Abstract
Saccharomyces cerevisiae has two independent transport systems for the removal of arsenite from the cytosol. Acr3p is a plasma membrane transporter that confers resistance to arsenite, presumably by arsenite extrusion from the cells. Ycf1p, a member of the ABC transporter superfamily, catalyzes the ATP-driven uptake of As(III) into the vacuole, also producing resistance to arsenite. Vacuolar accumulation requires a reductant such as glutathione, suggesting that the substrate is the glutathione conjugate, As(GS)3. Disruption of either the ACR3 or YCF1 gene results in sensitivity to arsenite and disruption of both genes produces additive hypersensitivity. Thus, Acr3p and Ycf1p represent separate pathways for the detoxification of arsenite in yeast.
Keywords: ACR3, YCF1, SbIII, resistance, ABC transporters
Exposure to drugs and metals frequently results in the acquisition of resistance mechanisms. Development of transport systems for the export or sequestration of heavy metals and drugs is one of the most commonly utilized strategies used by all cell types for producing resistance (1, 2). In bacteria efflux systems that produce resistance to salts of the metalloids arsenic and antimony have been characterized (3). The pathways of metalloid resistance in eukaryotes are less clear. In both Leishmania and Chinese hamster cells, resistance correlates with active extrusion of arsenite from resistant cells (4, 5), but the relevant genes or gene products have not been identified. Human multidrug resistance-associated protein (MRP), a member of the ABC transporter superfamily, also confers resistance to arsenite (6), but arsenite transport by MRP has not been demonstrated.
The yeast Saccharomyces cerevisiae can serve as a model system for the study of arsenite resistance in eukaryotes. Recently, three ACR (arsenic compounds resistance) genes related to resistance to arsenite and arsenate, ACR1, ACR2, and ACR3, were identified on chromosome XVI of S. cerevisiae (7). ACR1 encodes a putative transcription factor; insertional inactivation of ACR1 resulted in sensitivity to both arsenate and arsenite. ACR2 encodes a protein required for arsenate but not arsenite resistance that has been shown to be an arsenate reductase (8). ACR3 encodes a putative membrane protein. Disruption of ACR3 resulted in sensitivity to arsenite, leading to the hypothesis that it is a plasma membrane arsenite efflux transporter (9).
In this report, we demonstrate that there are two parallel pathways for removal of toxic arsenite from yeast cytosol. The first pathway is extrusion of arsenite into the medium. We show that cells with an ACR3 disruption exhibit not only increased sensitivity to arsenite but also that deletion of ACR3 was directly correlated with increased arsenite accumulation, most likely reflecting loss of efflux activity. Both resistance and ability to prevent arsenite accumulation was restored by expression of ACR3 on a plasmid. The second pathway is vacuolar sequestration of As(III) in a reaction catalyzed by the product of the YCF1 gene. Ycf1p (yeast cadmium factor protein), a close homologue of the human MRP, has been shown to be a vacuolar glutathione S-conjugate pump with a broad range of substrate specificity (10, 11) and confers cadmium resistance in S. cerevisiae by catalyzing sequestration of Cd(GS)2 in the vacuole (12). Both MRP and a homologue from Arabidopsis thaliana complement a YFC1-disrupted yeast strain (13, 14). We show here that vacuolar membrane preparations exhibit uptake of 73As(III) in the presence of MgATP and glutathione (GSH). No uptake was observed in vacuolar membrane vesicles prepared from a YCF1-disrupted strain. The YCF1-disrupted strain was sensitive to As(III), As(V), Sb(III), and Cd(II) whereas the ACR3-disrupted strain was sensitive to As(III) and As(V) but remained resistant to Sb(III) and Cd(II). An ACR3-YCF1 double disruption produced hypersensitivity to arsenite and arsenate. These results are consistent with ACR3 and YCF1 encoding independent transporters, either of which confers arsenite resistance.
MATERIALS AND METHODS
Strains and Plasmids.
Plasmids and S. cerevisiae strains are described in Table 1. E. coli strain JM109 [recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′ (traD36 proAB+ lacIq lacZΔM15)] was used for molecular cloning.
