Abstract
The erbB-2/HER2 oncogene is overexpressed in a significant fraction of human carcinomas of the breast, ovary, and lung in a manner that correlates with poor prognosis. Although the encoded protein resembles several receptors for growth factors, no high affinity ligand of ErbB-2 has so far been fully characterized. However, several lines of evidence have raised the possibility that ErbB-2 can augment signal transduction initiated by binding of certain growth factors to their direct receptors. Here, we contrasted these two models of ErbB-2 function: First, examination of a large series of epidermal growth factor (EGF)-like ligands and neuregulins, including virus-encoded ligands as well as related motifs derived from the precursor of EGF, failed to detect interactions with ErbB-2 when this protein was singly expressed. Second, by using antibodies that block inter-ErbB interactions and cells devoid of surface ErbB-2, we learned that signaling by all ligands examined, except those derived from the precursor of EGF, was enhanced by the oncoprotein. These results imply that ErbB-2 evolved as a shared receptor subunit of all ErbB-specific growth factors. Thus, oncogenicity of ErbB-2 in human epithelia may not rely on the existence of a specific ligand but rather on its ability to act as a coreceptor for multiple stroma-derived growth factors.
Cellular growth and fate determination are controlled by a large variety of extracellular ligands and specific cell surface receptors. The largest family of such receptors is that of the growth factor receptors with intrinsic tyrosine kinase activity (1). Type-1 tyrosine kinase receptors, also known as ErbB/HER proteins, comprise one of the better-characterized subfamilies of growth factor receptors, of which the epidermal growth factor (EGF) receptor (ErbB-1) is the prototype (reviewed in ref. 2). The four ErbB members form homo- and heterodimeric complexes on binding of EGF-like or neuregulin (NRG) ligands, and, thereby, their kinase activity is stimulated and intracellular signals are generated. Constitutive stimulation of these pathways through autocrine or other mechanisms is associated with several types of human cancer (3). Most relevant is the frequent overexpression, often as a result of gene amplification, of ErbB-2/HER2 in breast, ovary, lung, and other types of epithelial cancers (reviewed in refs. 4 and 5). In some tissues, this overexpression was correlated with poorer prognosis and a more aggressive tumor phenotype (6).
Although ErbB-2 shares extensive structural homology with other ErbBs both along the catalytic intracellular domain and in the extracellular putative ligand binding region, many attempts to identify stimulatory ligands specific to ErbB-2 have so far failed. For example, detection of an activity that enhances ErbB-2 phosphorylation led to molecular cloning of the Neu differentiation factor (NDF) and heregulin, two of a dozen isoforms of NRG1, all of which bind to ErbB-3 and ErbB-4 (7). Nevertheless, several observations imply that ErbB-2 homodimers, the plausible outcome of a direct ligand, may be functional in vivo. An oncogenic mutation that activates ErbB-2 phosphorylation apparently stabilizes such homodimers (8), and bivalent anti-ErbB-2 antibodies are mitogenic because they, like a direct ligand, dimerize ErbB-2 on the cell surface (9).
In parallel with attempts to isolate a direct ligand, several approaches culminated at the possibility that ErbB-2 functions, at least in part, as a coreceptor. Thus, coexpression of ErbB-2 together with ErbB-1 enhanced EGF-induced mitogenesis (10), and ErbB-2 presence reconstituted an extremely potent proliferative activity of ErbB-3, which is totally inactive when singly expressed (9). Consistent with its transactivating capability, ErbB-2 was found to act as the preferred partner of ligand-driven ErbB heterodimers (11, 12). The use of intracellular antibodies to ErbB-2 (13) has led to the conclusion that it can enhance signaling by two growth factors, EGF and NDF, through an ability to decelerate their release from the direct receptors, namely ErbB-1 and either ErbB-3 or ErbB-4, respectively (14).
Does ErbB-2 function as a high affinity receptor for a still unknown ligand of the EGF/NRG families, or could it act solely as a shared receptor subunit that amplifies signaling by prolonging the action of heterologous ligands? The present study addressed this question by using two strategies: First, we examined ligands that have not been previously tested for direct interaction with ErbB-2. On the other hand, we analyzed the generality of the transactivation ability of ErbB-2 by combining most existing ErbB ligands with mAbs that block a putative ligand binding site of ErbB-2. Our results strongly support the possibility that ErbB-2 evolved as a pan-EGF/NRG receptor rather than a high affinity receptor for a novel ligand. The implications of this scenario to epithelial tumors overexpressing ErbB-2 and to their inductive interactions with the underlying mesenchyme are discussed.
MATERIALS AND METHODS
Materials, Cell Lines, and Antibodies.
The construction and sources of recombinant and synthetic growth factors were as previously specified (15, 16, 17). Recombinant soluble extracellular domains of ErbB proteins fused to the Fc portion of human immmunoglobulin G (IgB) have been described (18). Antibodies directed against ErbB-2, used for receptor activation and immunoprecipitation, have been described (19), as have those against ErbB-3 and ErbB-4 (18). An antiphosphotyrosine mAb (PY-20) was purchased from Santa Cruz Biotechnology. A mAb to the active form of the mitogen-activated protein kinase (MAPK) (20) was a gift from R. Seger (Weizmann Institute). T47D human breast cancer cells and their derivative T47D-5R have been described (13). 32D myeloid cells that ectopically express ErbB receptors have been described (9).
