How have the factors required for transcription initiation (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, and RNA polymerase II [pol II]) evolved to accommodate the elaborate transcriptional programs required for growth, differentiation, and development of multicellular organisms? Here we present analysis of the recently completed Drosophila melanogaster genome sequence, as well as those of Caenorhabditis elegans, Saccharomyces cerevisiae, and humans, that sheds light on this well studied question in eukaryotic biology. All four organisms encode single isoforms of RNA pol II, TFIIB, TFIIE, TFIIF, and TFIIH components, but multiple, sequence-related isoforms of TFIID components (Fig. 1; Albright and Tjian 2000). In addition, Drosophila and humans encode multiple isoforms of TFIIA components (Upadhyaya et al. 1999; Ozer et al. 2000). Current evidence indicates that tissue- and cell type–specific transcription is directed by differentially expressed TFIID and possibly TFIIA isoforms (Zeidler et al. 1996; Upadhyaya et al. 1999; Albright and Tjian 2000; Ozer et al. 2000). Thus, in accord with experimental data, this analysis points to TFIIA and TFIID as the factors that help generate the broad transcriptional repertoire of multicellular organisms. The identification of the complete set of TFIIA and TFIID components in a genetically and biochemically tractable organism like Drosophila is an important step toward understanding the mechanisms governing developmentally regulated transcription not only in Drosophila but also in humans.
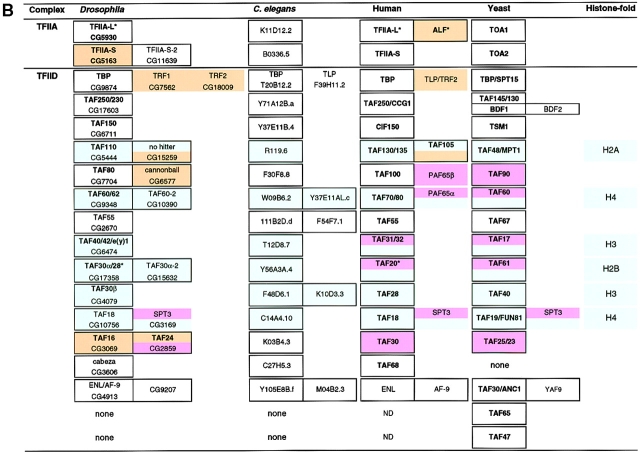
Figure 1.
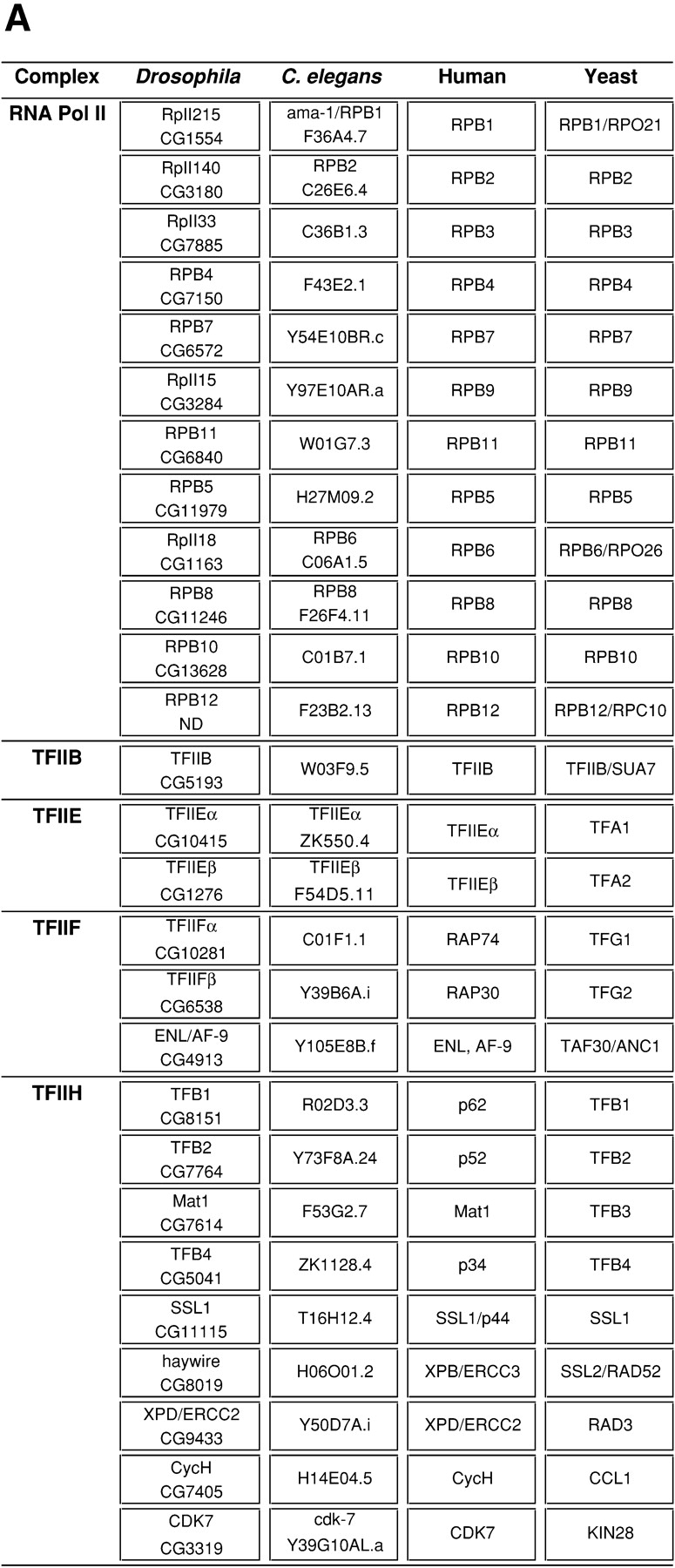
RNA pol II and GTF-encoding subunit genes from D. melanogaster, C. elegans, humans, and yeast are grouped by protein complex. Additional information about these genes can be accessed using the indicated gene name or identification number at web sites for the Drosophila (http://www.fruitfly.org/annot/), C. elegans (http://www.wormbase.org/), yeast (http://www.proteome.com/databases/index.html), or human genomes (http://www.ncbi.nlm.nih.gov/). No Gadfly identification has been assigned for Rpb12 because it was not identified by gene prediction programs and no expressed sequence tags have