Table 1.
Strains and plasmids
| Strains/plasmids | Genotype/description | Source |
|---|---|---|
| S. cerevisiae strains | ||
| W303-1B | MATα ade2-1 his3-11, 15 leu2-3, 112 ura3-1 trp-1 | Ref. 45 |
| MG100 | MATα ade2-1 his3-11, 15 leu2-3, 112 ura3-1 trp-1 acr3∷HIS3 | This study |
| DTY165 | MATα ura3-52 his6 leu2-3, 112 his3-Δ, 200 trp1-901 lys2-801 suc2-Δ | Ref. 10 |
| DTY167 | MATα ura3-52 his6 leu2-3, 112 his3-Δ, 200 trp1-901 lys2-801 suc2-Δ ycfl∷hisG | Ref. 10 |
| MG101 | MATα ura3-52 his6 leu2-3, 112 his3-Δ, 200 trp1-901 lys2-801 suc2-Δ acr3∷URA3 | This study |
| MG102 | MATα ura3-52 his6 leu2-3, 112 his3-Δ, 200 trp1-901 lys2-801 suc2-Δ ycfl∷hisG acr3∷URA3 | This study |
| Plasmids | ||
| pGEM-T | E. coli cloning vector, Apr | Promega |
| pGEM-T-ACR3 | 3.1-kbp PCR fragment containing ACR3, ACR2, and part of ACR1 cloned in pGEM-T | This study |
| pUC18-HIS31 | 1.7-kbp BamHI fragment containing HIS3 ligated into BamHI site of a pUC18 derivative in which the BamHI site proximal to the EcoRI site had been destroyed by a fill in reaction | S. Ackerman |
| pUC18-URA3212 | 1.17-kbp HindIII fragment cloned into the HindIII site of pUC18 | S. Ackerman |
| pGEM-T-ACR3-HIS3 | 1.1-kbp BamHI-KpnI fragment of pGEM-T-ACR3 replaced by 1.7-kbp BamHI-SmaI HIS3 fragment of pUC18 | This study |
| pGEM-T-ACR3-URA3 | 1.17-kbp HindIII fragment containing URA3 cloned into KpnI site of pGEM-T-ACR3 | This study |
| YEp352 | S. cerevisiae–E. coli shuttle vector, Apr, URA3 | Ref. 21 |
| YEpACR3 | 2.1-kbp EcoRI-PacI fragment containing ACR3 ligated into EcoRI-SmaI-digested YEp352 | This study |
Media.
S. cerevisiae strains were grown at 30°C in complete yeast extract/peptone/dextrose (15) medium supplemented with 2% glucose. Alternatively, the minimal selective (15) medium with 2% glucose supplemented with auxotrophic requirements was used. Escherichia coli cells were grown in Luria–Bertani medium (16) supplemented with 125 μg/ml ampicillin as required.
DNA Manipulations.
Cloning procedures were carried out as described (16, 17). Transformation of yeast cells was carried out by using a lithium acetate method (18). Yeast genomic DNA was isolated by using QIAamp spin column (Qiagen, Chatsworth, CA).
Disruption of the ACR3 Gene.
