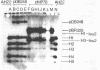
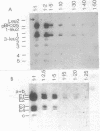
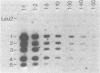
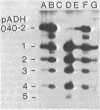
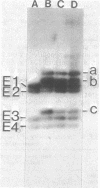
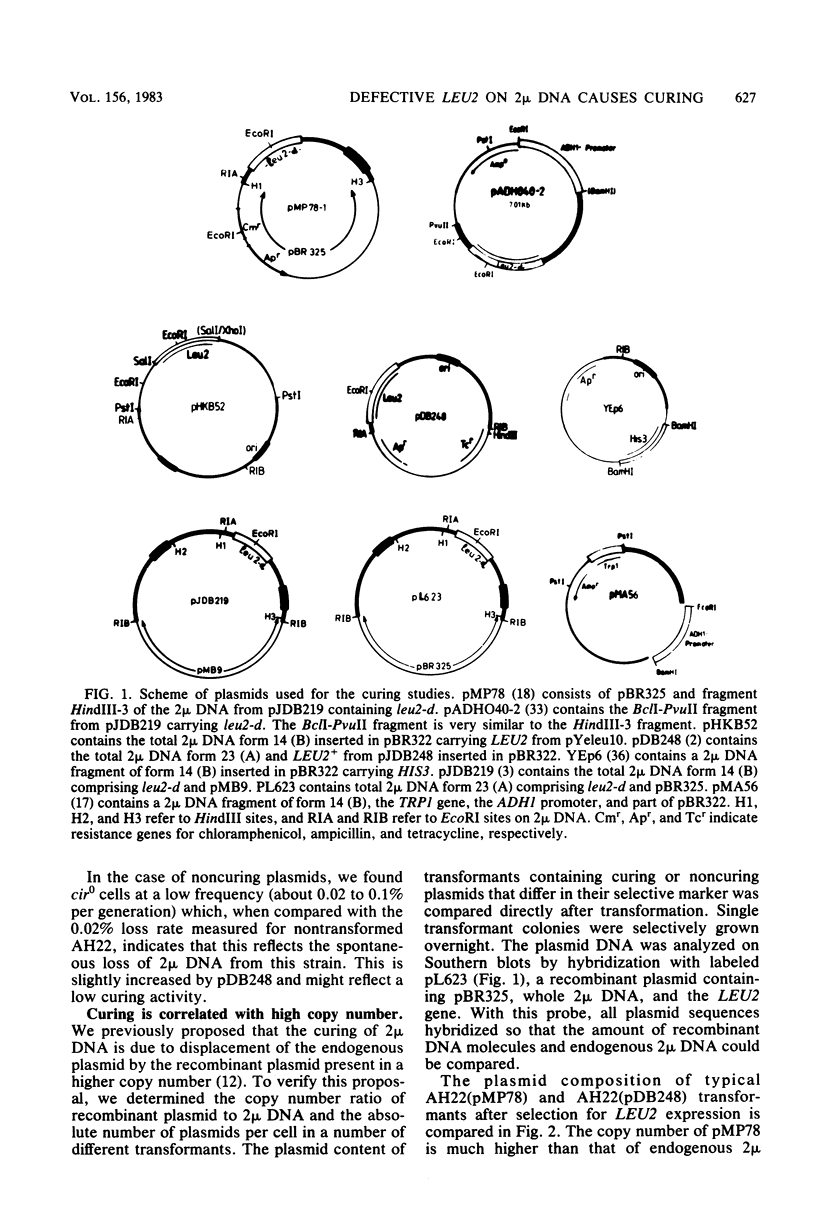
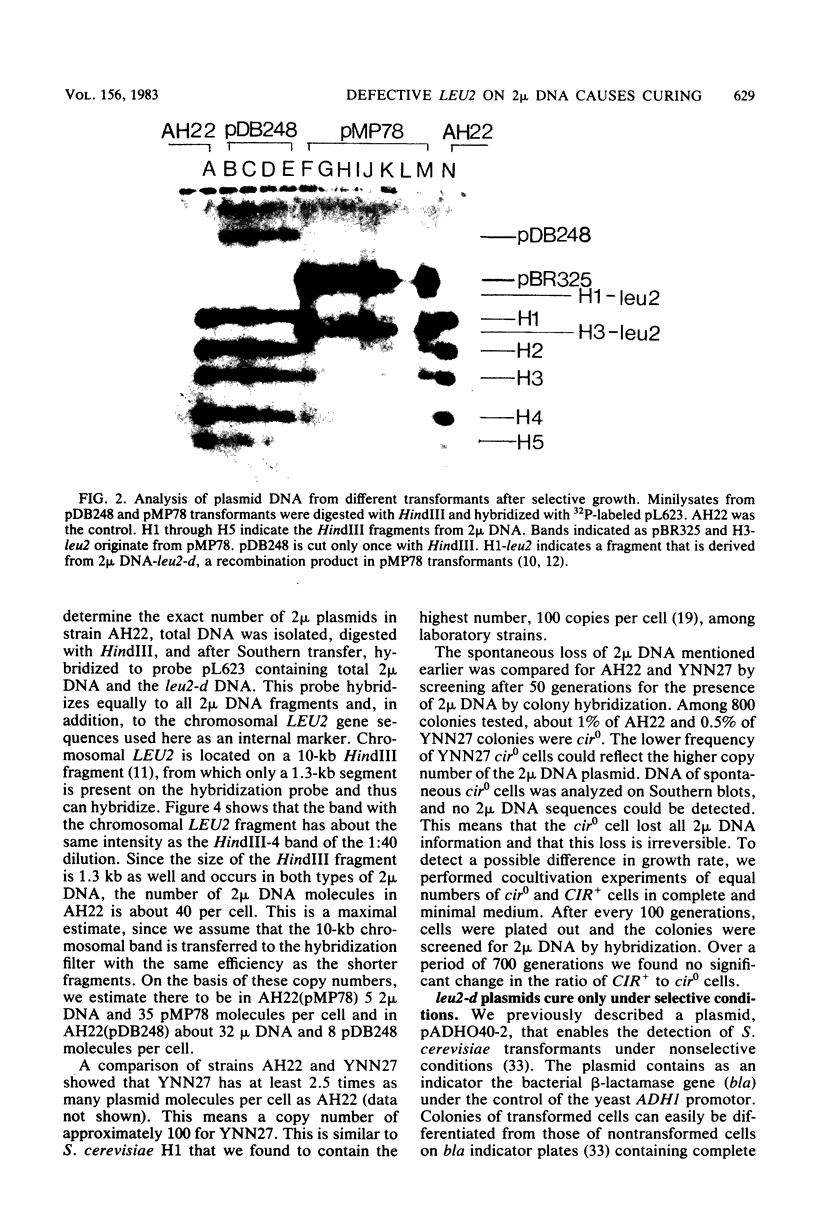
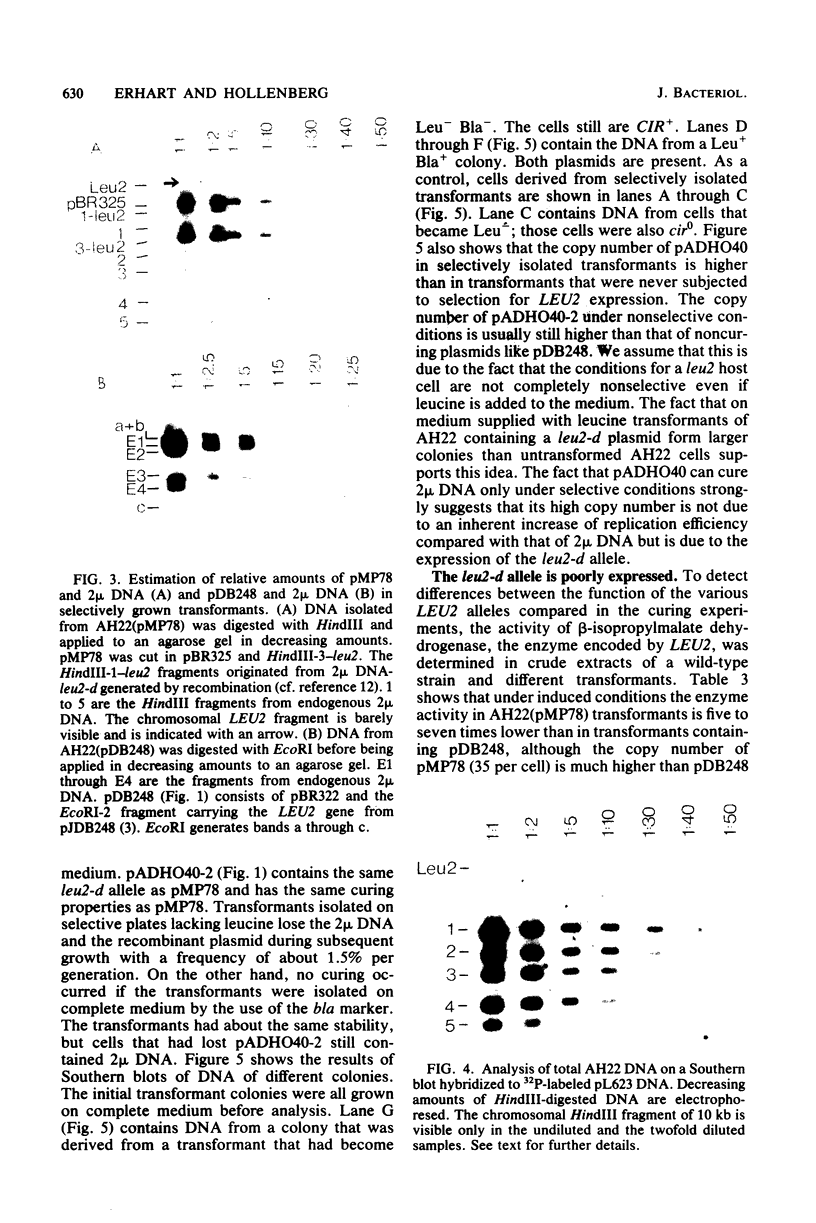
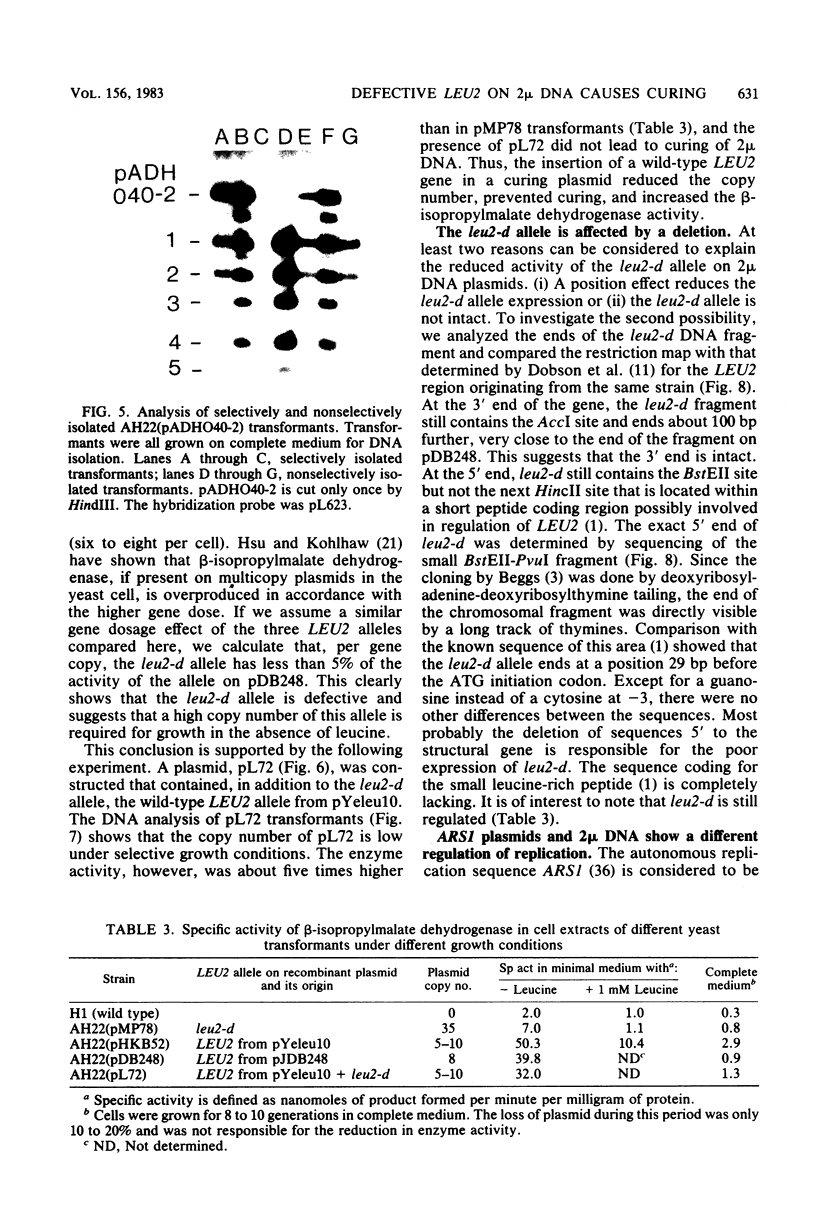
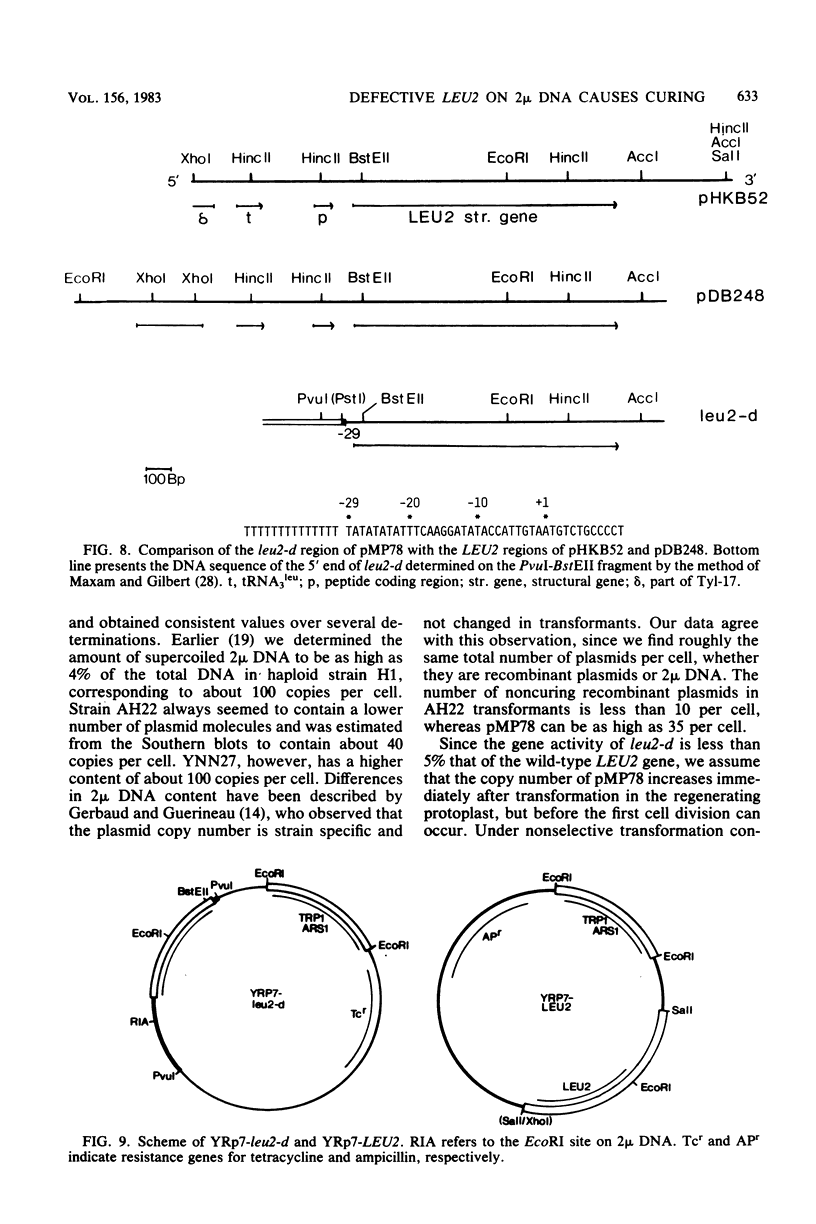
Abstract
The copy number of 2 mu DNA-derived plasmids in CIR+ Saccharomyces cerevisiae transformants is determined by its selective marker and is usually much lower than that of the endogenous plasmid. Only plasmids containing the leu2 allele of pJDB219, designated as leu2-d, under selective conditions displayed a higher copy number than did endogenous 2 mu DNA and by displacement generated cured cells. Spontaneous loss of 2 mu DNA occurred with a frequency of about 0.02% per generation. Curing plasmids, like pMP78, have copy numbers of 35; noncuring plasmids, like pDB248 or YEp6, have copy numbers of 4 to 8. The 2 mu DNA copy number in strains AH22 