been isolated. However, searches using the human and yeast Rpb12 homologues match translated genomic sequence from EMBL/GenBank/DDBJ accession No. AC017903, and further analysis of sequences surrounding this match reveal additional amino acid similarity that spans 3 exons (our unpublished observation). Each row contains homologous genes from each of the four organisms. An asterisk indicates that the gene is alternatively spliced. ND indicates information that has not been determined. The following identification numbers correlate to predicted mRNAs that in addition to encoding the indicated protein may also encode another protein, presumably due to a gene prediction error: Drosophila CG7150, CG6572, and C. elegans C36B1.3, F43E2.1, Y97E10AR.a, F39H11.2, and Y37E11AL.c. (A) Genes encoding components of RNA pol II, TFIIB, TFIIE, TFIIF, and TFIIH. (B) Genes encoding components of TFIIA and TFIID. Genes in bold typeface have been demonstrated experimentally to be a component of the indicated complex. Genes in normal typeface that are bordered in a black box may be a component of the indicated complex and await experimental evidence. Genes that are not bordered in a black box are components of other complexes. Genes that are expressed in a tissue- or cell type–specific manner are shaded orange, genes that encode components of HAT complexes are shaded pink, and genes that encode sequence similarity to the histone fold motif are shaded blue. A search program that identifies protein motifs (http://www.isrec.isb-sib.ch/software/PFSCAN_form.html) and visual comparison of sequences was used to find the histone fold motifs in TAFIIs and the transcription factors indicated in the text. Yeast BDF1, TAFII48, and TAFII65 are recently described components of TFIID (Sanders and Weil 2000; Matangkasombut et al. 2000; Reese et al. 2000). Yeast TAFII145 and BDF1 display functional and sequence similarity to the NH2 and COOH termini, respectively, of human TAFII250 and therefore are placed in the same box (Matangkasombut et al. 2000). Bdf1 and Bdf2 display sequence similarity but only Bdf1 has been demonstrated to associate with TFIID. TAFII30/ANC1 is also a component of NuA3, TFIIF, and SWI/SNF complexes (John et al. 2000).