Disruption of the ACR3 gene was carried out by a one-step gene replacement method (19). A 3.1-kilobase (kb) fragment of yeast genomic DNA containing ACR3, ACR2, and a portion of ACR1 was amplified by PCR using a forward primer 5′-CGACATAAGCTTATCTTGTCC-3′ that hybridizes with a region 533 bp downstream of ACR3 and a reverse primer 5′-GTCATTTGAGAGATTCGTACAGC-3′ that hybridizes to a region 483 bp inside of ACR1. The fragment was ligated into pGEM-T. The resulting plasmid pGEM-T-ACR3, carrying ACR3, ACR2, and a portion of ACR1, was digested with KpnI. The linearized plasmid was made blunt with T4 DNA polymerase, was digested with BamHI, which excised a 1,145-bp fragment that included all but the first 34 bp and last 35 bp of ACR3, and was ligated with a 1.7-kb HIS3 gene that had been obtained from plasmid pUC18-HIS3-1 as a 1,769-bp BamHI-SmaI fragment. The resulting plasmid was digested with EcoRI, and the 3.6-kbp fragment was isolated, purified, and transformed into yeast strain W303–1B, producing the ACR3-disrupted strain MG100 by homologous recombination. Recombinants were selected for growth in the absence of histidine and were screened for arsenite sensitivity. Disruption of ACR3 was verified by PCR using a forward primer 5′-AGATCTATGTCAGAAGATCAAAAAAGTG-3′ that introduces a BglII site immediately in front of the first ACR3 codon and hybridizes from that point. The reverse primer was 5′-GAATTCATTTCTATTGTTCCATATATAATATGGTTTAAGGATCCTCG-3′, which hybridizes at the last ACR3 codon and introduces an EcoRI site immediately following ACR3.
To construct a strain with both ACR3 and YCF1 disrupted, a similar strategy was carried out starting with strain DTY167, in which YCF1 had been deleted (20). Plasmid pGEM-T-ACR3 was digested at the unique KpnI site in ACR3, was made blunt with T4 DNA polymerase, and was ligated with URA3. URA3 was obtained from plasmid pUC18-URA3–212 as a 1.17-kb HindIII fragment and was made blunt. The resulting plasmid was digested with EcoRI, and a 4.2-kb fragment was isolated, purified, and transformed into yeast strain DTY167, producing the ACR3-YCF1 double-disrupted strain MG102 by homologous recombination. Strain MG101, in which ACR3 of DTY165 was disrupted, was constructed similarly. Disruption of ACR3 in the resulting uracil prototrophs was verified by PCR, as described above.
For complementation, ACR3 under control of its endogenous promoter was cloned from plasmid pGEM-T-ACR3 by digestion with PacI, was made blunt, and then was digested with EcoRI. The resulting 2.1-kb fragment was ligated with EcoRI-SmaI-digested yeast E. coli shuttle vector plasmid YEp352 (21). The ligation mixture was transformed into E. coli strain JM109, and the resulting plasmid, YEpACR3, was isolated and transformed into both wild-type and ACR3-disrupted W303–1B. The transformants were selected on minimal SD medium without uracil and histidine.
Metal Ion Resistance Assays.
Strains were grown overnight at 30°C in liquid minimal selective medium with 2% glucose and appropriate supplements. The cultures were diluted into minimal media to an OD600 of 0.1 in presence of varying concentrations of the indicated additions and were incubated for an additional 24 hr, after which the growth was estimated from OD600. For growth on solid media, cells were streaked from single colonies and were incubated for 3–4 days at 30°C.
Membrane Preparations.
For preparation of membrane vesicles, a 10-ml stationary phase culture of the appropriate yeast strain was diluted into 200 ml of fresh yeast extract/peptone/dextrose medium. The culture was grown at 30°C to an OD600 of ≈1.0. The cells were chilled on ice for 20 min, were harvested at 4,000 × g at 4°C, and were washed twice with a buffer consisting of 50 mM Tris⋅HCl (pH 7.5), containing 5 mM Na2EDTA, 1 mM Na2EGTA, 0.3 M sucrose, 5 mg/ml BSA, 20 mM β-mercaptoethanol, and 1 mM 4-(2-aminoethyl)benzenesulfonylfluoride (buffer A). After the final wash, cells were suspended in 3 ml of buffer A per gram of wet cells and were passed through a French pressure cell at 20,000 psi. Diisopropylfluorophosphate (2.5 μl/g of wet cells) was immediately added, and the cells were disrupted by grinding with acid-washed glass beads (150–212 μm) (10 g/g of wet cells) (22). The suspension was diluted with 30 ml of buffer B (10 mM Tris⋅HCl, pH 7.5/1 mM Na2EGTA/10% glycerol/20 mM β-mercaptoethanol/1 mM 4-(2-aminoethyl)benzenesulfonylfluoride/2 mM MgCl2) and was centrifuged at 3,000 × g for 10 min at 4°C to remove the glass beads, unbroken cells, and cell debris. The supernatant suspension was centrifuged at 17,000 × g for 40 min to remove organelles and then at 140,000 × g for 1 hr. The pelleted membranes were washed once and were suspended in 2 ml of buffer C (75 mM Hepes⋅KOH, pH 7.2/0.15 M KCl/2 mM MgCl2), all at 4°C. The suspension then was gently homogenized several times with a Wheaton Scientific homogenizer immersed in ice. The membrane vesicles were rapidly frozen in liquid nitrogen and were stored at −80°C until use. The protein content of the membrane vesicles was determined by an Amido Black filter assay (23).
