125 band in lanes 5 and 6 of Fig. 2). This AMPPNP-mediated protection of Cys-125 against cyanylation also was evident from a similar experiment with the single Cys-125 mutant protein (Fig. 2, lanes 7 and 8). Extensive NEM treatment apparently reduces the electrophoretic mobilities of the polypeptides. When the same markers run in lane 1 were treated with excess NEM and analyzed in lane 4, a general reduction of the mobilities and broadening of the bands were apparent. Band broadening is particularly pronounced for the radiolabeled band corresponding to cleavage at Cys-125. Side reactions between NEM and other amino acid residues, especially lysines, are likely to contribute to these changes in electrophoretic properties.
Figure 2.
The use of NEM as a sulfhydryl blocking agent in protein footprinting by NTCB. The signs across the row denoted AMPPNP specify whether the ATP analog was absent (−) or present (+) during partial cysteine cyanylation by NTCB, and those across the row denoted NEM similarly specify whether excess NEM was added at the time of unfolding the cyanylated protein at pH 9. All proteins were 32P-labeled at their N-terminal HMK tag and, after electrophoresis, the gel was dried for autoradiography. Lane 1, size markers obtained by partial cleavage at the 10 cysteinyl residues of the L125C protein; lane 4, the size markers in lane 1 treated with NEM; lanes 2 and 3, NTCB footprinting of the L125C mutant protein without NEM treatment; lanes 5 and 6, NTCB footprinting of the L125C mutant protein with NEM treatment; lanes 7 and 8, NTCB footprinting of the single Cys-125 mutant with the incorporation of NEM treatment. The amount of NTCB used in the treatment of this single cysteine protein was one-half of that used for the proteins with multiple cysteines.
Direct Test of the Transfer of a Cyano Group from a Modified to an Unmodified Cysteine.
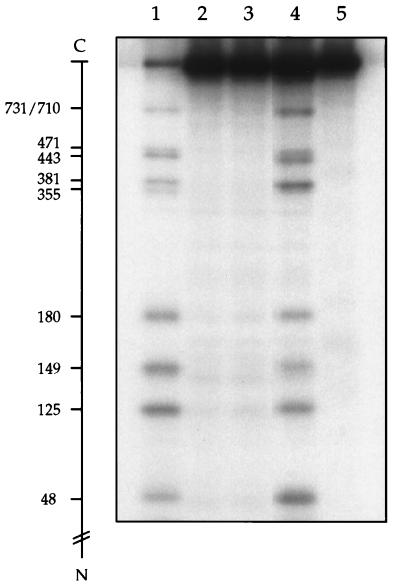
To test more directly whether shuffling of the cyano groups can occur under the peptide cleavage conditions, untagged wild-type yeast DNA topoisomerase II first was denatured and treated with excess NTCB to cyanylate the majority of the cysteinyl residues. The reaction was terminated by the addition of acetic acid, dialyzed thoroughly against 15% (vol/vol) acetic acid to remove unreacted NTCB, and dried under reduced pressure to remove acetic acid. An equal amount of HMK-tagged L125C protein was denatured in 8 M urea and 0.2 M N-[2-hydroxyethyl]piperazine-N′-[3-propanesulfonic acid], pH 9.0, and mixed with the dried cyanylated protein. The mixture was incubated for 15 h at 37°C to effect peptide cleavage at the S-cyanocysteines and then dialyzed and treated with protein kinase A in the presence of [γ-32P]ATP to radiolabel the HMK-tagged L125C mutant but not the untagged wild-type protein fragments. Fig. 3 depicts an autoradiogram of the radiolabeled bands after their resolution by gel electrophoresis. The sample described above was loaded in lane 4 of the gel. Clearly, cleavage of the tagged protein had occurred, and the extent of cleavage at each cysteine of the tagged protein was approximately the same. For the other lanes in Fig. 3, lane 1 contained the size markers, and lanes 2 and 3 contained two control samples. In the lane 3 sample, NTCB was omitted; in the lane 2 sample, NTCB was added after acidifying the denatured untagged protein. Neither of the controls showed significant peptide cleavage at the cysteines of the tagged protein. The absence of cleavage products of the tagged protein in lane 2 demonstrated that the cleavage of the tagged protein in lane 4 was not a result of trace NTCB left after dialysis. Thus, cleavage at the cysteines in the lane 4 sample was a consequence of incubating the uncyanylated tagged protein with the cyanylated untagged protein at alkaline pH, which, in turn, points to intermolecular cyano transfer from the untagged polypeptide to the tagged polypeptide during peptide cleavage at the S-cyanocysteines. For the sample run in lane 5 of Fig. 3, NEM was added to 10 mM at the time of mixing the denatured tagged protein and the cyanylated untagged protein; otherwise, the mixture was processed in the same way as the lane 4 sample. In contrast to the pattern seen in lane 4, little polypeptide cleavage of the tagged protein was seen in lane 5. The results of these experiments show that intermolecular transfer of a cyano group can occur and that such shuffling can be virtually eliminated by the addition of NEM as a blocking agent. The occurrence of intermolecular cyano transfer suggests that intramolecular cyano shuffling also can occur; at low protein concentrations, intramolecular transfer is probably the major route.
