Abstract
Germ-line mutations in the BRCA1 tumor-suppressor gene are associated with an increased susceptibility to breast and ovarian cancer. BRCA1 contains a carboxyl-terminal domain (BRCT) that is shared with several other proteins involved in maintaining genome integrity. In an effort to understand the function of BRCA1, we sought to isolate proteins that interact with the BRCT domain. Purified BRCT polypeptide was used as a probe to screen a human placenta cDNA expression library by Far Western analysis. Here we report that BRCA1 interacts in vivo and in vitro with the Rb-binding proteins, RbAp46 and RbAp48, as well as with Rb. Moreover, the BRCT domain associates with the histone deacetylases HDAC1 and HDAC2. These results demonstrate that BRCA1 interacts with components of the histone deacetylase complex, and therefore may explain the involvement of BRCA1 in multiple processes such as transcription, DNA repair, and recombination.
More than half of families with inherited breast and ovarian cancer susceptibility are thought to harbor germ-line mutations in the BRCA1 gene. Frequent loss of the wild-type allele in tumors of mutation carriers suggests that BRCA1 acts as a tumor-suppressor gene. Surprisingly, mutations in BRCA1 in sporadic breast and ovarian cancer are extremely rare (1–3). To date, more than 600 different mutations in the BRCA1 gene have been reported (Breast Cancer Information Core: www.nhgri.nih.gov/Intramural_research/Lab_transfer/Bic/). The majority of these are truncation mutations distributed over the entire length of the gene. Several missense mutations have also been shown to segregate with cancer susceptibility (1, 4, 5).
The BRCA1 gene was isolated and mapped to human chromosome 17q21 (6). The gene encodes an 1,863-aa protein with an apparent molecular mass of 220 kDa. Only a few conserved sequence motifs have been identified in the BRCA1 protein: an amino-terminal RING finger, a carboxyl-terminal region that contains two repeats of a newly identified motif, designated BRCT (BRCA1 carboxyl terminus) domain (7), and three nuclear localization signals in the central portion of the molecule (8). However, much of the biochemical function of BRCA1 is unknown.
BRCA1 is found in nuclear foci that form in a cell cycle-dependent manner (9, 10). Several lines of evidence suggest that BRCA1 expression is cell cycle regulated and plays a role in cell cycle checkpoints. BRCA1 mRNA is highly expressed during embryonic development and is increased in breast epithelia during pregnancy and in adult testis during the final stages of meiosis and spermatogenesis (11, 12), suggesting a role in terminal differentiation. Brca1−/− mouse embryos die early in development from cell proliferation defects, including cell cycle arrest (13, 14). In human cell lines, BRCA1 expression suppresses cell growth (15–17). The presence of the BRCT motif in BRCA1 also links it to cell cycle control. Many other cell cycle checkpoint proteins, such as the p53-binding protein (53BP1), fission yeast replication checkpoint proteins, (RAD9 and RAD4), the DNA repair proteins XRCC1 and XRCC4, and all members of the retinoblastoma protein family (Rbp107 and Rbp130) contain a BRCT motif (7, 18).
A number of observations also link BRCA1 to transcription regulation. The carboxyl-terminal region has an intrinsic trans-activation activity in cells when fused to the GAL4 DNA-binding domain (19, 20). BRCA1 activates transcription of the cell cycle regulators p21WAF/CIP and MDM2 when cotransfected with p53 (21, 22), supporting a role in the control of the cell cycle and proliferation. Other evidence suggests that BRCA1 plays a role in DNA repair. BRCA1 colocalizes with the double-strand break-repair, homologous recombination protein, RAD51, the human homolog of Escherichia coli RecA (9, 10, 23, 24). After exposure to ionizing radiation and other DNA-damaging agents, BRCA1 becomes hyperphosphorylated, disperses from nuclear foci, and accumulates in proliferating cell nuclear antigen-containing structures (10). Recently, it was reported that embryonic stem cells lacking BRCA1 are hypersensitive to ionizing radiation and are unable to mediate transcription-coupled repair after DNA damage (25).
Several proteins are reported to bind and interact directly with BRCA1. Among them are components of the nuclear import pathway (8) that bind to the nuclear localization signals; a component of the ubiquitin pathway (26); and a novel RING finger/BRCT domain-containing protein, BARD1 (27), binding to the RING finger motif. Recently, p53, RNA helicase A, and CtIP were reported to bind BRCA1, supporting its role in transcriptional regulation (21, 22, 28–30).
We hypothesized that the carboxyl terminus of BRCA1, harboring a trans-activation function and consisting of two BRCT domains, would interact with other proteins that mediate tumor suppression, transcription regulation and, DNA repair. We screened a human placental cDNA expression library by a Far Western method (31) to identify proteins that interact with the carboxyl terminus of BRCA1. We found that the retinoblastoma-binding protein, RbAp46, interacts in vitro with the BRCT domain as well as with full-length BRCA1 in vivo. RbAp46, together with its homolog RbAp48, were first isolated based on interaction with the carboxyl terminus of the retinoblastoma protein, Rb (32–34). These proteins, members of the WD (Trp-Asp) repeat family, are highly similar in sequence and are functionally related (34). Recently, RbAp48 was found to be one of the three subunits of chromatin assembly factor 1 (35, 36). Both RbAp46 and RbAp48 are components of histone deacetylase complexes and are involved in chromatin remodeling (37, 38). Additionally, we found that the carboxyl terminus of BRCA1 associates with the histone deacetylases HDAC1 and HDAC2, implying that BRCA1 is a component of a histone deacetylase complex.