Membranes were separated into plasma membrane-enriched and vacuolar membrane-enriched fractions by sucrose density gradient centrifugation (24). The membranes were layered on a four-step gradient containing 3 ml each of four different sucrose solutions [18%, 26%, 34%, and 68% (wt/wt)] in buffer C. After centrifugation for 2 hr at 49,000 × g in a Sorvall TH641 swinging bucket rotor at 4°C, two turbid sections were observed, one in the region of 26% sucrose and one at the interface of 34 and 68% sucrose. The turbid fractions were individually isolated, were diluted 3- to 10-fold with buffer C, and were centrifuged for 1 hr at 140,000 × g. The pellet was suspended in 0.5 ml of buffer C containing 2 mM MgCl2 and was stored at −80°C until use.
The membranes were analyzed for localization markers by polyacrylamide gel electrophoresis and immunoblotting. Samples were dissolved in 0.1 ml of SDS sample buffer, were incubated for 5 min at room temperature, and were analyzed by SDS/PAGE (25) by using a 10% polyacrylamide gel. Proteins were electrophoretically transferred to a nitrocellulose membrane (0.2-μm pore size) and were immunoblotted overnight at 25 mV and 4°C with either of an antibody directed against Pma1p or Pho8p, each at a dilution of 1:2,000. A chemiluminescent assay was used to detect the antigen–antibody reaction. The filter was incubated with 10 ml of the enhanced chemiluminescence solution (New England Nuclear) and was exposed on x-ray film for 1 min at room temperature.
Transport Assays.
For uptake assays in vivo, cells were grown in yeast extract/peptone/dextrose medium containing 2% glucose at 30°C to exponential phase, followed by induction with 0.1 mM sodium arsenite for an additional 24 hr. The cells were washed twice with cold degassed water and once with degassed buffer C and were suspended at a density of 1 × 108 cells/ml in buffer C. To initiate the assay, 0.1 ml of cells was diluted with 0.9 ml of buffer C containing 0.1 M glucose. After 30 min at 30°C, Na273AsO2 was added to a final concentration of 5 μM. Portions (0.1 ml) were withdrawn at intervals and were filtered through nitrocellulose filters (0.2-μm pore size; Whatman). The filters were washed with 5 ml of room temperature buffer C and were dried, and the radioactivity was quantified in liquid scintillation counter. Na273AsO2 was prepared by reduction of radioactive arsenate (26).
For uptake assays in vitro, frozen membrane vesicles were thawed rapidly and were stored on ice until use. Unless otherwise noted, the transport assay contained 5 mM MgCl2, 0.1 mM Na273AsO2, and 0.4 mM GSH in buffer C. Vesicles were added at 0.5 mg/ml of membrane protein. After 5 min at room temperature, the reaction was initiated by addition of ATP to a final concentration of 10 mM in a total volume of 0.3 ml. At intervals, samples (50 μl) were withdrawn and filtered on wet nitrocellulose filters. The filters were washed with 5 ml of cold buffer C and were dried, and radioactivity was measured. In control experiments, ATP was replaced with AMP at a final concentration of 10 mM.
RESULTS
ACR3 Confers Arsenite Resistance in S. cerevisiae.