Figure 3.
Intermolecular transfer of cyano groups from a cyanylated protein to an uncyanylated protein. See text for detailed description of the experiment. Heavily cyanylated untagged wild-type yeast DNA topoisomerase II was freed of unreacted NTCB and mixed with uncyanylated, HMK-tagged L125C mutant protein. The mixed proteins were processed for peptide cleavage at the cyanylated cysteines without the presence of NEM. After 32P-labeling of the tagged protein fragments, the mixture was analyzed in lane 4. The radiolabeled bands in lane 4 indicated cleavage at the cysteinyl residues of the tagged protein, which was not treated with NTCB but was exposed to a cyanylated protein under peptide cleavage conditions. Lane 1 contained size markers obtained by cleavage at the cysteinyl residues of the L125C mutant protein. Lanes 2 and 3 contained, respectively, the control in which NTCB was added after acidification of the denatured, untagged protein and the control in which no NTCB was added. The sample analyzed in lane 5 was the same as that in lane 4, except that NEM was added to 10 mM at the time of mixing the tagged uncyanylated protein and the untagged cyanylated protein to block free sulfhydryls needed for cyano transfer.
ATP-Modulated Dimer Interface in Yeast DNA Topoisomerase II.
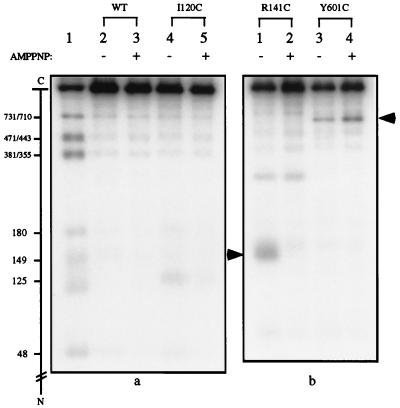
The above results indicated that cysteine footprinting by NTCB was feasible if a sulfhydryl blocking agent was added at the time of peptide cleavage at the S-cyanocysteines. When purified I120C and R141C mutant proteins were reexamined with the inclusion of NEM addition (Fig. 4), peptide cleavage at these positions was found to be much reduced when cyanylation was carried out in the presence of AMPPNP (Fig. 4a, compare lanes 4 and 5, and Fig. 4b, compare lanes 1 and 2). The cleavage pattern of R141C also showed an unexpected band corresponding to cleavage around residue 270 (Fig. 4b, lanes 1 and 2). Nucleotide sequencing of the R141C mutant showed that the codon for Glu-268 of the wild-type enzyme had been changed to one for a cysteine. The peptide cleavage patterns of R141C in the absence and presence of AMPPNP indicate that the binding of AMPPNP had no significant effect on cyanylation at Cys-268.
Figure 4.