and YNN27 were determined to be 40 and 100, respectively. The high copy number of leu2-d-containing plasmids can be explained by its weak expression of less than 5% that of the wild-type LEU2 gene. The leu2-d allele has a deletion of the 5'-end sequence starting from 29 base pairs before the ATG initiation codon, but surprisingly, its expression is still regulated. On YRp7, which contains the chromosomal autonomic replication sequence ARS1, the defective leu2-d allele could not complement a leu2 host strain. This suggests a more stringent control of replication of ARS1-containing plasmids than of 2 mu-containing plasmids.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreadis A., Hsu Y. P., Kohlhaw G. B., Schimmel P. Nucleotide sequence of yeast LEU2 shows 5'-noncoding region has sequences cognate to leucine. Cell. 1982 Dec;31(2 Pt 1):319–325. doi: 10.1016/0092-8674(82)90125-8. [DOI] [PubMed] [Google Scholar]
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Breunig K. D., Mackedonski V., Hollenberg C. P. Transcription of the bacterial beta-lactamase gene in Saccharomyces cerevisiae. Gene. 1982 Nov;20(1):1–10. doi: 10.1016/0378-1119(82)90082-8. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Hicks J. B. Replication and recombination functions associated with the yeast plasmid, 2 mu circle. Cell. 1980 Sep;21(2):501–508. doi: 10.1016/0092-8674(80)90487-0. [DOI] [PubMed] [Google Scholar]
- CALVO J. M., KALYANPUR M. G., STEVENS C. M. 2-Isopropylmalate and 3-isopropylmalate as intermediates in leucine biosynthesis. Biochemistry. 1962 Nov;1:1157–1161. doi: 10.1021/bi00912a029. [DOI] [PubMed] [Google Scholar]
- Dobson M. J., Kingsman S. M., Kingsman A. J. Sequence variation in the LEU2 region of the saccharomyces cerevisiae genome. Gene. 1981 Dec;16(1-3):133–139. doi: 10.1016/0378-1119(81)90069-x. [DOI] [PubMed] [Google Scholar]
- Gerbaud C., Fournier P., Blanc H., Aigle M., Heslot H., Guerineau M. High frequency of yeast transformation by plasmids carrying part or entire 2-micron yeast plasmid. Gene. 1979 Mar;5(3):233–253. doi: 10.1016/0378-1119(79)90080-5. [DOI] [PubMed] [Google Scholar]