MATERIALS AND METHODS
Plasmid DNA Constructs.
The plasmid pET-BRCT encoding histidine-tagged fusion BRCT was constructed by subcloning the SacI and XhoI fragment from the full-length BRCA1 in-frame into pET28 (Novagen). Shorter fragments of BRCT-wild type and the mutant BRCT-Y1853X were PCR-amplified from pET-BRCT with Pfu polymerase (Stratagene) by using the forward primer TTGCCAAGGCAAGAGCTCGAGGGAACCCCTTAC with either of the following reverse primers: GCCCTCTAGACTCGAGCGTCAGTAGAGGCTGTG (wild type); CTCTAGACTCGAGCGCTAGGTGTCCAG (Y1853X). The mutant BRCT-M1775R, described elsewhere (39), was also amplified by using the same primer pair as the BRCT-wild type. PCR products were cloned into the pCR-Blunt vector (Invitrogen), verified by sequencing, digested with SacI and XhoI, and subcloned in-frame into pET28 (Novagen) and pcDNAhis (Invitrogen). pGST-BRCT was constructed by subcloning BamHI-XbaI fragment from pET-BRCT into pGEX4T. pGST-NH2-BRCA1 was constructed by subcloning BamHI-EcoRI fragment of full-length BRCA1 cDNA (encoding the amino terminus, amino acids 1–304) into pGEX5X.
The RbAp46-binding domain was excised from the pTriplEX vector of the cDNA library (CLONTECH) with EcoRI and XbaI and subcloned into both PGEX-4T and pcDNAhis. Full-length RbAp46 and RbAp48 were reverse transciption–PCR-amplified from HeLa total RNA with primer pairs with BamHI site at the 5′ end and XbaI at the 3′ end. All primers read from 5′ to 3′ AAGGTACCCGGATCCATGGCGAGTAAAGAGATGTTT (sense) and CAGCGGTTCTAGATCTTAAGATCCTTGTCCCTCCAGTTC (antisense) for RbAp46 and ACATGCGGATCCATGGCCGACAAGGAAGCAGCC (sense) CAGAGGTCTAGACTAGGACCCTTGTCCTTCTGG (antisense) for RbAp48. PCR products were cloned into pCR-Blunt (Invitrogen), verified by sequencing, and digested with BamHI and XbaI and subcloned in-frame into pGEX-4T (Amersham Pharmacia) and into pcDNA3his (Invitrogen). pGT-Rb, pGT-RbΔEx21 (deletion of amino acids 703–737) and pGT-RbΔEx22 (deletion of amino acids 735–775) were kindly provided by William Kaelin (Dana– Farber Cancer Institute, Boston).
cDNA Expression Library Screening.
The carboxyl-terminal segment of BRCA1 (amino acids 1,553–1,863) was expressed as a histidine-tagged recombinant protein from pET-BRCT in BL21 (DE3) cells and affinity-purified on Ni-NTA agarose columns according to the manufacturer’s instructions (Qiagen, Chatsworth, CA). Eluted protein was dialyzed against PBS. This protein, at a concentration of 3 μg/ml in TBST (10 mM Tris⋅HCl, pH7.5/150 mM NaCl/0.5% Tween-20) plus 0.3% BSA was used as a probe on the filters for interacting proteins screen. E. coli XL-1 Blue cells (CLONTECH) were infected with the human placenta cDNA library in λpTriplEX phage (CLONTECH). Protein expression from the library was induced by incubation with 10 mM isopropyl β-d-thiogalactoside-presoaked filters for 4 hours at 37°C. Filters were washed with TBST and blocked with 5% nonfat dry milk in TBST. Filters then were incubated with recombinant histidine-tagged BRCT polypeptide or recombinant histidine-tagged CBFβ-SMMHC (a gift from N. Adya, National Human Genome Research Institute) followed by incubation with affinity-purified rabbit polyclonal antibody directed against the histidine tag (Santa Cruz Biotechnology). Positive clones were visualized with horseradish peroxidase-conjugated anti-rabbit IgG (Amersham Pharmacia) and chemiluminesence (Pierce). Purified plaques were converted to pTripleX plasmids and sequenced.
Glutathione S-transferase (GST) Fusion Proteins and GST Pull-Down Experiments.
GST and GST fusion proteins were expressed in E. coli DH5α (GIBCO/BRL) or TOP10 (Invitrogen) cells transformed with pGEX4T, pGST-BRCT, pGST-NH2-BRCA1, pGSTp46-BD, pGSTp46-FL, pGSTp48, pGT-Rb, pGT-RbΔEx21, or pGT-RbΔEx22. GST fusion proteins were purified by affinity chromatography with glutathione-Sepharose (Amersham Pharmacia) beads as described (40), and protein concentration was estimated on a Coomassie-blue stained SDS/PAGE gel. Purified pGST-HD (huntingtin) was a gift of H. Reddy (National Human Genome Research Institute). Approximately equal amounts of different GST fusion proteins were mixed with 35S-labeled proteins synthesized by an in vitro reticulocyte coupled transcription/translation system TNT (Promega) according to manufacturer’s instructions and diluted in binding buffer (50 mM Tris⋅HCl, pH7.5/150 mM NaCl/1 mM EDTA/0.3 mM DTT/0.1% Nonidet P-40/0.5 mM PMSF/1 μg/ml leupeptin/2 μg/ml aprotinin and pepstatin). Beads were extensively washed 3–5 times with wash buffer (50 mM Tris⋅HCl, pH7.5/150 mM NaCl/1 mM EDTA/0.3 mM DTT/0.5% Nonidet P-40 and mixture of protease inhibitors as described above). Bound proteins were separated on SDS/10% PAGE and visualized by using autoradiography.