Expression of Recombinant Precursor of EGF (proEGF) Fusion Proteins.
Four fragments containing the EGF-like domains of proEGF were constructed by PCR reactions on the full-length cDNA sequence of human proEGF in the pHEGF502 vector (kindly provided by Graeme I. Bell, Howard Hughes Medical Institute, Chicago) (21). The fragments, denoted pro1–4 (amino acids 314–479), pro5–8 (amino acids 741–952), pro5–9 (amino acids 741-1023), and EGF (amino acids 970-1023) were inserted into the pGEX expression vector (Amersham Pharmacia). Bacteria transformed with the constructs were induced to express the proteins and were harvested and lysed. Centrifugation-cleared lysates were mixed with glutathione-agarose beads and were incubated at 4°C while gently shaking. Elution of the bound proteins was carried out with 15 mM reduced glutathione and was followed by dialysis against PBS.
Cell Lysate Preparation.
Cells grown as monolayers were solubilized as described (19). Proteins were separated electrophoretically either directly or after immunoprecipitation, were transferred to a nitrocellulose membrane, and were detected by immunoblotting.
Determination of Tyrosine Phosphorylation and MAPK Activation.
Cells were incubated in PBS containing various ligands or mAbs at 37°C for the indicated time intervals. The treatment was ended by washing with ice-cold PBS. Whole cell lysates or immunoprecipitates were immunoblotted with an antiphosphotyrosine antibody (PY-20) or with a mAb that recognizes the doubly phosphorylated form of MAPK (20).
Cell Proliferation Assays.
Proliferation of IL-3-dependent 32D cells expressing ErbB proteins was determined as described (9).
RESULTS
ErbB Ligands Cannot Activate a Singly Expressed ErbB-2, but Multiple Growth factors Can Activate it in Epithelial Cancer Cells.
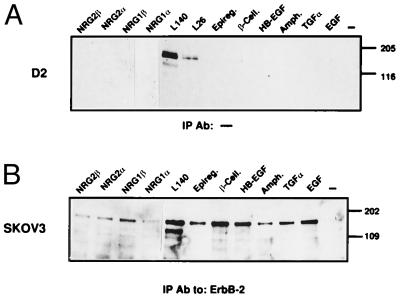
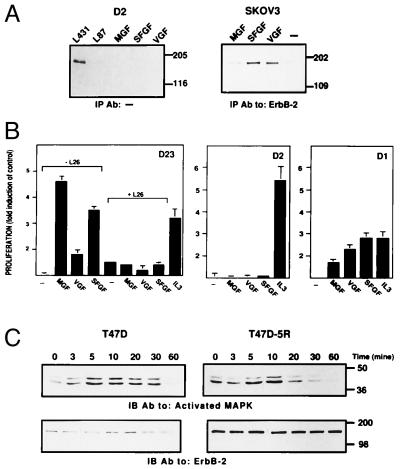
The ability of ErbB-2 to serve as a surrogate receptor when coexpressed with other family members, as well as the so-far unsuccessful search for a specific ErbB-2-binding ligand, suggest that its importance may reside in an intrinsic capacity to enhance signaling by a vast majority of ErbB-stimulating ligands. To experimentally test this scenario, we used an engineered 32D myeloid cell line that originally expresses no ErbB protein (9) and a large variety of known ErbB ligands (either EGF-like or NRGs). 32D cells that singly express ErbB-2 (D2) were incubated with growth factors, and the stimulation of ErbB-2 was followed by examining its phosphorylation on tyrosine residues (Fig. 1A). None of the 10 ligands tested was able to stimulate ErbB-2. That the protein is stimulatable under these conditions was evident from the ability of a mAb to ErbB-2 [L140 (19)] to stimulate tyrosine autophosphorylation. Antibody bivalency is essential for kinase stimulation (19), indicating that homodimerization of ErbB-2, a bona fide attribute of a direct ErbB-2 ligand, is functional in D2 cells. By contrast with their inability to stimulate a singly expressed ErbB-2, all 10 ligands we examined stimulated ErbB-2 phosphorylation to different extents in SKOV-3 ovarian cancer cells (Fig. 1B), which express ErbB-2 along with ErbB-1 and ErbB-3. To exclude dependence on cell type, we also examined Chinese hamster ovary cells, which express ErbB-2 in the absence of other ErbB members, and T47D breast cancer cells that express all four ErbBs. Similar to the results presented in Fig. 1, none of the growth factors tested was able to activate ErbB-2 in the former, but all ligands were active on the latter cell type (data not shown). In conclusion, although homodimeric stimulation of ErbB-2 is achievable, its activation by hitherto identified ErbB ligands strictly depends on coexpression of other receptor partners.
Figure 1.
ErbB-2 activation depends on coexpression of other ErbB proteins. ErbB-2 phosphorylation was determined in cells expressing the receptor singly (A, D2) or in combination with ErbB-1 and ErbB-3 (B, SKOV3). The indicated ligands (100 ng/ml) or antibodies (20 μg/ml) were used to treat the cells for 5 min at 37°C. Receptor activation in whole cell lysates (A) or immunoprecipitates of ErbB-2 (B) was determined by an antibody directed against phosphorylated tyrosine.