The Biology of Transcription Initiation
Biochemical fractionation of Drosophila embryos, human cells, and yeast cells has defined a set of multiprotein complexes termed general transcription factors (GTFs; TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) required for mRNA transcription initiation in vitro (Orphanides et al. 1996; Hampsey 1998). Transcription is initiated by recognition of core promoter elements by TFIID and sequential or concerted assembly of the other GTFs and RNA pol II to form the preinitiation complex (PIC). Although GTFs play essential roles during transcription initiation, it is the factors that regulate the ability of the GTFs to assemble and stably bind a core promoter that are probably major determinants of gene-specific transcription levels. For example, activators and coactivators are thought to stimulate transcription by recruiting GTFs to a promoter, thereby accelerating PIC assembly.
The GTF TFIID is composed of TATA-binding protein (TBP) and coactivator subunits termed TBP-associated factors (TAFIIs; Burley and Roeder 1996; Green 2000; Albright and Tjian 2000). TAFIIs not only function as “conventional” coactivators by serving as physical links between DNA-binding activator proteins and the PIC but also possess enzymatic or promoter recognition activities that presumably enhance the efficiency of PIC assembly. TFIIA has also been described as a coactivator and displays a number of TAFII-like properties: it binds to TBP and TAFIIs; it interacts with specific transcriptional activators; it is generally required for activated transcription in vitro; and it contributes to promoter selectivity (Orphanides et al. 1996; Hampsey 1998).
TAFII, TBP, and TFIIA Components Mediate Gene-specific Transcription
Inactivation of individual TAFIIs in Drosophila, mammalian, and yeast cells has demonstrated that TAFIIs are not required for the transcription of all RNA pol II genes, and in fact there is great variation in regard to the identity and number of gene targets for individual TAFIIs (Green 2000; Albright and Tjian 2000). Furthermore, different domains within a single TAFII can play gene-specific roles in transcription (O'Brien and Tjian 2000). The isolation of a human B cell–specific isoform of TAFII130 (TAFII105) raised the possibility that substoichiometric subunits of TFIID mediate tissue- or cell type–specific transcription and that additional components of TFIID may have escaped detection because of their low abundance (Dikstein et al. 1996, Yamit-Hezi et al. 2000). These possibilities have been born out in Drosophila where isoforms of TAFII110 and TAFII80 (No hitter [Nht] and Cannonball [Can], respectively) are expressed exclusively in testis and regulate transcription of a subset of genes required for spermatogenesis, and isoforms of TBP (TBP-related factors [TRF1 and TRF2]) are expressed in a tissue-specific manner and bind different genes in salivary gland cells (Hansen et al. 1997; Rabenstein et al. 1999; Hiller, M., T.-Y. Lin, and M. Fuller, personal communication). Similarly, analysis of the human TFIIA-L isoform ALF (TFIIAα/β-like factor) reveals that its expression is restricted to the testis; however, it remains to be determined if it is used for the transcription of testis-specific genes (Upadhyaya et al. 1999; Ozer et al. 2000). In Drosophila, TFIIA-S is expressed in a dynamic pattern during eye development and is transiently upregulated in photoreceptor precursor cells before their fate is determined (Zeidler et al. 1996). Therefore, the role of TFIIA and TFIID in transcription initiation is governed by the expression patterns and activities of their varied components.