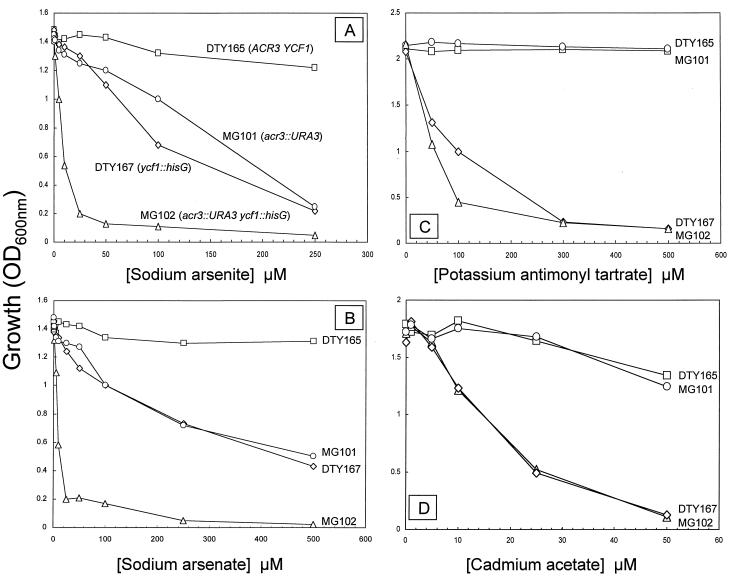
The 1,212-bp ACR3 gene in S. cerevisiae strains W303–1B and DTY167 was disrupted, producing strains MG100 and MG101, respectively. Disruption of ACR3 had no effect on growth of cells in the absence of arsenite. Cultures derived from single colony isolates with the ACR3 disruption exhibited sensitivity to both arsenite (Fig. 1A) and arsenate (Fig. 1B). MG101 was as resistant to Sb(III) (Fig. 1C) and Cd(II) (Fig. 1D) as its parent DTY165, indicating that Acr3p is specific for As(III). To confirm that arsenite sensitivity was the result of disruption of ACR3, the wild-type and disrupted strains were separately transformed with the ACR3-bearing plasmid YEpACR3. The plasmid complemented the arsenite and arsenate sensitive phenotypes (data not shown). It is interesting to note that cells expressing ACR3 on this multicopy plasmid exhibited increased resistance to both arsenite and arsenate compared with parental strains. If the amount of the putative positive transcription factor Acr1p were limiting for expression of the ACR genes, an increase in resistance with ACR3 alone would not occur. If Acr2p were limiting for arsenate resistance, then ACR3 alone would not produce an increase in resistance to arsenate as well as arsenite. These results indicate that the amount of Acr3p is rate-limiting for resistance to both oxidized and reduced arsenic salts.
Figure 1.
Metal ion sensitivity of S. cerevisiae strains with disruptions of ACR3 and/or YCF1. Cells were grown in liquid selective minimal medium with the indicated concentrations of sodium arsenite [As(III)] (A), sodium arsenate [As(V)] (B), potassium antimonyl tartrate [Sb(III)] (C), or Cd(OAc)2 [Cd(II)] (D). Strains were DTY165 (parental) (□); MG101 (acr3∷URA3) (○); DTY167 (ycf1∷hisG) (⋄); and MG102 (acr3∷URA3 ycf1∷hisG) (▵).
YCF1 Confers Arsenite Resistance in S. cerevisiae.
MRP, an ABC transporter, has been reported to confer resistance to arsenite in human cells (6). Similarly PgpA, a Leishmania MRP homologue, has been associated with arsenite resistance (27). In S. cerevisiae, YCF1, which confers cadmium resistance, encodes a close homologue of MRP (10, 11). The YCF1-disrupted strain DTY167 (20) was examined for metal ion sensitivity (Fig. 1). Disruption of either ACR3 or YCF1 resulted in equivalent sensitivity to arsenite (Fig. 1A). These results are consistent with ACR3 and YCF1 encoding independent pathways for arsenite detoxification. Double disruption of both genes in strain MG102 resulted in additive hypersensitivity. Similar results were found for arsenate (Fig. 1B). This was anticipated because Acr2p reduces arsenate to arsenite (8), which then can be transported by either system. In contrast, only YCF1 conferred resistance also to Sb(III) and Cd(II) (Fig. 1 C and D). Considering the chemical similarity of As(III) and Sb(III), it is surprising that ACR3 does not produce antimonite resistance. However, the results demonstrate that the substrate specificity of Acr3p and Ycf1p differ; the former is a specific arsenic transporter whereas the latter has a broader range of metal ion substrates.