ErbB-2 Augments Stimulation of Mitogenesis by Multiple ErbB Ligands.
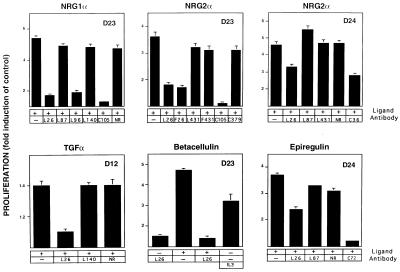
Because ErbB-2 can enhance signaling by NDF and EGF (14) and it is the preferred heterodimerizing partner of the respective receptors (11, 12), we hypothesized a similar role for this receptor in the transmission of signals by the majority of ErbB ligands. To examine the involvement of ErbB-2 in signaling by additional ligands, we applied mAbs that can inhibit ErbB-2 interactions with its family members [class II mAbs (19)] and 32D myeloid cells expressing defined ErbB combinations (9, 15). When deprived of IL-3, these cells totally depend on exogenous growth factors for survival. Cells expressing ErbB-2 with either ErbB-1 (D12), ErbB-3 (D23), or ErbB-4 (D24) were stimulated by EGF-like ligands in the presence of ErbB-specific mAbs. NRGs of several isoforms (NRG1α, NRG1β, and NRG2α) induced cellular proliferation by promoting complexes containing ErbB-2 in combination with either ErbB-3 or ErbB-4 (Fig. 2; data not shown). This effect could be significantly decreased by anti-ErbB-2 antibodies capable of heterodimer destabilization (L26 and L96), as well as by their monovalent fragments (F26). mAbs directed against different epitopes (L87, L140, and L431) were incapable of exerting a similar effect, suggesting that interreceptor interactions, stimulated by all of the examined ligands, depend on a similar domain of ErbB-2. Inhibition of mitogenicity stimulated in cells coexpressing ErbB-2 with ErbB-3 was marked and similar in extent to that achieved by a ligand-displacing antibody directed against ErbB-3 [mAb C105 (18)]. Mitogenic stimulation by ligands that primarily stimulate ErbB-1 exhibited a similar pattern of ErbB-2 dependency (Fig. 2, lower panels). As previously demonstrated for EGF (19), the L26 antibody inhibited proliferation induced by transforming growth factor α in D12 cells. Both betacellulin and epiregulin, which benefit from ErbB-2 participation in their signaling (15, 22), induced a decreased mitogenicity in the presence of mAb L26 in D23 and in D24 cells, respectively. Taken together, the results shown in Fig. 2 indicate that ErbB-2 is capable of increasing ligand-stimulated mitogenicity without discriminating between the heterodimerizing ErbBs and their respective ligands.
Figure 2.
ErbB-2-dependency of growth stimulation by EGF-like ligands. 32D cells expressing ErbB-2 with either ErbB-1 (D12), ErbB-3 (D23), or ErbB-4 (D24) were tested for cell proliferation. Cells deprived of IL-3 were treated with the indicated ligands. Anti-ErbB-2 mAbs belonging to class I (L431), class II (L26, L96), class III (L140), and class IV (L87) or their respective Fab fragments (F26, F431) were added simultaneously. Alternatively, control antibodies were used, including an unrelated mAb (NR), mAbs capable of ligand displacement from ErbB-3 (C105) or ErbB-4 (C72, C36), or an antibody against ErbB-3 that is incapable of displacing NRGs (C379). The extent of cell proliferation was determined 24 h after the addition of stimulating factors by using the colorimetric 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay. The results are presented as fold induction over control untreated cells and are the mean ± SD of eight determinations. Note that most mAbs (e.g., L26) have a weak agonist activity of their own.
ErbB-2 Enhances and Prolongs Signal Transduction by Multiple Growth Factors.
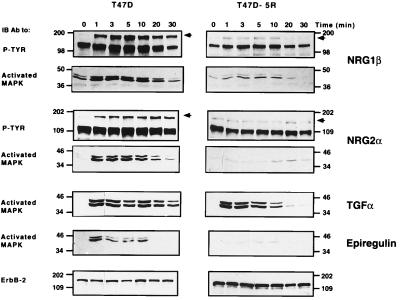
Despite complexity of the ErbB signaling network, achieved by multiplicity of both ligands and receptors, signaling is funneled into a major cascade involving activation of the MAPK pathway. Recruitment of this pathway by an oncogenic ErbB-2 is essential for transformation (23), and ErbB-2 is known to augment signaling by EGF and NDF through MAPK (14). To pursue whether ErbB-2 involvement is a common cardinal element in signals promoted by ErbB ligands other than EGF and NDF, we used a breast cancer cell line, T47D, expressing all ErbB receptors and its derivative, T47D-5R, devoid of ErbB-2 surface expression due to intracellular entrapment (13). As demonstrated in Fig. 3, the parental cell line is induced, by different ligands, to activate the MAPK cascade, as determined by the detection of its two activated forms (20). Concomitant phosphorylation of a 180-kDa protein ensured the correlation between ErbB activation and subsequent events (shown for NRG1 and NRG2α). Comparing the kinetics of activation to that in cells lacking surface ErbB-2 revealed a significant inhibition of intracellular activation in the latter. Both receptor phosphorylation and MAPK activation were affected. Stimulation by NRGs was decreased in duration as well as in intensity in cells lacking surface ErbB-2. Likewise, transforming growth factor α, although capable of inducing a similar increase in MAPK phosphorylation to that in the parental cells, showed a significant reduction in activation kinetics in T47D-5R cells. Stimulation by an additional ErbB-1-activating ligand, epiregulin, was affected in a similar manner to that of NRGs, decreasing to a barely detectable level in the absence of surface ErbB-2. To validate adequate expression of ErbB receptors in the 5R derivative, their amount was compared with that in the parental strain (data not shown): ErbB-1, ErbB-3, and ErbB-4 exhibited unaltered expression in T47D-5R cells. ErbB-2, in these cells, showed a characteristic faster electrophoretic migration, confirming its retention in the endoplasmic reticulum (13). In conclusion, expression of ErbB-2 at the cell surface can significantly prolong signaling by several growth factors, suggesting a pan-ErbB stimulatory effect that is independent on ligand identity.