Protein Extract Preparation and Coimmunoprecipitation.
Nuclear extracts were prepared from subconfluent HeLa cells for coimmunoprecipitation as described (41), with some modifications. Briefly, cells were harvested and washed twice with ice-cold PBS and once with buffer A [20 mM Hepes, pH 7.9/1.5 mM MgCl2/10 mM KCl/0.5 mM DTT/0.1 mM EDTA/0.1 mM EGTA supplemented with mixture of protease inhibitors and phosphatase inhibitors (NaVO3, NaPPi, NaF)]. Cell pellets were incubated with buffer A on ice with for 15 min and then Dounce homogenized with 30–40 strokes. Samples were centrifuged, and pellets were resuspended in buffer C (20 mM Hepes, pH 7.9/420 mM NaCl/1.5 mM MgCl2/0.2 mM EDTA/25% glycerol supplemented with protease and phosphatase inhibitors) for 15 min on ice. The samples were sonicated and centrifuged in an microcentrifuge at 16,000 × g rpm for 15 min. Protein concentrations were determined by using the BCA protein assay (Pierce), and aliquots of nuclear extracts were stored at −80°C.
For coimmunoprecipitation, HeLa extracts (1.5 mg) diluted in binding buffer (supplemented with protease and phosphatase inhibitors) were incubated with the following mAbs: AB-3 against BRCA1 (Oncogene Science); RBBP against RbAp proteins (Transduction Laboratories, Lexington, KY); RbAp48–3-255 against RbAp (GeneTex, San Antonio, TX); or KM-1 against c-Jun (Santa Cruz Biotechnology) in the presence of protein A/G Plus (Santa Cruz Biotechnology) overnight at 4°C on a rotator. The immune complexes were washed five times in wash buffer and loaded onto SDS/6% PAGE. After electrophoresis, proteins were transferred to nitrocellulose membranes. Blots were blocked with 5% nonfat dry milk in TBST and incubated with 2 μg/ml mAb Ab-1 antibody against BRCA1 (Oncogene Science), or 1 μg/ml rabbit polyclonal antibody Ab-D (PharMingen). Proteins were visualized with horseradish peroxidase-conjugated secondary antibody and chemiluminescence (Pierce).
Whole-cell extracts from MCF-7 and HeLa cells were prepared for GST in vivo binding assays. Cells were harvested in RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS, supplemented with protease and phosphatase inhibitors). Cellular debris was cleared by using centrifugation, and protein concentration was determined by the BCA protein assay (Pierce). For in vivo associations, approximately equal amounts of GST fusion proteins coupled to beads were incubated with 300 μg of MCF-7 and HeLa whole-cell extracts diluted in binding buffer for 3 hr at 4°C on a rotator. Beads were extensively washed in washing buffer and bound proteins were separated on SDS/10% PAGE, transferred into nitrocellulose membrane, and followed by Western blot analysis with goat polyclonal antibodies directed against HDAC1 and HDAC2 (Santa Cruz Biotechnology, and alkaline phosphatase detection.
Immunofluorescence Staining.
HeLa cells or Saos2 cells grown in chamber slides to subconfluency were fixed for 30 min in 3% paraformaldehyde in PBS, and then rendered permeable with 0.1% Triton X-100 in PBS. Cells were washed with PBS, blocked with 10% goat serum in PBS, and then stained with 1:100 rabbit polyclonal antibody to BRCA1 (I-20, Santa Cruz Biotechnology), 1:100 mouse mAb to BRCA1 (Ab-1, Oncogene Science), 1:100 mouse mAb to RbAps (RBBP, Transduction Laboratories, Lexington, KY or 3–255, GeneTex, San Antonio, TX), 1:100 mouse mAb IF8 (Santa Cruz Biotechnology) to Rb or in combinations in humidified chamber for 30 min at 37°C. BRCA1, RbAps, and Rb were visualized with 1:200 anti-rabbit IgG conjugated to Texas red (Kirkegaard & Perry Laboratories) and 1:200 anti-mouse IgG conjugated to Alexa 488 (Molecular Probes), respectively. Immunofluorescence was recorded by using microscope and charge-coupled device camera.
RESULTS
Screening for Interacting Proteins.