Finally, it is critical to note that analysis of the function of TAFIIs is complicated by the fact that they are components of at least two other complexes that lack TBP, p300/CBP-associated factor (PCAF) and TBP-free TAFII-containing complex (TFTC) (Struhl and Moqtaderi 1998; Bell and Tora 1999). The human PCAF histone acetyltransferase (HAT) complex contains three TAFIIs that are shared with TFIID (TAFII31/32, TAFII20/15, and TAFII30) and three TAFII isoforms (PCAF-associated factor 65β [PAF65β], PAF65α, and SPT3) related to TAFII100, TAFII70/80, and TAFII18, respectively (Birck et al. 1998; Ogryzko et al. 1998). Yeast possess an analogous complex, Spr-Ada-Gcn5-acetyltransferase (SAGA), containing TFIID TAFIIs and the Gcn5 HAT, and Drosophila may also, as it contains a Gcn5/PCAF homologue that interacts with TAFII24 (Smith et al. 1998; Brown et al. 2000; Georgieva et al. 2000).
The Genomics of Transcription Initiation
Searches of the completed Drosophila, C. elegans, and yeast genomes and the partial human genome for sequence homologues of biochemically identified components of the general transcription machinery have led to the following conclusions. First, all of the components of RNA pol II, TFIIB, TFIIE, TFIIF, and TFIIH are encoded by single copy genes in Drosophila, C. elegans, and yeast (Fig. 1 A). Second, multiple isoforms of TFIID components are encoded in Drosophila, C. elegans, humans, and yeast, and multiple isoforms of TFIIA components are encoded in Drosophila and humans (Fig. 1 B). Third, each organism encodes isoforms of different sets of TFIIA and TFIID components, some which are unique to a particular organism.
Sequence comparisons uncovered Drosophila homologues of TAFIIs previously identified in yeast or humans by biochemical means but which had not been described in Drosophila (yeast TAFII67/human TAFII55, yeast TAFII30/ human ENL/AF-9, and yeast TAFII19/human TAFII18; Green 2000). Thus, all TAFIIs present in both yeast and humans are present in Drosophila, as well as C. elegans. In contrast, yeast TAFII47 and TAFII65 are absent from Drosophila, C. elegans, and apparently from humans, suggesting that these TAFIIs perform a yeast-specific role, such as serving as coactivators for DNA-binding activators that are not present in metazoans. Finally, there are TAFIIs present in Drosophila, C. elegans, and humans that are absent from yeast (human TAFII68/Drosophila Cabeza and multiple TAFII isoforms). In addition to Can and Nht, there are alternatively spliced forms of TAFII30α, two genes (TAFII24 and TAFII16) that encode Drosophila homologues of human TAFII30, and TAFII60 and TAF30α isoforms (TAFII60-2 and TAF30α-2, respectively) (Kokubo et al. 1994; Georgieva et al. 2000). TFIIA-S and TFIIA-L are the only other GTF components in Drosophila and humans, respectively, that are expressed in multiple isoforms (Upadhyaya et al. 1999; Ozer et al. 2000). The fact that these proteins are unique to multicellular organisms suggests that they play cell-specific roles.
A number of TAFIIs contain a common structural motif called the histone fold that was originally shown to drive folding and association of each of the core histones (H2A, H2B, H3, and H4) and subsequently shown to play a similar role in association of TAFIIs (Xie et al. 1996; Wolffe 1998). TAFII pairs, such as Drosophila TAFII40 and TAFII60, form heterotetramers, analogous to H3 and H4, and numerous other TAFII–TAFII and TAFII–nonTAFII interactions have been shown to involve histone fold motifs (Gangloff et al. 2000). The demonstrated histone fold interaction of human TAFII135 and TAFII20, predicts that Drosophila isoforms of these proteins, Nht and TAFII30α-2, respectively, may heterodimerize and hints at the existence of a human TAFII20 isoform that would heterodimerize with the TAFII135 isoform, TAFII105. B cell–specific expression of the hypothetical TAFII20 isoform may explain why TAFII105 associates with TFIID in B cells but not in other cell types (Dikstein et al. 1996).