- Hicks J. B., Hinnen A., Fink G. R. Properties of yeast transformation. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1305–1313. doi: 10.1101/sqb.1979.043.01.149. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzeman R. A., Hagie F. E., Levine H. L., Goeddel D. V., Ammerer G., Hall B. D. Expression of a human gene for interferon in yeast. Nature. 1981 Oct 29;293(5835):717–722. doi: 10.1038/293717a0. [DOI] [PubMed] [Google Scholar]
- Hollenberg C. P., Borst P., van Bruggen E. F. Mitochondrial DNA. V. A 25 micron closed circular duplex DNA molecule in wild-type yeast mitochondria. Stucture and genetic complexity. Biochim Biophys Acta. 1970 May 21;209(1):1–15. [PubMed] [Google Scholar]
- Hollenberg C. P., Degelmann A., Kustermann-Kuhn B., Royer H. D. Characterization of 2-mum DNA of Saccharomyces cerevisiae by restriction fragment analysis and integration in an Escherichia coli plasmid. Proc Natl Acad Sci U S A. 1976 Jun;73(6):2072–2076. doi: 10.1073/pnas.73.6.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y. P., Kohlhaw G. B., Niederberger P. Evidence that alpha-isopropylmalate synthase of Saccharomyces cerevisiae is under the "general" control of amino acid biosynthesis. J Bacteriol. 1982 May;150(2):969–972. doi: 10.1128/jb.150.2.969-972.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y. P., Kohlhaw G. B. Overproduction and control of the LEU2 gene product, beta-isopropylmalate dehydrogenase, in transformed yeast strains. J Biol Chem. 1982 Jan 10;257(1):39–41. [PubMed] [Google Scholar]
- Kohlhaw G. B., Hsu Y. P., Lemmon R. D., Petes T. D. Transposed LEU2 gene of Saccharomyces cerevisiae is regulated normally. J Bacteriol. 1980 Nov;144(2):852–855. doi: 10.1128/jb.144.2.852-855.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Livingston D. M., Kupfer D. M. Control of Saccharomyces cerevisiae 2microN DNA replication by cell division cycle genes that control nuclear DNA replication. J Mol Biol. 1977 Oct 25;116(2):249–260. doi: 10.1016/0022-2836(77)90215-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Williamson D. H. Replicating circular DNA molecules in yeast. Cell. 1975 Mar;4(3):249–253. doi: 10.1016/0092-8674(75)90172-5. [DOI] [PubMed] [Google Scholar]
- Ratzkin B., Carbon J. Functional expression of cloned yeast DNA in Escherichia coli. Proc Natl Acad Sci U S A. 1977 Feb;74(2):487–491. doi: 10.1073/pnas.74.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb D. T., Thomas M., Kelly J., Selker E., Davis R. W. Eukaryotic DNA segments capable of autonomous replication in yeast. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4559–4563. doi: 10.1073/pnas.77.8.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storms R. K., McNeil J. B., Khandekar P. S., An G., Parker J., Friesen J. D. Chimeric plasmids for cloning of deoxyribonucleic acid sequences in Saccharomyces cerevisiae. J Bacteriol. 1979 Oct;140(1):73–82. doi: 10.1128/jb.140.1.73-82.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- Zakian V. A., Brewer B. J., Fangman W. L. Replication of each copy of the yeast 2 micron DNA plasmid occurs during the S phase. Cell. 1979 Aug;17(4):923–934. doi: 10.1016/0092-8674(79)90332-5. [DOI] [PubMed] [Google Scholar]