Figure 3.
The effect of surface-expressed ErbB-2 on the kinetics of ligand-induced tyrosine phosphorylation and MAPK activation. ErbB ligands were used to stimulate T47D breast cancer cells and their derivative, T47D-5R, which lacks surface expression of ErbB-2. A comparable number of cells was stimulated at 37°C by the indicated ligands (at 100 ng/ml) for various time intervals. Receptor activation, in whole cell lysates, was detected by immunoblotting (IB) with an antibody directed against phosphorylated tyrosine (P-TYR). MAPK activation in the same preparations was determined by using an antibody against the active doubly phosphorylated form of Erk proteins (Activated MAPK). For control of equal gel loading, the upper part of membranes used to detect MAPK was used to determine the amount of ErbB-2. Note that the 5R cells exhibited up-regulation of the cell-retained ErbB-2.
proEGF-Derived Units Are Unable to Recognize ErbB-2.
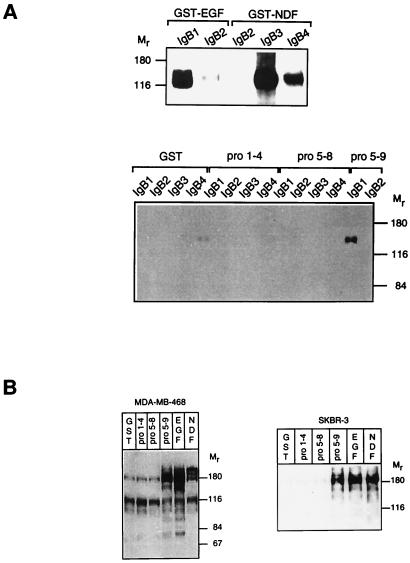
Because the extracellular domain of ErbB-2 is homologous to the ligand-binding domains of other ErbB proteins and because all ErbB ligands share an EGF-like motif (24), an ErbB-2-specific ligand, if it exists, may include an EGF-like domain. Other possibilities, such as binding of a non-EGF-like ligand to a distinct site of ErbB-2, cannot, however, be excluded. The precursor of EGF, which shares transmembrane topology with most other precursors of ErbB ligands, includes nine EGF-like motifs, of which only the membrane proximal unit is an established growth factor (i.e., EGF). To examine whether other proEGF domains might harbor a capacity to recognize ErbB-2, we studied their functionality as separate fragments. Four recombinant fragments were designed: EGF-like domains I–IV (pro1–4), domains V–VIII (pro5–8), domains V–IX (pro5–9), and domain IX. The latter corresponds to the active unit, namely EGF, and served as a positive control. These protein fragments, as well as the analogous functional domain of NRG1α (NDF) were expressed in bacteria in the form of glutathione S-transferase (GST) fusion proteins. To ensure correct expression and folding of the putative ligands, the functional domains of both EGF (GST-EGF) and NDF (GST-NDF) were tested for binding in vitro to soluble ErbB receptors [IgBs (18)]. Binding of the soluble receptors, denoted IgB1 through IgB4, to glutathione agarose-immobilized ligands confirmed that both GST-EGF and GST-NDF retained their receptor specificity (Fig. 4A Upper). Examining domains of proEGF in a similar manner could not reveal any novel recognition (Fig. 4A Lower), although the recombinant proteins exhibited the correct molecular weights and reacted with antibodies directed to respective peptides (data not shown). That failure to detect interaction in vitro was not caused by protein misfolding was implied by the retention of IgB1 binding by the pro5–9 recombinant protein consisting of the functional domain IX (Fig. 4A Lower). The absence of this domain, as in the case of pro1–4 and pro5–8 proteins, abolished recognition, reinforcing its sufficiency for receptor binding. Moreover, none of the fragments could recognize any other ErbB protein, although IgB3 and IgB4 bound NRGs, and IgB2 bound all tested mAbs to ErbB-2 (Fig. 4A; data not shown).
Figure 4.
Activation of ErbB receptors by EGF-like motifs of human proEGF. (A) GST fusion proteins containing EGF-like motifs 1–4, 5–8, or 5–9 of the EGF precursor were immobilized on glutathione-agarose beads. For control, GST fusion proteins containing EGF or NDF were used. The beads were incubated for 1 h at 4°C with conditioned media containing 1 μg of the indicated IgB protein. Protein complexes were immunoblotted with an anti-human Fc antiserum for detection of bound IgBs. (B) Monolayers of the indicated human breast cancer cell lines were incubated, for 10 min at 37°C, in the presence of 100 ng/ml GST fusion proteins or 5 ng/ml ligands (EGF or NDF). Receptor activation was detected by an antiphosphotyrosine antibody.