To identify proteins that collaborate with BRCA1 in mediating tumor suppression, we sought to isolate proteins that physically interact with BRCA1. The intrinsic transcriptional activity of the BRCT domain precluded the use of the yeast two-hybrid system. Therefore, a Far Western assay was developed using histidine-tagged BRCT fusion protein as a probe to screen a human placenta cDNA expression library (Fig. 1A). A histidine-tagged CBFβ-SMMHC fusion protein was used as a negative control to eliminate clones that bound to the histidine tag. The sequence of one of the positive clones was identical to a portion (nucleotides 959–1,584) of the retinoblastoma-binding protein, RbAp46 (GenBank accession no. U35143). This clone, RY22, has a 1.0-kb insert and encodes 202 aa of the carboxyl terminus between amino acids 223 and 425 of RbAp46 followed by the 3′-untranslated region. Given the potential role of RbAp46 in chromatin remodeling, histone modifications, and gene expression, we further characterized this interaction between the BRCT domain of BRCA1 and RbAp46.
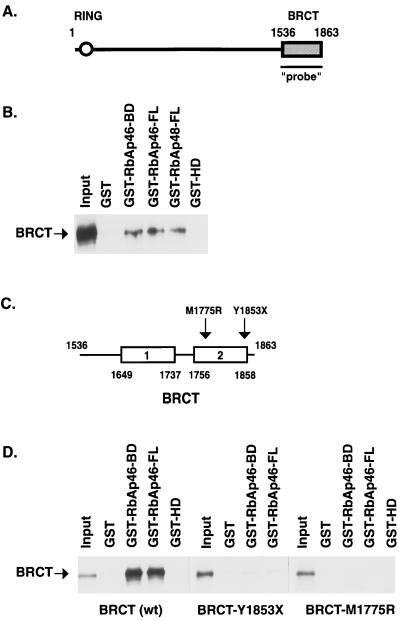
Figure 1.
The BRCT domain binds to Rb-binding proteins in vitro. (A) Schematic representation of BRCA1 gene. The region used as a probe in the library screen is indicated. (B) GST pull-down assays demonstrating that BRCT domain interacts with partial polypeptide of RbAp46 and full-length RbAp46 and RbAp48 proteins. In vitro-translated, 35S-labeled BRCT (10 μl) was incubated with 20 μl of glutathione-Sepharose beads and an equal amount of GST or GST-fusion proteins, as indicated. After extensive washing, bound proteins were eluted, resolved on SDS/10% PAGE, and visualized by using autoradiography. A portion of the in vitro-translated, 35S-labeled BRCT, corresponding to ≈20% of the labeled protein in the binding reaction, was loaded as “Input”. (C) Schematic representation of the BRCT repeats. Mutations analyzed for in vitro binding are indicated. (D) Mutations in BRCT domain interfere with interaction between RbAp46 and BRCT. GST pull-down assays were performed as in A with the wild type or mutation-containing BRCT polypeptides as indicated. Approximately 5% of each in vitro-translated protein in the binding reactions were loaded as Input.
The BRCT Domain Interacts with RbAp46 and RbAp48 in Vitro.
We tested whether the BRCT domain interacts in vitro with the RbAp46 carboxyl terminus, as identified by the library screen (designated BD), as well as with the full-length RbAp46 protein (FL). Purified GST proteins fused to either the RbAp46-BD or RbAp46-FL cDNA were incubated with in vitro-transcribed and -translated BRCT domain. Both GST-RbAp46-BD and GST-RbAp46-FL bound 35S-labeled BRCT, whereas GST alone and GST-HD did not (Fig. 1B). These results demonstrate that RbAp46 specifically binds the carboxyl terminus of BRCA1. Furthermore, these results indicate that the carboxyl terminus of RbAp46 between amino acids 223 and 425 is sufficient to bind the carboxyl terminus of BRCA1.
Because RbAp48 is 90% identical to RbAp46 at the amino acid level and is functionally related (34), we tested whether RbAp48 also interacts with 35S-labeled BRCT. Results shown in Fig. 1B confirm that RbAp48 binds specifically to the carboxyl terminus of BRCA1.
Several mutations in the BRCT domain segregate with inherited breast cancer in families. We investigated the effect of two BRCT mutations (Fig. 1C) on in vitro binding to RbAp46. The mutation Y1853X is a nonsense mutation that introduces a stop codon 10 aa before the native stop codon. M1775R is a missense mutation found in several unrelated breast cancer families (1, 5). Results shown in Fig. 1D show that the truncation mutation, Y1853X, has dramatically reduced binding to RbAp46 compared with wild-type BRCT, and the missense mutation M1775R does not bind at all. These results suggest that interaction with RbAp46 is important for BRCA1 function.
BRCA1 Associates with Rb-Binding Proteins in Vivo.