Footprinting of three additional cysteine-substitution mutants of yeast DNA topoisomerase II. Peptide cleavage at S-cyanocysteines was carried out in the presence of excess NEM. The absence or presence of AMPPNP during cyanylation by NTCB is indicated by a minus or plus sign in the top margin. In the autoradiogram shown in a, lane 1 of the gel contained size markers obtained by cleavage of the HMK-tagged yeast DNA topoisomerase II and treatment of the cleavage products with NEM. The proteins analyzed in lanes 2–5 are specified in the upper margin. (b) The autoradiogram of a similar gel. The lane containing the size markers for the gel on the right was not shown, and the arrows in the left and right margins mark the positions of the radiolabeled bands corresponding to cleavage at Cys-141 in the R141C mutant and Cys-601 of the Y601C mutant, respectively.
An attempt also was made to examine the effect of AMPPNP binding on the formation of a separate dimer interface, the B′-B′ interface observed in the crystal structure of a 92-kDa fragment of yeast DNA topoisomerase II (36). This B′-B′ interface contains two symmetry-related hydrophobic patches, each includes Ile-539, Leu-550, and Leu-556 of one protomer and Tyr-601 of the other protomer (36). Three cysteine-substitution mutants, I539C, L556C, and Y601C, therefore were constructed to assess whether this interface existed in solution and how AMPPNP might affect it. The I539C and L556C mutants failed to complement the top2–4 mutation, however, and these proteins were not studied further. Examination of cyanylation and peptide cleavage of purified Y601C indicated that Cys-601 was readily accessible to cyanylation by NTCB, whether in the absence or presence of AMPPNP (Fig. 4b, lanes 3 and 4).
DISCUSSION
The results presented above indicate that NTCB can be used as a cysteine-specific protein footprinting reagent. For a protein that contains a single cysteine, cyanylation by NTCB occurs readily under physiological conditions if the cysteinyl residue is not buried, and efficient peptide cleavage at the cyanylated cysteine subsequently can be carried out by unfolding and incubating the protein at pH 9. For a protein with multiple cysteines, partial cyanylation of the native protein also occurs readily at the accessible cysteinyl residues, but the cyano groups apparently can redistribute among all cysteines upon unfolding of the protein at pH 9. Therefore, in the latter case meaningful footprints are obtainable only if the shuffling of the cyano groups is prevented by blocking all uncyanylated cysteines with a second thiol reagent during peptide cleavage at the S-cyanocysteines.
We have not examined the mechanism of transferring a cyano group from an S-cyanocysteine to a free sulfhydryl. It is well documented that a reversible reaction can occur between a cysteinyl sulfhydryl and an S-cyanocysteinyl group to form a disulfide link and a cyanide anion (30–33). Because the cyanide anion can attack either sulfur of the disulfide link in the reverse reaction, this reaction can lead to the transfer of a cyano group. Under conditions disfavoring the reverse reaction, the forward direction of this reaction also may lead to a net loss of cyano groups from the S-cyanocysteinyl groups and, hence, a reduction of the overall efficiency of protein cleavage for a given level of initial cyanylation. Inspection of several pairs of NEM-treated and untreated samples analyzed in Figs. 2 and 4 revealed that the extent of peptide cleavage generally was higher for the NEM-treated ones, suggesting that cyano transfer and cyano loss might occur in parallel. Direct intermolecular or intramolecular transfer of a cyano group from an S-cyanocysteinyl residue to a cysteinyl residue is also plausible, and additional measurements are needed to distinguish these possibilities.
As mentioned in the Introduction, the potential of cysteine footprinting with NTCB is much enhanced in combination with cysteine-scanning mutagenesis. In principle, a pool of cysteine-scanning mutants can be examined as a single sample, especially if the protein being examined contains no exposed cysteinyl groups to add strong cyanylation sites in all members of the pool. In practice, the resolution of the cleavage products needs to be improved if a pool containing many different cysteine-substitution mutants is to be used as a single sample. Extensive treatment with NEM, for example, is not ideal in view of its effects on the mobilities and bandwidths of the protein fragments during SDS/PAGE. Treatment with a lower concentration of NEM or for a shorter period to minimize its reaction with amino groups might also help to optimize the procedure.