The inability of proEGF-derived units to act as ErbB-binding ligands was evident also from experiments performed with living breast cancer cells (Fig. 4B). By detecting phosphorylation of proteins on tyrosine residues in whole cell lysates, we could demonstrate a pattern of receptor activation which is in accordance with the above binding. Only fragments containing domain IX could activate phosphorylation of proteins corresponding to ErbB receptors. Moreover, comparing an ErbB-2 overexpressing cell line (SKBR-3) with one devoid of the receptor (MDA-MB-468) revealed a similar specificity of stimulation, namely the dependence of activation on the ninth EGF-like domain. Collectively, these results indicate that no other EGF-like domain derived from the precursor molecule could serve as an ErbB-2-specific ligand.
ErbB-2 Is Activated by Three Viral Ligands only when Coexpressed with Other Family Members.
Three EGF-like ligands encoded by poxviruses have been shown to resemble ErbB-activating molecules in structure as well as in activity. These ligands, including the vaccinia virus growth factor, the Shope fibroma virus growth factor (SFGF), and the Myxoma virus growth factor, harness the proliferation-inducing activity of ErbB receptors for the enhancement of their virulence (25). Synthetic analogs of these three viral ligands revealed specific patterns of ErbB specificity. For example, SFGF acts as a pan-ErbB ligand whereas Myxoma virus growth factor is more specific to the ErbB-2/ErbB-3 complex (17). Because evolutionarily the ErbB family evolved from a single protein whose ortholog in nematodes is Let-23 (26), and because it is likely that poxviruses coevolved with their vertebrate hosts (25), we assumed that an ErbB-2-specific ligand, if it ever existed, may have been retained in the genome of this large family of DNA viruses. To examine direct interaction between ErbB-2 and the three viral ligands, we used 32D cells singly expressing the protein (D2). As demonstrated in Fig. 5, none of the three known viral ligands promoted homodimerization of ErbB-2 and the consequent kinase activation (Fig. 5A Left) or mitogenic effect (Fig. 5B Center), although both activities were displayed by a mAb specific to ErbB-2 (Fig. 5A; data not shown).
Figure 5.
Viral peptides recruit ErbB-2. (A) Phosphorylation of ErbB-2 by viral peptides [vaccinnia virus growth factor (VGF), Myxoma virus growth factor (MGF), and SFGF] and antibodies (L87, L431) was examined as described in the legend to Fig. 1. (B) IL-3-deprived D23 cells were stimulated by viral peptides in the presence (+L26) or absence (−L26) of a class II mAb to the human ErbB-2 (Left). Cells singly expressing ErbB-2 (D2) or ErbB-1 (D1) served as negative and positive controls for ligand activity, respectively. Proliferation induction was determined by the 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay as described in the legend to Fig. 2. For control of endogenous proliferation signals, we incubated cells with IL-3. (C) The effect of ErbB-2 on downstream activation by SFGF was examined in cells that do (T47D) or do not (T47D-5R) express ErbB-2 on their surface. A time response of activation was detected in whole cell lysates by immunoblotting with an antibody against activated MAPK. The amount of ErbB-2 was verified by immunoblotting the upper part of the membrane with an antibody against the receptor.
Nevertheless, by using similar approaches to those presented above (Figs. 2 and 3), we learned that the three viral ligands, like their mammalian counterparts, depend on ErbB-2 for cellular activation. All three ligands could induce phosphorylation of ErbB-2 in SKOV3 cells (Fig. 5A Right), suggesting that the viral growth factors can recruit ErbB-2 into heterodimeric complexes. The involvement of ErbB-2 was also manifested biologically by a mitogenic assay (Fig. 5B); although none of the viral ligands was active on cells singly expressing the kinase-defective ErbB-3 receptor (data not shown), all three ligands potently stimulated cells coexpressing it with the ligand-less ErbB-2 (Fig. 5B Left). Recruitment of ErbB-2 by the viral ligands in these cells was evident also from the inhibitory activity of a class II mAb (L26) to ErbB-2 (Fig. 5B). Lastly, by using SFGF on T47D cells and the engineered 5R derivative, we observed an ErbB-2-mediated prolongation and enhancement of MAPK activation (Fig. 5C). Thus, although this ligand is capable of activating various ErbB complexes (17), it seems that SFGF, like the corresponding mammalian growth factors, depends on ErbB-2 as a coreceptor rather than as a direct high-affinity receptor.
DISCUSSION
Despite extensive investigation and a wealth of clinical data, the biochemical role of ErbB-2 in human cancer remains an enigma (4, 5): Although the structure and enzymatic function of the oncoprotein suggest that it is stimulated by a specific growth factor, in vitro studies along with the continuous failure to isolate a direct ligand imply a nonconventional receptor function (reviewed in ref. 27). This possibility has been strengthened by gene targeting experiments indicating cooperation between ErbB-2 and the neuregulin receptor ErbB-4 (28). By using a variety of ErbB ligands, our present study weakens the commonly held scenario arguing that ErbB-2 functions as an orphan receptor. Instead, a cooperative role in signal transduction is strongly supported.