In addition to the TAFIIs indicated in Fig. 1 B, other Drosophila transcription factors contain histone fold motifs: Prodos (Drosophila genome project Gadfly accession number CG7128), NF-YB-like (CG10477), NF-YC-like (CG3075, CG11301), CHRAC-14 (CG13399), CHRAC-16 (CG15736), Dr1 (CG4185), NC2α (CG10318), and BIP2 (CG2009). It is interesting to speculate that these factors may be unidentified TAFII components of TFIID or binding partners for known TAFIIs in complexes that lack TBP.
Putting It All Together
Analysis of eukaryotic genomes has defined sets of proteins that are similar in sequence to known components of TFIIA and TFIID. Since known components of TFIIA and TFIID have been shown to play key roles in developmentally regulated transcription, it is exciting to speculate that the newly identified genes will play similar roles and that TFIIA and TFIID components have evolved to support tissue- or cell type–specific transcriptional requirements of individual eukaryotic organisms.
The challenge now is to determine if TAFIIs that have been identified on the basis of their sequence are components of TBP-containing complexes or other TAFII-containing complexes, whether TAFIIs and TFIIA isoforms are differentially expressed during development, and how differentially expressed TBP, TAFII, and TFIIA isoforms function in concert with the ubiquitously expressed form of TFIID and TFIIA to regulate gene expression. The subunit composition of human PCAF complex leads to the prediction that Drosophila TAFII60-2 and Can and C. elegans Y37E11AL.c are components of PCAF/SAGA and not TFIID. On the other hand, protein isoforms that are unique to a particular organism, such as Drosophila TAFII30α-2 and C. elegans F54F7.1 and K10D3.3, may be tissue- or cell type–specific components of TFIID and not PCAF/SAGA.
Drosophila may be the most appropriate organism for these studies since the biochemical activities of these factors can be determined using established TFIIA and TFIID purification schemes and in vitro transcription systems, and developmental requirements for these factors can be determined using existing mutants or mutants generated by traditional mutagenesis schemes, P-element insertion, RNA interference (RNAi), or homologous recombination (Kennerdell and Carthew 1998; Rörth et al. 1998; Spradling et al. 1999; Rong and Golic 2000).
In terms of the RNA pol II transcriptional machinery, this review has covered only the tip of the iceberg. Detailed analysis of Drosophila genes encoding DNA-binding transcription factors, coactivators, corepressors, chromatin remodeling factors, and other trans-acting regulators of transcription remains to be tackled. However, completion of the Drosophila genome sequence has set the stage for biochemical, molecular, and genetic studies in Drosophila that should lead to advances in our understanding of developmentally regulated RNA pol II transcription.
In addition to being able to identify new components of the transcription machinery, the Drosophila genome project has provided several valuable tools for studying RNA pol II transcription. First, it has led to the identification of fly stocks containing P-element insertions that disrupt GTF genes, providing the opportunity to investigate developmental and possibly mechanistic roles for the encoded factors (Rörth et al. 1998; Spradling et al. 1999). Second, sequencing of full-length expressed sequence tags (i.e., cDNAs) has helped define RNA pol II transcription start sites that may lead to the identification of novel core promoter elements or provide insight into how different combinations of core promoter elements contribute to transcription initiation. The recent description of a TC-rich sequence (TC-box) that is specifically bound by Drosophila TRF1 and the identification of isoforms (i.e., TAFII60-2) of known TAFIIs (i.e., TAFII60) that recognize core promoter elements hints at the existence of additional core promoter elements (Burke et al. 1998; Holmes and Tjian 2000). Finally, the description of the ∼13,600 Drosophila genes allows for construction of DNA microarrays (i.e., gene chips) that can be used to identify gene targets for individual components of the transcription machinery (Adams et al. 2000).