Application of cysteine footprinting by NTCB to yeast DNA topoisomerase II supports the notion that the binding of ATP induces the formation of a dimer interface, which buries a number of residues including Ile-120, Leu-125, and Arg-141 (corresponding to Ile-94, His-99, and Leu-115, respectively, of E. coli GyrB protein). An alternative interpretation of the cysteine footprinting data, that a bound AMPPNP directly contacts these residues and thus affects their cyanylation, is less attractive in view of the crystal structure of the AMPPNP-bound bacterial gyrase ATPase domain. The retention of functionality of the yeast enzyme upon substitutions of Ile-120, Leu-125, or Arg-141 by cysteine also argues against a crucial role of any one of these residues in ATP binding.
Our results indicate that cyanylation of Tyr-601, which is located in a hydrophobic patch in the B′-B′ dimer interface in the crystal structure of a 92-kDa fragment of yeast DNA topoisomerase II, does not depend on the presence of AMPPNP. Thus, at least in the absence of DNA the B′-B′ contacts, if present, appear to be unaffected by AMPPNP binding. It is also significant that Cys-601 in the Y601C mutant enzyme seems fairly exposed. As shown in lanes 3 and 4 of Fig. 4b, cyanylation occurs more readily at Cys-601 than at any of the nine cysteinyl residues of wild-type yeast DNA topoisomerase II. It is plausible that contacts between the two B′ subfragments, as revealed in the crystal structure of the 92-kDa fragment (36), are absent in other conformations of the yeast enzyme. Extensive studies have suggested that there are a multitude of conformational states of the type IIA DNA topoisomerases during its catalytic cycle and that interconversion between these conformational states may involve large, interdomainal movements (reviewed in ref. 37; see also ref. 38).
Acknowledgments
We thank Wei Li for his initial efforts in testing the feasibility of using NTCB as a footprinting reagent and his assistance during the course of this study, Qiyong Liu for his help in enzyme preparation and in the assay of AMPPNP-mediated close-clamp formation of yeast DNA topoisomerase II, Daniel Gschwend and Qiyong Liu for their help with computer graphics, and Guido Guidotti and Roger Lundblad for helpful discussions. This work was supported by National Institutes of Health Grant GM24544 and summer fellowships (to B.T.) from Harvard University and the Burroughs Wellcome Fund.
ABBREVIATIONS
- NTCB
2-nitro-5-thiocyanobenzoic acid
- AMPPNP
5′-adenylyl-β,γ-imidodiphosphate
- NEM
N-ethylmaleimide
- GyrB
the B subunit of DNA gyrase
- HMK
heart muscle kinase phosphorylation site
References
- 1.Johnson A, Meyer B J, Ptashne M. Proc Natl Acad Sci USA. 1978;75:1783–1787. doi: 10.1073/pnas.75.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galas D J, Schmitz A. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siebenlist U, Simpson R B, Gilbert W. Cell. 1980;20:269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- 4.Tullius T D. Annu Rev Biophys Biomol Struct. 1989;18:213–237. doi: 10.1146/annurev.bb.18.060189.001241. [DOI] [PubMed] [Google Scholar]
- 5.Kennelly P J, Krebs E G. J Biol Chem. 1991;266:15555–15558. [PubMed] [Google Scholar]
- 6.Matsudaira P, Jakes R, Cameron L, Atherton E. Proc Natl Acad Sci USA. 1985;82:6788–6792. doi: 10.1073/pnas.82.20.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shuman J D, Vinson C R, McKnight S L. Science. 1990;249:771–774. doi: 10.1126/science.2202050. [DOI] [PubMed] [Google Scholar]
- 8.Manak J R, Prywes R. Mol Cell Biol. 1991;11:3652–3659. doi: 10.1128/mcb.11.7.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jay D G. J Biol Chem. 1984;259:15572–15578. [PubMed] [Google Scholar]