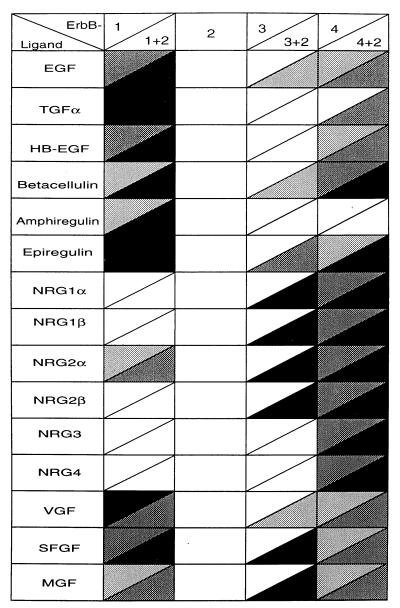
The orphan receptor scenario predicts that an ErbB-2-specific ligand exists and that it contributes to tumor virulence by promoting homodimerization of the overexpressed ErbB-2 protein. However, contrary to this prediction, ErbB-2 homodimers that are driven either by a bivalent antibody (9) or by a point mutation (29) induce a mitogenic response that is weaker than that generated by ErbB-2-containing heterodimeric complexes. Another prediction made by the orphan receptor hypothesis is that the ErbB-2 ligand, if it exists, contains an EGF-like motif of six cysteine residues. However, it seems that no known EGF-like motif can directly bind to ErbB-2 with high affinity. For example, our most recent search for such an element in newly cloned EST databases identified one candidate, which we denoted NRG4 because the encoded protein exclusively binds ErbB-4 as its primary receptor (30). The EGF-like motif is found not only in ligand growth factors but also in cell adhesion proteins. For example, multiple copies of this domain are included in the extracellular matrix proteins laminin, tenascin, and thrombospondin, as well as in two Drosophila cell fate-determining proteins: Notch and Delta (reviewed in ref. 31). Our present results (Fig. 4), imply that all of the motifs included in proEGF, except the membrane proximal domain, belong to the second category of function. Indeed, modeling of the eight other motifs of proEGF, according to the published three-dimensional structure of EGF (32), indicated that domains 1–4 and 5–8 fall into distinct groups but that both groups significantly differ from the structures of EGF and NDF (M. Eisenstein, S.G., and Y.Y., unpublished results). Another important conclusion that emerged from the analysis of proEGF motifs is that the Gly-Xxx-Arg-Cys motif common to all ErbB ligands, but absent in nonligand motifs, may predict ErbB binding. Table 1 lists all of the currently known molecules that contain this motif, in the context of the EGF-like domain, along with their ErbB activating preference. Although it is clear that none binds to ErbB-2, it is also evident that signaling by all known ErbB ligands is enhanced by ErbB-2. This conclusion, along with the observation that certain anti-ErbB-2 antibodies can inhibit signaling by several NRGs and EGF-like ligands (Fig. 2), reinforces the possibility that ErbB-2 acts as a heterodimer partner rather than a direct receptor. Also supportive is the observation that each of the three other ErbB proteins serves as a direct receptor for more than one ligand (Table 1). It is therefore conceivable that, if ErbB-2 were able to bind a direct ligand, such a molecule would have been discovered, at least once.
Table 1.
Receptor specificity of EGF-like ligands and neuregulins
All of the ErbB-stimulatory ligands are presented along with their ErbB preference. Interactions with the indicated ErbB homodimers (above diagonals) and the corresponding heterodimers with ErbB-2 (below diagonals) are indicated by using a color code: The most mitogenic interactions of each ligand are shown in black whereas white areas indicate absence of mitogenic signals. Note that ErbB-2 homodimers respond to no known ligand but that the mitogenic action of practically all growth factors can be augmented in the presence of ErbB-2. The data represent compliation of previous results obtained primarily with IL-3-dependent cells and the following ligands: NRG1s (9, 35), NRG2s (16), NRG3 (36), NRG4 (30), EGF (9, 22, 35, 37), transforming growth factor α (22, 37), epiregulin (15, 38), betacellulin (22, 39, 40), amphiregulin, and the viral ligands (17).
In the absence of an ErbB-2-specific ligand, it may not be practical to test the prediction that ErbB-2 acts solely as a receptor subunit. However, the presence of genes encoding EGF-like ligands in the genome of poxviruses provided us an attractive opportunity to test this possibility. Like ErbB-2-overexpressing human carcinomas, the skin lesions induced by poxviruses display epithelial hyperproliferation and a transformed phenotype (25). Because poxviruses underwent coevolution with their mammalian hosts and were selected for efficient induction of epithelial lesions, it is reasonable to assume that an ErbB-2 ligand, if it existed, would have conferred a significant selective advantage to poxviruses that encoded it. Therefore, the observation that none of the three known viral growth factors can directly interact with ErbB-2 (Fig. 5) implies that this receptor may not be able to accommodate a specific ligand. On the other hand, ErbB-2 seems to fulfil a similar role in viral infection to that played in human carcinomas; the observed specificity of SFGF and especially Myxoma virus growth factor to the most mitogenic heterodimer, namely the ErbB-2/ErbB-3 combination (Table 1), suggests that poxviruses, much like carcinogenic mechanisms, gained the ability to harness the signal amplification ability of ErbB-2.