Acknowledgments
We thank M. Hiller, T.-Y. Lin, and M. Fuller for providing unpublished data on Can and Nht; Celera Genomics and the Berkeley Drosophila Genome Project for allowing us to search the Drosophila genome sequence before publication; Y. Lei for assistance performing sequence searches; and R. Kamakaka, L. Pile, P. Wade, and K. Wassarman for providing comments on the manuscript.
This work was supported by the Intramural Program in the National Institute of Child Health and Human Development.
Footnotes
Abbreviations used in this paper: GTF, general transcription factor; PIC, preinitiation complex; TAFII, TBP-associated factor; TBP, TATA-binding protein.
References
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F. The genome sequence of Drosophila melanogaster . Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Albright S.R., Tjian R. TAFs revisitedmore data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- Bell D., Tora L. Regulation of gene expression by multiple forms of TFIID and other TAFII-containing complexes. Exp. Cell Res. 1999;246:11–19. doi: 10.1006/excr.1998.4294. [DOI] [PubMed] [Google Scholar]
- Birck C., Poch O., Romier C., Ruff M., Mengus G., Lavigne A.-C., Davidson I., Moras D. Human TAFII28 and TAFII18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell. 1998;94:239–249. doi: 10.1016/s0092-8674(00)81423-3. [DOI] [PubMed] [Google Scholar]
- Brown C.E., Lechner T., Howe L., Workman J.L. The many hats of transcriptional coactivators. Trends Biochem. Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- Burke T.W., Willy P.J., Kutach A.K., Butler J.E.F., Kadonaga J.T. The DPE, a conserved downstream core promoter element that is functionally analogous to the TATA box. Cold Spring Harbor Symp. Quant. Biol. 1998;63:75–82. doi: 10.1101/sqb.1998.63.75. [DOI] [PubMed] [Google Scholar]
- Burley S.K., Roeder R.G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu. Rev. Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- Dikstein R., Zhou S., Tjian R. Human TAFII105 is a cell type-specific TFIID subunit related to hTAFII130. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. [DOI] [PubMed] [Google Scholar]
- Gangloff Y.-G., Werten S., Romier G., Carre L., Poch O., Moras D., Davidson I. The human TFIID components TAFII135 and TAFII20 and yeast SAGA components ADA1 and TAFII68 heterodimerize to form histone-like pairs. Mol. Cell Biol. 2000;20:340–351. doi: 10.1128/mcb.20.1.340-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva S., Kirschner D.B., Jagla T., Nabirochkina E., Hanke S., Schenkel H., de Lorenzo C., Sinha P., Jagla K., Mechler B., Tora L. Two novel Drosophila TAFIIs have homology with human TAFII30 and are differentially regulated during development. Mol. Cell Biol. 2000;20:1639–1648. doi: 10.1128/mcb.20.5.1639-1648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M.R. TBP-associated factors (TAFIIs)multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 2000;25:59–63. doi: 10.1016/s0968-0004(99)01527-3. [DOI] [PubMed] [Google Scholar]
- Hampsey M. Molecular genetics of the RNA polymerase II general transcription machinery. Microbiol. Mol. Biol. Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S.K., Takada S., Jacobson R.H., Lis J.T., Tjian R. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell. 1997;91:71–83. doi: 10.1016/s0092-8674(01)80010-6. [DOI] [PubMed] [Google Scholar]
- Holmes M.C., Tjian R. Promoter-selective properties of the TBP-related factor TRF1. Science. 2000;288:867–870. doi: 10.1126/science.288.5467.867. [DOI] [PubMed] [Google Scholar]
- John S., Howe L., Tafrow S.T., Grant P.A., Sternglanz R., Workman J.L. The something about silencing protein, sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 2000;14:1196–1208. [PMC free article] [PubMed] [Google Scholar]