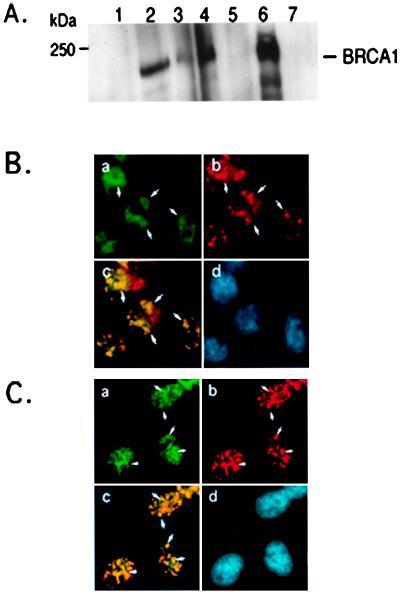
To determine whether BRCA1 and the retinoblastoma-binding proteins can interact and form a complex in vivo, we performed coimmunoprecipitation assays. Immunoprecipitation of the endogenous Rb-binding proteins from extracts of HeLa cells by two different mAbs (RBBP, Transduction Laboratories, Lexington, KY; RbAp48–15G12, GeneTex, San Antonio, TX) revealed that BRCA1 coprecipitates with Rb-binding proteins but not with c-jun (Fig. 2A; ref. 42). Similar results were observed with MCF-7 cells (data not shown). To further confirm the in vivo association between Rb-binding proteins and BRCA1, we asked whether BRCA1 and Rb-binding proteins colocalize in the cell. We first validated the I-20 rabbit polyclonal antibody (Santa Cruz Biotechnology) for BRCA1 staining in immunofluorescence assays. We performed two-color immunostaining of Saos2 cells with the commonly used mouse mAb (Ab-1, Oncogene Science) and the I-20 rabbit polyclonal antibody. Nearly identical staining patterns were produced by these two BRCA1 antibodies (data not shown). Two-color immunostaining assays with the BRCA1 rabbit polyclonal Ab (I-20, Santa-Cruz Biotechnology) and a Rb-binding protein mouse mAb (RbAp 3–225, GenTex, San Antonio, TX or RBBP, Transduction Laboratories, Lexington, KY) were performed with HeLa cells and Saos2 osteosarcoma cells. Results shown in Fig. 2B reveal that BRCA1 and Rb-binding proteins colocalize to the nucleus in a punctate pattern. In addition to the punctate spots, RbAps staining also has a granular appearance, particularly in the nucleus of Saos2 cells. Nevertheless, areas of intense staining for signal for RbAps are observed in both HeLa and Saos2 cell types. These dense areas overlap and colocalize with the punctuate pattern of BRCA1.
Figure 2.
In vivo association of BRCA1 with Rb-binding proteins. (A) Coimmunoprecipitation of BRCA1 and RbAp46/48. Endogenous BRCA1 protein was immunoprecipitated by anti-BRCA1 mAb SG11 (lane 2), anti-RbAp46/48 mAbs: RbBp (lane 3); RbAp48–3-255 (lane 4) and controls: normal mouse IgG (NMIgG, lane 1) and anti-c-jun mAb KM-1 (lane 5) from 1.5 mg of HeLa cell extract. Total cell lysate (100 μg) was run in lane 6. Protein extract (1.5 mg) from the BRCA1-null cell line, HCC1937 (42), were immunoprecipitated with anti-BRCA1 mAb SG11 (lane 7). Samples were separated on SDS/6% PAGE and immunoblotted with the mAb anti-BRCA1 antibody (Ab-1, Oncogene Science). (B) Colocalization of BRCA1 and RbAp46/48 in HeLa cells. Cells were prepared as described in Materials and Methods, stained with a mouse mAb against RbAp48/46, 3–225, (green in a), a rabbit polyclonal Ab against BRCA1, I-20, (red in b), and visualized by using microscope and charge-coupled device camera. The regions of overlap between red and green signals appear as yellow (c), indicating colocalization of BRCA1 and RbAps. The nucleus of each cell are shown by 4′,6-diamidino-2-phenylindole (DAPI) staining (d). White arrowheads indicate some regions of overlap of BRCA1 and RbAps staining. (C) Colocalization of BRCA1 and RbAp46/48 in Saos2 cells. Cells were prepared as described in Materials and Methods and stained as in B.
BRCT Domain Interacts with Rb in Vitro and in Vivo.
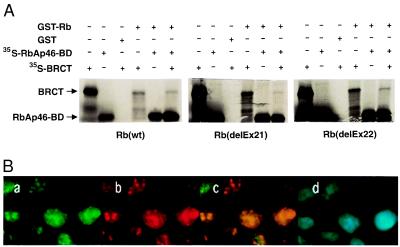
Because RbAp46 and RbAp48 are reported to interact with Rb (33, 34), we asked whether binding of BRCA1 and RbAp46 exist in a complex with Rb. Data shown in Fig. 3A demonstrate that Rb associates with RbAp46 (full-length and partial polypeptide, BD) in the presence of BRCT. Moreover, Rb binds the BRCT polypeptide in the absence of RbAp46, albeit less efficiently. Two mutations in the pocket domain of Rb [deletion of exon 21 and deletion of exon 22 (ref. 40)] do not affect binding to BRCT and RbAp46 (Fig. 3A). These results define the region of RbAp46 between amino acids 223 and 425 as sufficient to interact with Rb, and suggest that sequences other than exons 21 and 22 in the carboxyl fragment of Rb pocket region are important for binding RbAp46 and BRCT. Comparable results were observed with full-length RbAp46 (data not shown). We then tested whether BRCA1 and Rb are part of a complex in vivo. Two-color immunostaining of Rb and BRCA1 with a rabbit polyclonal antibody against BRCA1 (I-20) and a mouse mAb against Rb (IF8, Santa Cruz Biotechnology), revealed BRCA1 and Rb colocalized in nuclear dots in HeLa cells (Fig. 3B) but not in Saos2 cells that contain a mutated Rb (data not shown). Taken together, these results suggest that BRCA1 and Rb interact in vivo; however, the interaction between RbAps and BRCA1 is independent of Rb.