Perhaps the best exemplification of the capacity of ErbB-2 to transactivate signaling initiated by ligands binding to other ErbBs is the ability to reconstitute an extremely strong mitogenic activity of ErbB-3, a receptor whose homodimers are inactive (9). Because ErbB-3 is expressed by many carcinomas at moderately high levels and ErbB-2 is ubiquitously expressed, the cooperation between the two receptors is thought to drive or maintain the transformed phenotype of epithelial tumor cells (33). Examination of the molecular mechanism underlying ligand-induced formation of this heterodimer may provide an explanation to the role played by ErbB-2 (34): Apparently, ErbB-2 can bind at very low affinity ligands like NRG1, but only when they are presented to it by their primary receptors. This model predicts that ErbB ligands are endowed with two binding sites and that the lower affinity site preferentially recognizes the putative binding cleft of ErbB-2, which may be the target of class II mAbs (19).
In conclusion, ErbB-2 emerges as a master coordinator of a signaling network rather than as a receptor that mediates the action of one specific ligand. The relative topology of ErbB proteins, which are situated primarily on the basolateral face of epithelial cells, and their respective ligands, which are synthesized by the underlying stromal cells, implies that ErbB-2 can act as an amplifier of signaling by all of the stromal ligands listed in Table 1. Complete sequencing of the human genome and characterization of the remaining EGF motif-containing genes will ultimately answer the question whether this is the only function of ErbB-2 or whether a still-unknown ligand that binds to it with high affinity does exist.
Acknowledgments
We thank Graeme Bell for human EGF cDNA and Roni Seger for anti-MAPK antibodies. This research was supported in part by the Bristol-Myers Squibb Foundation Cancer Grant Award, by the U.S. Department of the Army (Grant DAMD 17-97-1-7290), by a grant from the National Institutes of Health (Grant CA 72981 to Y.Y.), and by the Ovarian Cancer Research Fund, Inc.
ABBREVIATIONS
- EGF
epidermal growth factor
- NDF
Neu differentiation factor
- NRG
neuregulin
- SFGF
Shope fibroma virus growth factor
- proEGF
precursor of the epidermal growth factor
- GST
glutathione S-transferase
- MAPK
mitogen-activated protein kinase
- IgB
ErbB extracellular domains fused to an Fc portion of human Ig G
References
- 1.van der Geer P, Hunter T, Lindberg R A. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- 2.Burden S, Yarden Y. Neuron. 1997;18:847–855. doi: 10.1016/s0896-6273(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 3.Salomon D S, Brandt R, Ciardiello F, Normanno N. Crit Rev Oncol Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 4.Hynes N E, Stern D F. Biochim Biophys Acta. 1994;1198:165–184. doi: 10.1016/0304-419x(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 5.Klapper, L. N., Kirschbaum, M. H., Sela, M. & Yarden, Y. (1999) Adv. Cancer Res., in press. [PubMed]
- 6.Slamon D J, Godolphin W, Jones L A, Holt J A, Wong S G, Keith D E, Levin W J, Stuart S G, Udove J, Ullrich A, et al. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 7.Tzahar E, Levkowitz G, Karunagaran D, Yi L, Peles E, Lavi S, Chang D, Liu N, Yayon A, Wen D, Yarden Y. J Biol Chem. 1994;269:25226–25233. [PubMed] [Google Scholar]
- 8.Weiner D B, Liu J, Cohen J A, Williams W V, Greene M I. Nature (London) 1989;339:230–231. doi: 10.1038/339230a0. [DOI] [PubMed] [Google Scholar]
- 9.Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin B J, Sela M, et al. EMBO J. 1996;15:2452–2467. [PMC free article] [PubMed] [Google Scholar]
- 10.Kokai Y, Myers J N, Wada T, Brown V I, LeVea C M, Davis J G, Dobashi K, Greene M I. Cell. 1989;58:287–292. doi: 10.1016/0092-8674(89)90843-x. [DOI] [PubMed] [Google Scholar]
- 11.Tzahar E, Waterman H, Chen X, Levkowitz G, Karunagaran D, Lavi S, Ratzkin B J, Yarden Y. Mol Cell Biol. 1996;16:5276–5287. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graus-Porta D, Beerli R R, Daly J M, Hynes N E. EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beerli R R, Wels W, Hynes N E. J Biol Chem. 1994;269:23931–23936. [PubMed] [Google Scholar]
- 14.Karunagaran D, Tzahar E, Beerli R R, Chen X, Graus-Porta D, Ratzkin B J, Seger R, Hynes N E, Yarden Y. EMBO J. 1996;15:254–264. [PMC free article] [PubMed] [Google Scholar]