- Kennerdell J.R., Carthew R.W. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Kokubo T., Gong D.-W., Wootton J.C., Horikoshi M., Roeder R.G., Nakatani Y. Molecular cloning of Drosophila TFIID subunits. Nature. 1994;367:484–487. doi: 10.1038/367484a0. [DOI] [PubMed] [Google Scholar]
- Matangkasombut O., Buratowski R.M., Swilling N.W., Buratowski S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- O'Brien T., Tjian R. Different functional domains of TAFII250 modulate expression of distinct subsets of mammalian genes. Proc. Natl. Acad. Sci. USA. 2000;97:2456–2461. doi: 10.1073/pnas.97.6.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko V.V., Kotani T., Zhang X., Schiltz R.L., Howard T., Yang X.-J., Howard B.H., Qin J., Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Lagrange T., Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Ozer J., Moore P.A., Lieberman P.M. A testis-specific transcription factor IIA (TFIIAτ) stimulates TATA-binding protein-DNA binding and transcription activation. J. Biol. Chem. 2000;275:122–128. doi: 10.1074/jbc.275.1.122. [DOI] [PubMed] [Google Scholar]
- Rabenstein M.D., Zhou S., Lis J.T., Tjian R. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc. Natl. Acad. Sci. USA. 1999;96:4791–4796. doi: 10.1073/pnas.96.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese J.C., Zhang Z., Kurpad H. Identification of a yeast transcription factor IID subunit, TSG2/TAF48. J. Biol. Chem. 2000;275:17391–17398. doi: 10.1074/jbc.M001635200. [DOI] [PubMed] [Google Scholar]
- Rong Y.S., Golic K.G. Gene targeting by homologous recombination in Drosophila . Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- Rörth P., Szabo K., Bailey A., Laverty T., Rehm J., Rubin G.M., Weigmann K., Milan M., Benes V., Ansorge W., Cohen S.M. Systematic gain-of-function genetics in Drosophila . Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- Sanders S.L., Weil P.A. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem. 2000;275:13895–13900. doi: 10.1074/jbc.275.18.13895. [DOI] [PubMed] [Google Scholar]
- Smith E.R., Belote J.M., Schiltz R.L., Yang X.J., Moore P.A., Berger S.L., Nakatani Y., Allis C.D. Cloning of Drosophila GCN5conserved features among metazoan GCN5 family members. Nucleic Acids Res. 1998;26:2948–2954. doi: 10.1093/nar/26.12.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A.C., Stern D., Beaton A., Rhem E.J., Laverty T., Mozden N., Misra S., Rubin G.M. The Berkeley Drosophila Genome Project gene disruption projectsingle P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Moqtaderi Z. The TAFs in the HAT. Cell. 1998;94:1–4. doi: 10.1016/s0092-8674(00)81213-1. [DOI] [PubMed] [Google Scholar]
- Upadhyaya A.B., Lee S.H., Dejong J. Identification of a general transcription factor TFIIAα/β homologue selectively expressed in testis. J. Biol. Chem. 1999;274:18040–18048. doi: 10.1074/jbc.274.25.18040. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. Chromatin structure & function 1998. Academic Press; New York: pp. 7–157 [Google Scholar]
- Xie X., Kokubo T., Cohen S., Mirza U., Hoffman A., Chait B., Roeder R., Nakatani Y., Burley S. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- Yamit-Hezi A., Nir S., Wolstein O., Dikstein R. Interaction of TAFII105 with selected p65/RelA dimers is associated with activation of subset of NF-kappa B genes. J. Biol. Chem. 2000;275:18180–18187. doi: 10.1074/jbc.275.24.18180. [DOI] [PubMed] [Google Scholar]
- Zeidler M.P., Yokomori K., Tjian R., Mlodzik M. Drosophila TFIIA-S is up-regulated and required during Ras-mediated photoreceptor determination. Genes Dev. 1996;10:50–59. doi: 10.1101/gad.10.1.50. [DOI] [PubMed] [Google Scholar]