Figure 3.
Rb interacts with the BRCT domain. (A) GST pull-down experiments show that Rb interacts with BRCT domain in the presence or absence of partial RbAp46 polypeptide. In vitro-translated, 35S-labeled BRCT (12.5 μl) and partial RbAp46 polypeptides were incubated individually or in combination with 20 μl of glutathione-Sepharose beads with equal amount of GST-Rb or GST-Rb pocket mutation fusion proteins as indicated. GST-coated beads were incubated with each polypeptide separately. After extensive washing, bound proteins were eluted, resolved on SDS/10% PAGE and visualized by using autoradiography. Approximately 20% of labeled protein in the binding reaction were loaded as Input. (B) Colocalization of BRCA1 and Rb. HeLa cells were prepared as described in Material and Methods, stained with a mouse mAb against Rb, IF8, (green in a), a rabbit polyclonal antibody against BRCA1, I-20, (red in b). The regions of overlap between red and green signals appear as yellow (c), indicating colocalization of BRCA1 and Rb. The nucleus of each cells are shown by DAPI staining (d).
BRCA1 Is a Component of Histone Deacetylases.
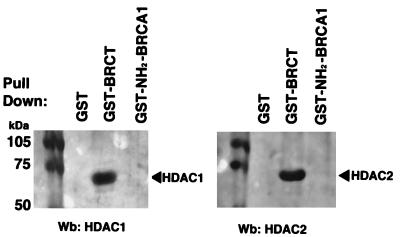
Because RbAp46 and RbAp48 are components of histone deacetylase complexes, we investigated whether BRCA1 associates with other components of this complex. HeLa and MCF-7 whole-cell extracts were incubated with GST alone, GST-BRCT, or GST-NH2BRCA1 (amino acids 1–304). Bound proteins were probed with antibodies for HDAC1 and HDAC2, the catalytic subunits of the histone deacetylase complex. Results shown in Fig. 4 indicate that BRCA1 associates with HDAC1 and HDAC2 via the BRCT domain either directly or indirectly in both cell lines. This interaction is specific, because GST alone or GST fused to the amino terminus of BRCA1 failed to bind HDAC1 and HDAC2.
Figure 4.
The BRCT domain associates with HDAC1 and HDAC2. Western blot analysis of GST pull-down assays using antibodies against HDAC1 and HDAC2. HeLa whole-cell lysates (300 μg) were incubated with 20 μl of glutathione-Sepharose beads covered with equal amounts of GST, GST-BRCT (amino acids 1,536–1,863), or GST-NH2-BRCA1 (amino acids 1–304) fusion proteins. Bound proteins were resolved on SDS/10% PAGE, transferred to nitrocellulose membranes, and blotted with HDAC1 or HDAC2 Abs.
DISCUSSION
We present evidence that BRCA1, via its carboxyl terminus, interacts with components of the complex containing human histone deacetylases HDAC1 and/or HDAC2. We also demonstrate that BRCA1 directly interacts with Rb-binding proteins RbAp46 and RbAp48 as shown by in vivo associations and in vitro binding assays. Additionally, Rb binds to the BRCT domain of BRCA1, suggesting that these proteins may form a complex.
The association of BRCA1 with RbAp46 depends on the carboxyl terminus of RbAp46 and the BRCT domain in the carboxyl terminus of BRCA1. This region contains two BRCT motifs also found in other proteins involved in cell cycle checkpoints (7). Two mutations in the second BRCT repeat associated with an elevated breast cancer risk, Y1853X and M1775R, disrupted the binding of BRCT to RbAp46, suggesting that the interaction between BRCA1 and RbAps is important for BRCA1 function. Moreover, these mutations interfere with the transcriptional activity of BRCA1 and reverse the BRCA1-mediated growth suppression in yeast (19, 20, 39). Of interest, Rb contains a diverged version of the BRCT consensus (18), raising the possibility that Rb-binding proteins may interact with other BRCT-containing proteins as well.
Recent evidence demonstrates that RbAp46 and RbAp48 are components of histone deacetylase complexes (37, 38). RbAp48 is also a component of the chromatin assembly factor, CAF-1 (35, 36), and RbAp46 has recently been identified as a component of human acetyltransferase HAT1 activity (43). Thus, both RbAp46 and RbAp48 may serve as targeting molecules whose function is to bring HDAC, HAT1, or CAF-1 enzymes, to their histone substrates. Our results further support the notion that BRCA1 is involved in processes that require chromatin remodeling, such as DNA repair or recombination, in addition to transcription regulation. Association of the BRCT domain of BRCA1 with Rb suggests not only that both proteins may exist in a complex, but also that BRCA1 and Rb may act in concert. In light of recent reports that Rb acts through histone deacetylases to repress transcription (44–46), our results point to a potential mechanism by which Rb and BRCA1 mediate tumor suppression. It would be of interest to examine the role of BRCA1 in regulation of transcription from E2F-repressed promoters. The association of BRCT with Rb does not involve exon 21 and exon 22 of the “pocket region” of Rb because these Rb mutants still bind the BRCT domain. Similarly, RbAp46 does not bind to these sequences in the pocket region of Rb. As HDAC was reported to interact with the pocket region (44, 46), it is likely that BRCA1 and RbAps will occupy distinct binding sites on Rb and can potentially form a multiprotein complex.