Acr3p Is a Plasma Membrane Arsenite Exporter.
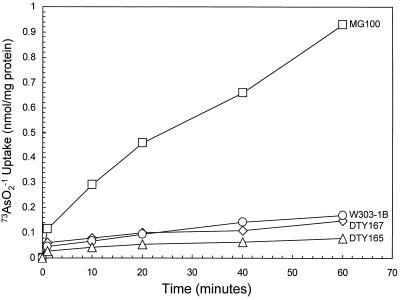
Decreased accumulation of arsenite from cells of E. coli expressing arsenite resistance genes reflects active extrusion; cells resistant to arsenite do not accumulate radiolabeled arsenite whereas sensitive cells accumulate the metalloid (28). Accumulation of 73AsO2−1 into cells of S. cerevisiae was examined (Fig. 2). In the parental strains W303–1B and DTY165 exclusion of radiolabeled arsenite was observed, consistent with active efflux. Cells of MG100, with an ACR3 disruption, accumulated 10-fold more arsenite over a 60-min period, consonant with inability to extrude the metalloid. In contrast, disruption of YCF1 had no effect on accumulation; cells of DTY167 were able to exclude arsenite as efficiently as its parent. These results are consistent with Acr3p catalyzing arsenite efflux through the plasma membrane. In these experiments, the cells were induced with sodium arsenite; even wild-type cells were unable to exclude arsenite without induction (data not shown).
Figure 2.
ACR3-encoded arsenite exclusion from cells of S. cerevisiae. 73AsO2− accumulation was measured in cells of W303–1B (parental) (○); MG100 (acr3∷HIS3) (□); DTY165 (parental) (▵); and DTY167 (ycf1∷hisG) (⋄).
Ycf1p Is a Vacuolar Arsenite Pump.
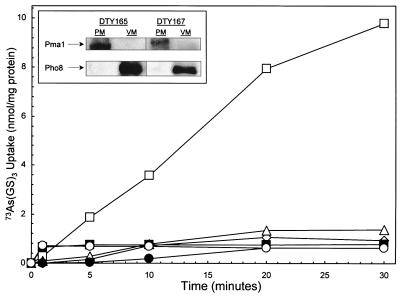
Since Ycf1p has been shown to catalyze ATP-coupled Cd(GS)2 transport into vacuoles (12), crude membranes prepared from S. cerevisiae were fractionated by sucrose density gradient centrifugation. By Western blot analysis, a dense fraction was found to react with an antibody against Pma1p, the plasma membrane H+-translocating ATPase (29) (Fig. 3 Inset). A light fraction reacted with an antibody directed against Pho8p, a vacuolar membrane enzyme (30). Vacuolar membrane vesicles prepared from cells of S. cerevisiae DTY165 accumulated radiolabeled arsenite (Fig. 3). In contrast to cellular extrusion, in vitro transport activity did not depend on induction with arsenite (data not shown). Accumulation required reduced glutathione under conditions in which arsenite spontaneously forms an As(GS)3 conjugate (31). This result is consistent with the fact that many of the substrates of MRP and its homologues are S-glutathione conjugates (10, 32). Of interest, tris(2-carboxyethyl)phosphine, a strong nonthiol reductant, was as effective as GSH (data not shown). Phosphine complexes of Tc(III) are substrates of P-glycoprotein (33), suggesting that phosphines form adducts with As(III) that are recognized by Ycf1p. Accumulation in vacuolar membrane vesicles required MgATP (Fig. 3). MnATP, CaATP, or other Mg2+-nucleoside triphosphates could substitute to varying degrees (data not shown). 73As(GS)3 transport was totally inhibited by equimolar amounts of Cd(II), Sb(III), Hg(II), or the organoarsenical phenylarsine oxide (data not shown). In those experiments, GSH was added in excess over both As(III) and competitor, which would result in spontaneous formation of their respective S-glutathione conjugates. These results are consistent with those metal ions being competitive substrates. The effect of the ionophores valinomycin and carbonyl cyanide-p-trifluoromethoxyphenylhydrazone was examined. Neither inhibited (data not shown), consistent with 73As(GS)3 transport being catalyzed by a primary ATP-coupled pump.