- 15.Shelly M, Pinkas-Kramarski R, Guarino B C, Waterman H, Wang L-M, Lyass L, Alimandi M, Kuo A, Bacus S S, Pierce J H, et al. J Biol Chem. 1998;273:10496–10505. doi: 10.1074/jbc.273.17.10496. [DOI] [PubMed] [Google Scholar]
- 16.Pinkas-Kramarski R, Shelly M, Guarino B C, Wang L M, Lyass L, Alroy I, Alimandi M, Kuo A, Moyer J D, Lavi S, et al. Mol Cell Biol. 1998;18:6090–6101. doi: 10.1128/mcb.18.10.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tzahar E, Guarino B C, Waterman H, Levkowitz G, Shelly M, Pinkas-Kramarski R, Wang L-M, Alimandi M, Kuo A, Moyer J D, et al. EMBO J. 1998;17:5948–5963. doi: 10.1093/emboj/17.20.5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Levkowitz G, Tzahar E, Karunagaran D, Lavi S, Ben Baruch N, Leitner O, Ratzkin B J, Bacus S S, Yarden Y. J Biol Chem. 1996;271:7620–7629. [PubMed] [Google Scholar]
- 19.Klapper L N, Vaisman N, Hurwitz E, Pinkas-Kramarski R, Yarden Y, Sela M. Oncogene. 1997;14:2099–2109. doi: 10.1038/sj.onc.1201029. [DOI] [PubMed] [Google Scholar]
- 20.Yung Y, Dolginov Y, Yao Z, Rubinfeld H, Michael D, Hanoch T, Roubini E, Lando Z, Zharhari D, Seger R. FEBS Lett. 1997;408:292–296. doi: 10.1016/s0014-5793(97)00442-0. [DOI] [PubMed] [Google Scholar]
- 21.Mroczkowski B, Reich M, Chen K, Bell G I, Cohen S. Mol Cell Biol. 1989;9:2771–2778. doi: 10.1128/mcb.9.7.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinkas-Kramarski R, Lenferink A E, Bacus S S, Lyass L, van de Poll M L, Klapper L N, Tzahar E, Sela M, van Zoelen E J, Yarden Y. Oncogene. 1998;16:1249–1258. doi: 10.1038/sj.onc.1201642. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Levy R, Paterson H F, Marshall C J, Yarden Y. EMBO J. 1994;13:3302–3311. doi: 10.1002/j.1460-2075.1994.tb06632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groenen L C, Nice E C, Burgess A W. Growth Factors. 1994;11:235–257. doi: 10.3109/08977199409010997. [DOI] [PubMed] [Google Scholar]
- 25.Buller R M L, Palumbo G J. Microbiol Rev. 1991;55:80–122. doi: 10.1128/mr.55.1.80-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aroian R V, Lesa G M, Sternberg P W. EMBO J. 1994;13:360–366. doi: 10.1002/j.1460-2075.1994.tb06269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzahar E, Yarden Y. Biochim Biophys Acta. 1998;1377:M25–M37. doi: 10.1016/s0304-419x(97)00032-2. [DOI] [PubMed] [Google Scholar]
- 28.Lee K F, Simon H, Chen H, Bates B, Hung M C, Hauser C. Nature (London) 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 29.Cohen B D, Kiener P K, Green J M, Foy L, Fell H P, Zhang K. J Biol Chem. 1996;271:30897–30903. doi: 10.1074/jbc.271.48.30897. [DOI] [PubMed] [Google Scholar]
- 30.Harari, D., Tzahar, E., Romano, J., Shelly, M., Pierce, J. H., Andrews, G. C. & Yarden, Y. (1999) Oncogene, in press. [DOI] [PubMed]
- 31.Engel J. FEBS Lett. 1989;251:1–7. doi: 10.1016/0014-5793(89)81417-6. [DOI] [PubMed] [Google Scholar]
- 32.Kohda D, Inagaki F. Biochemistry. 1992;31:677–685. doi: 10.1021/bi00118a007. [DOI] [PubMed] [Google Scholar]
- 33.Wallasch C, Weiss F U, Niederfellner G, Jallal B, Issing W, Ullrich A. EMBO J. 1995;14:4267–4275. doi: 10.1002/j.1460-2075.1995.tb00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzahar E, Pinkas-Kramarski R, Moyer J D, Klapper L N, Alroy I, Levkowitz G, Shelly M, Henis S, Eisenstein M, Ratzkin B J, et al. EMBO J. 1997;16:4938–4950. doi: 10.1093/emboj/16.16.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riese D J, van Raaij T M, Plowman G D, Andrews G C, Stern D F. Mol Cell Biol. 1995;15:5770–5776. doi: 10.1128/mcb.15.10.5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang D, Sliwkowski M X, Mark M, Frantz G, Akita R, Sun Y, Hillan K, Crowley C, Brush J, Godowski P J. Proc Natl Acad Sci USA. 1997;94:9562–9567. doi: 10.1073/pnas.94.18.9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alimandi M, Wang L-M, Bottaro D, Lee C-C, Angera K, Frankel M, Fedi P, Tang F, Tang C, Lippman M, et al. EMBO J. 1997;16:5608–5617. doi: 10.1093/emboj/16.18.5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riese D J, Komurasaki T, Plowman G D, Stern D F. J Biol Chem. 1998;273:11288–11294. doi: 10.1074/jbc.273.18.11288. [DOI] [PubMed] [Google Scholar]
- 39.Riese D J, Bermingham Y, van Raaij T M, Buckley S, Plowman G D, Stern D F. Oncogene. 1996;12:345–353. [PubMed] [Google Scholar]
- 40.Wang L M, Kuo A, Alimandi M, Very M C, Lee C C, Kapoor V, Ellmore N, Chen X H, Pierce J H. Proc Natl Acad Sci USA. 1998;95:6809–6814. doi: 10.1073/pnas.95.12.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]