Changes in chromatin structure have been shown to be important in modulating the activity of multiprotein complexes responsible for transcription, replication, recombination, and DNA repair (47–51). Histone acetylation is generally associated with gene activation. Many transcriptional coactivators such as p300/CBP, pCAF, and SRC-1 possess intrinsic histone acetyltransferase activity (reviewed in refs. 52 and 53). Recently, BRCA2 has been shown to have acetyltransferase activity (54). Histone deacetylases are often associated with nucleosomal condensation and transcriptional repression (55, 56).
To date, BRCA1 has been reported to activate transcription of the cell cycle regulator p21waf/cip, thought in part to mediate the cell cycle arrest effect of BRCA1 when ectopically expressed (16). BRCA1-mediated activation of specific genes could occur by the sequestration of histone deacetylases from DNA promoters. Such a mechanism has been suggested for the E1A carboxyl-terminal domain, which activates transcription by disrupting a promoter bound complex of CtBP (E1A carboxyl terminus binding protein) and HDAC1 (57).
Conversely, BRCA1 may potentially repress transcription of genes that promote cell proliferation. Dual action of both inducing and repressing gene expression has been described for other tumor-suppressor genes. Rb represses transcription of E2F-dependent promoters involved in DNA metabolism, such as TK and DHFR genes, and activates expression of terminal differentiation markers such as NF-IL6 and BRG1. p53 activates gene expression of proteins involved in cell cycle arrest such as p21waf/cip and MDM, whereas repressing transcription from enhancers and promoters of DNA viruses. Therefore, the possibility that the association of BRCA1 with histone deacetylase directly represses transcription of a subset of genes involved in growth cannot be excluded. Finally, BRCA1 has been implicated in DNA repair (9, 10, 25). Given that DNA repair requires chromatin remodeling before and after removal of lesions, association of BRCA1 with multiprotein complex-involved chromatin dynamics could contribute to the execution of these processes.
Acknowledgments
We thank Drs. P. Liu, J. Koh, S. Danoff, and F. Collins for helpful comments on the manuscript. We thank Dr. A. Gazdar for providing the HCC1937 cell line, Dr. P. H. Reddy for the GST-HD protein, M. Erdos for BRCA1 plasmids, and S. King for technical assistance.
ABBREVIATIONS
- BRCT
BRCA1 carboxyl terminus
- GST
glutathione S-transferase
References
- 1.Futreal P A, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett L M, Haugen-Strano A, Swensen J, Miki Y, et al. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 2.Merajver SD, Pham T M, Caduff R F, Chen M, Poy E L, Cooney K A, Weber B L, Collins F S, Johnston C, Frank T S. Nat Genet. 1995;9:439–443. doi: 10.1038/ng0495-439. [DOI] [PubMed] [Google Scholar]
- 3.Langston A A, Malone K E, Thompson J D, Daling J R, Ostrander E A. N Engl J Med. 1996;334:137–142. doi: 10.1056/NEJM199601183340301. [DOI] [PubMed] [Google Scholar]
- 4.Castilla L H, Couch F J, Erdos M R, Hoskins K F, Calzone K, Garber J E, Boyd J, Lubin M B, Deshano M L, Brody L C, et al. Nat Genet. 1994;8:387–391. doi: 10.1038/ng1294-387. [DOI] [PubMed] [Google Scholar]
- 5.Friedman L S, Ostermeyer E E, Szabo C I, Dowd P, Lynch E D, Rowell S E, King M-C. Nat Genet. 1994;8:399–404. doi: 10.1038/ng1294-399. [DOI] [PubMed] [Google Scholar]
- 6.Miki Y, Swensen J, Shattuck-Eidens D, Futreal P A, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett L M, Ding W, et al. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 7.Koonin E V, Altschul S F, Bork P. Nat Genet. 1996;13:266–268. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- 8.Chen C F, Li S, Chen Y, Chen P L, Sharp Z D, Lee W H. J Biol Chem. 1996;271:32863–32868. doi: 10.1074/jbc.271.51.32863. [DOI] [PubMed] [Google Scholar]
- 9.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston D M. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 10.Scully R, Chen J, Ochs R L, Keegan K, Hoekstra M, Feunteun J, Livingston D M. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 11.Marquis S T, Rajan J V, Wynshaw-Boris A, Xu J, Yin G Y, Abel K J, Weber B L, Chodosh L A. Nat Genet. 1995;11:17–26. doi: 10.1038/ng0995-17. [DOI] [PubMed] [Google Scholar]
- 12.Zabludoff S D, Wright W W, Harshman K, Wold B J. Oncogene. 1996;13:649–653. [PubMed] [Google Scholar]
- 13.Hakem R, de la Pompa J L, Sirard C, Mo R, Woo M, Hakem A, Wakeham A, Potter J, Reitmair A, Billia F, et al. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 14.Gowen L C, Johnson B L, Latour A M, Sulik K K, Koller B H. Nat Genet. 1996;12:91–194. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- 15.Holt J T, Thompson M E, Szabo C, Robinson-Benion C, Arteaga C L, King M C, Jensen R A. Nat Genet. 1996;12:298–302. doi: 10.1038/ng0396-298. [DOI] [PubMed] [Google Scholar]
