Abstract
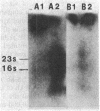
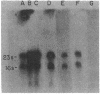
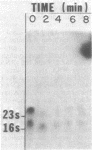
Total Escherichia coli RNA was separated by electrophoresis on methyl mercury agarose gels, transferred to diazobenzyloxymethyl-paper, and hybridized to various DNA probes containing different segments of the nrd genes to determine the organization of these genes. A 3.2-kilobase polycistronic mRNA transcript which hybridizes to both the nrdA and nrdB genes indicated that the nrdA and nrdB genes are organized in an operon. The polycistronic transcript contained the nrdA gene at the 5' end and the nrdB gene at the 3' end. The size of the polycistronic mRNA was sufficient to code for the 80,000-molecular-weight B1 protein and the 40,000-molecular-weight B2 protein. The results also indicated that the nrdA and nrdB genes are the only genes in E. coli that code for ribonucleoside diphosphate reductase. Two smaller RNA species that hybridized to nrd DNA were observed and probably overlap with the 3.2-kilobase nrd mRNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achord D., Kennell D. Metabolism of messenger RNA from the gal operon of Escherichia coli. J Mol Biol. 1974 Dec 15;90(3):581–599. doi: 10.1016/0022-2836(74)90236-8. [DOI] [PubMed] [Google Scholar]
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. C., Canellakis Z. N., Lundin B., Reichard P., Thelander L. Ribonucleoside diphosphate reductase. Purification of the two subunits, proteins B1 and B2. Eur J Biochem. 1969 Jul;9(4):561–573. doi: 10.1111/j.1432-1033.1969.tb00646.x. [DOI] [PubMed] [Google Scholar]
- Brown N. C., Reichard P. Role of effector binding in allosteric control of ribonucleoside diphosphate reductase. J Mol Biol. 1969 Nov 28;46(1):39–55. doi: 10.1016/0022-2836(69)90056-4. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Filpula D., Fuchs J. A. Regulation of ribonucleoside diphosphate reductase synthesis in Escherichia coli: increased enzyme synthesis as a result of inhibition of deoxyribonucleic acid synthesis. J Bacteriol. 1977 Apr;130(1):107–113. doi: 10.1128/jb.130.1.107-113.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filpula D., Fuchs J. A. Regulation of the synthesis of ribonucleoside diphosphate reductase in Escherichia coli: specific activity of the enzyme in relationship to perturbations of DNA replication. J Bacteriol. 1978 Aug;135(2):429–435. doi: 10.1128/jb.135.2.429-435.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J. A. Coordinate control of the synthesis of ribonucleoside diphosphate reductase components in Escherichia coli. J Bacteriol. 1977 May;130(2):957–959. doi: 10.1128/jb.130.2.957-959.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J. A., Karlström H. O. Mapping of nrdA and nrdB in Escherichia coli K-12. J Bacteriol. 1976 Dec;128(3):810–814. doi: 10.1128/jb.128.3.810-814.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke P. D., Fuchs J. A. Regulation of ribonucleoside diphosphate reductase mRNA synthesis in Escherichia coli. J Bacteriol. 1983 Jun;154(3):1040–1045. doi: 10.1128/jb.154.3.1040-1045.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidecker G., Messing J., Gronenborn B. A versatile primer for DNA sequencing in the M13mp2 cloning system. Gene. 1980 Jun;10(1):69–73. doi: 10.1016/0378-1119(80)90145-6. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Lim L. W., Kennell D. Evidence for random endonucleolytic cleavages between messages in decay of Escherichia coli trp mRNA. J Mol Biol. 1980 Aug 5;141(2):227–233. doi: 10.1016/0022-2836(80)90388-5. [DOI] [PubMed] [Google Scholar]
- Lim L. W., Kennell D. Models for decay of Escherichia coli lac messenger RNA and evidence for inactivating cleavages between its messages. J Mol Biol. 1979 Dec 5;135(2):369–390. doi: 10.1016/0022-2836(79)90442-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Platz A., Sjöberg B. M. Construction and characterization of hybrid plasmids containing the Escherichia coli nrd region. J Bacteriol. 1980 Aug;143(2):561–568. doi: 10.1128/jb.143.2.561-568.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Summers W. C. A simple method for extraction of RNA from E. coli utilizing diethyl pyrocarbonate. Anal Biochem. 1970 Feb;33(2):459–463. doi: 10.1016/0003-2697(70)90316-7. [DOI] [PubMed] [Google Scholar]
- Thelander L. Physicochemical characterization of ribonucleoside diphosphate reductase from Escherichia coli. J Biol Chem. 1973 Jul 10;248(13):4591–4601. [PubMed] [Google Scholar]
- Thelander L., Reichard P. Reduction of ribonucleotides. Annu Rev Biochem. 1979;48:133–158. doi: 10.1146/annurev.bi.48.070179.001025. [DOI] [PubMed] [Google Scholar]
- Yamada M., Takeda Y., Okamoto K., Hirota Y. Physical map of the nrdA-nrdB-ftsB-glpT region of the chromosomal DNA of Escherichia coli. Gene. 1982 Jun;18(3):309–318. doi: 10.1016/0378-1119(82)90169-x. [DOI] [PubMed] [Google Scholar]
- Yang R., Lis J., Wu R. Elution of DNA from agarose gels after electrophoresis. Methods Enzymol. 1979;68:176–182. doi: 10.1016/0076-6879(79)68012-6. [DOI] [PubMed] [Google Scholar]
- von Döbeln U., Reichard P. Binding of substrates to Escherichia coli ribonucleotide reductase. J Biol Chem. 1976 Jun 25;251(12):3616–3622. [PubMed] [Google Scholar]