- 10.Heyduk E, Heyduk T. Biochemistry. 1994;33:9643–9650. doi: 10.1021/bi00198a033. [DOI] [PubMed] [Google Scholar]
- 11.Wu J, Perrin D M, Sigman D S, Kaback H R. Proc Natl Acad Sci USA. 1995;92:9186–9190. doi: 10.1073/pnas.92.20.9186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greiner D P, Hughes K A, Gunasekeron A H, Meares C F. Proc Natl Acad Sci USA. 1996;93:71–75. doi: 10.1073/pnas.93.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyduk T, Heduk E, Severinov K, Tang H, Ebright R H. Proc Natl Acad Sci USA. 1996;93:10162–10166. doi: 10.1073/pnas.93.19.10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanai R, Wang J C. Proc Natl Acad Sci USA. 1991;88:10485–10489. doi: 10.1073/pnas.88.23.10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Arruda M V, Bazar H, Wallek M, Matsudaira P. J Biol Chem. 1992;267:13079–13085. [PubMed] [Google Scholar]
- 16.Jacobson G R, Schaffer M H, Stark G R, Vanaman T C. J Biol Chem. 1973;248:6583–6591. [PubMed] [Google Scholar]
- 17.Degani Y, Patchornik A. Biochemistry. 1974;13:1–11. doi: 10.1021/bi00698a001. [DOI] [PubMed] [Google Scholar]
- 18.Kuramitsu S, Hamaguchi K, Tachibana H, Hori T, Ogawa T, Ogawa H. Biochemistry. 1984;23:2363–2367. doi: 10.1021/bi00306a006. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita H, Nakatsuka T, Hirose M. J Biol Chem. 1995;270:29806–29812. doi: 10.1074/jbc.270.50.29806. [DOI] [PubMed] [Google Scholar]
- 20.Liu Q, Wang J C. J Biol Chem. 1998;273:20252–20260. doi: 10.1074/jbc.273.32.20252. [DOI] [PubMed] [Google Scholar]
- 21.Holm C, Goto T, Wang J C, Botstein D. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 22.Li W, Wang J C. J Biol Chem. 1997;272:31190–31195. doi: 10.1074/jbc.272.49.31190. [DOI] [PubMed] [Google Scholar]
- 23.Roca J, Berger J M, Harrison S C, Wang J C. Proc Natl Acad Sci USA. 1996;93:4057–4062. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roca J, Wang J C. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 25.Wigley D B, Davies G J, Dodson E J, Maxwell A, Dodson G. Nature (London) 1991;351:624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]
- 26.Ali J A, Jackson A P, Howells A J, Maxwell A. Biochemistry. 1993;32:2717–2724. doi: 10.1021/bi00061a033. [DOI] [PubMed] [Google Scholar]
- 27.Ali J A, Orphanides G, Maxwell A. Biochemistry. 1995;34:9801–9808. doi: 10.1021/bi00030a018. [DOI] [PubMed] [Google Scholar]
- 28.Caron P, Wang J C. In: DNA Topoisomerases and Their Applications in Pharmacology. Liu L F, editor. San Diego: Academic; 1994. pp. 271–297. [DOI] [PubMed] [Google Scholar]
- 29.Lindsley J E. Proc Natl Acad Sci USA. 1996;93:2975–2980. doi: 10.1073/pnas.93.7.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aldridge W N. Biochem J. 1951;48:271–276. doi: 10.1042/bj0480271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker A J, Kharasch N. J Am Chem Soc. 1960;82:3071–3075. [Google Scholar]
- 32.Catsimpoolas N, Wood J L. J Biol Chem. 1964;239:4132–4137. [PubMed] [Google Scholar]
- 33.Catsimpoolas N, Wood J L. J Biol Chem. 1966;241:1790–1796. [PubMed] [Google Scholar]
- 34.Gregory J D. J Am Chem Soc. 1955;77:3922–3923. [Google Scholar]
- 35.Smyth D G, Blumenfeld O O, Konigsberg W. Biochem J. 1964;91:589–595. doi: 10.1042/bj0910589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger J M, Gamblin S J, Harrison S C, Wang J C. Nature (London) 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 37.Wang J C. Q Rev Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 38.Berger J M, Fass D, Wang J C, Harrison S C. Proc Natl Acad Sci USA. 1998;95:7876–7881. doi: 10.1073/pnas.95.14.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]