- 16.Somasundaram K, Zhang H, Zeng Y-X, Houvras Y, Peng Y, Zhang H, Wu G-S, Licht J D, Weber B L, El-Deiry W S. Nature (London) 1997;389:187–190. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- 17.Thompson M E, Jensen R A, Obermiller P S, Page D L, Holt J T. Nat Genet. 1995;9:444–450. doi: 10.1038/ng0495-444. [DOI] [PubMed] [Google Scholar]
- 18.Bork P, Hofmann K, Bucher P, Neuwald A F, Altschul S F, Koonin E V. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 19.Chapman M S, Verma I M. Nature (London) 1996;382:678–679. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- 20.Monteiro A N, August A, Hanafusa H. Proc Natl Acad Sci USA. 1996;93:13595–13599. doi: 10.1073/pnas.93.24.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouchi T, Monteiro A N, August A, Aaronson S A, Hanafusa H. Proc Natl Acad Sci USA. 1998;95:2302–2306. doi: 10.1073/pnas.95.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Somasundaram K, Peng Y, Tian H, Zhang H, Bi D, Weber B L, El-Deiry W S. Oncogene. 1998;16:1713–1721. doi: 10.1038/sj.onc.1201932. [DOI] [PubMed] [Google Scholar]
- 23.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 24.Baumann P, Benson F E, West S C. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 25.Gowen L C, Avrutzkaya A V, Latour A M, Koller B H, Leadon S A. Science. 1998;281:1009–1012. doi: 10.1126/science.281.5379.1009. [DOI] [PubMed] [Google Scholar]
- 26.Jensen D E, Proctor M, Maquis S T, Gardner H P, Ha S I, Chodosh L A, Ishov A M, Tommerup N, Vissing H, Sekido Y, et al. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 27.Wu L C, Wang Z W, Tsan J T, Spillman M A, Phung A, Xu X L, Yang M C, Hwang L Y, Bowcock A M, Baer R. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 28.Anderson S F, Schlegel B P, Nakajima T, Wolpin E S, Parvin J D. Nat Genet. 1998;19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 29.Yu X, Wu LC, Bowcock A M, Aronheim A, Baer R. J Biol Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 30.Wong A K, Ormonde P A, Pero R, Chen Y, Lian L, Salada G, Berry S, Lawrence Q, Dayananth P, Ha P, et al. Oncogene. 1998;17:2279–2285. doi: 10.1038/sj.onc.1202150. [DOI] [PubMed] [Google Scholar]
- 31.Blackwood E M, Eiseman R N. Methods Enzymol. 1995;254:229–240. doi: 10.1016/0076-6879(95)54017-2. [DOI] [PubMed] [Google Scholar]
- 32.Huang S, Lee W H, Lee E Y. Nature (London) 1991;350:160–162. doi: 10.1038/350160a0. [DOI] [PubMed] [Google Scholar]
- 33.Qian Y W, Wang Y C, Hollingsworth R E, Jr, Jones D, Ling N, Lee E Y. J Biol Chem. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- 34.Qian Y W, Lee E Y. J Biol Chem. 1995;270:25507–25513. doi: 10.1074/jbc.270.43.25507. [DOI] [PubMed] [Google Scholar]
- 35.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 36.Tyler J K, Bulger M, Kamakaka R T, Kobayashi R, Kadonaga J T. J Cell Biol. 1996;16:6149–6159. doi: 10.1128/mcb.16.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 38.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 39.Humphrey J S, Salim A, Erdos M R, Collins F S, Brody L C, Klausner R D. Proc Natl Acad Sci USA. 1997;94:5820–5825. doi: 10.1073/pnas.94.11.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaelin W G, Jr, Pallas D C, DeCaprio J A, Kaye F J, Livingston D M. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 41.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomlinson G E, Chen T T-L, Stastny V A, Virmani A K, Spillman M A, Tonk V, Blum J L, Schneider N R, Wistuba I I, Shay J W, et al. Cancer Res. 1998;58:3237–3242. [PubMed] [Google Scholar]
- 43.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Curr Biol. 1998;15:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 44.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Nature (London) 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 45.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Nature (London) 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 46.Luo R X, Postigo A A, Dean D C. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 47.Kornberg R D, Lorch Y. Curr Opin Cell Biol. 1995;7:371–375. doi: 10.1016/0955-0674(95)80092-1. [DOI] [PubMed] [Google Scholar]
- 48.Hassig C A, Schreiber S L. Curr Opin Chem Biol. 1997;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 49.Smith S, Stillman B. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Almouzni G, Wolffe A P. Genes Dev. 1993;7:2033–2047. doi: 10.1101/gad.7.10.2033. [DOI] [PubMed] [Google Scholar]
- 51.Gaillard P H, Martini E M, Kaufman P D, Stillman B, Moustacchi E, Almouzni G. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 52.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 53.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 54.Siddique H, Zou J P, Rao V N, Reddy E S. Oncogene. 1998;16:2283–2285. doi: 10.1038/sj.onc.1202003. [DOI] [PubMed] [Google Scholar]
- 55.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 56.Wolffe A P. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 57.Sundqvist A, Sollerbrant K, Svensson C. FEBS Lett. 1998;429:183–188. doi: 10.1016/s0014-5793(98)00588-2. [DOI] [PubMed] [Google Scholar]