Figure 3.
YCF1-encoded 73As(GS)3 accumulation in vacuoles of S. cerevisiae. As(GS)3 accumulation was assayed in the plasma membrane-enriched (PM) and vacuolar membrane-enriched (VM) fractions from cells of DTY165 or DTY167. 73As(GS)3 was prepared by mixing 0.1 mM Na273AsO2 and 0.4 mM GSH. Accumulation was assayed with 5 mM MgCl2 and 10 mM ATP, unless otherwise noted. Curves: (□), DTY165 VM; (○), DTY165 VM - ATP; (■), DTY165 VM - GSH; (⋄), DTY165 PM; (▵), DTY167 VM; (●), DTY167 PM. (Inset) Membranes from strains DTY165 and DTY167 fractionated by sucrose density gradient centrifugation were analyzed by immunoblotting with antibody against the plasma membrane Pma1p or the vacuolar membrane Pho8p. The arrows indicate the predicted position of Pma1p and Pho8p estimated from the migration of proteins of known molecular mass.
Arsenite accumulation into vacuoles was not affected by disruption of ACR3 (data not shown), indicating that this ATP-coupled transport reaction is not catalyzed by Acr3p. Because disruption of YCF1 produced arsenite sensitivity, it was reasonable to consider that the transport activity is caused by the activity of the vacuolar Ycf1p pump. Although the vacuolar membranes from the parental strain DTY165 accumulated 73As(GS)3, vacuolar membranes from the YCF1-disrupted strain DTY167 exhibited no accumulation, nor did the plasma membrane fractions from either strain (Fig. 3). Thus, the in vitro transport activity reflects catalysis by Ycf1p and not Acr3p.
DISCUSSION
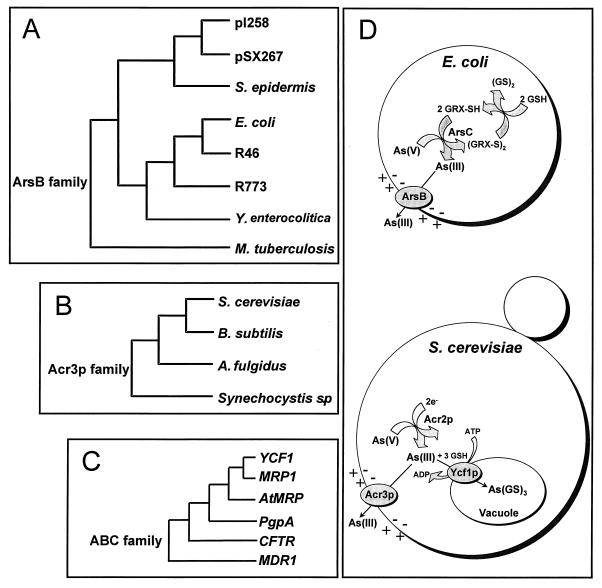
The pressure of environmental exposure to arsenic has resulted in the independent evolution of three distinct arsenite transporters. In both prokaryotic and eukaryotic organisms, resistance to arsenicals has been correlated with active extrusion (4, 5, 28). In prokaryotes, arsenite resistance is primarily linked to the ArsB family of arsenite efflux proteins (Fig. 4A) (3). However, no eukaryotic ArsB orthologues have been identified to date. Recently, an arsenite resistance gene encoding a putative membrane protein was identified in Bacillus subtilis (34). Homologues of this protein have been identified in other bacteria (35), in archaea (36), and in S. cerevisiae (7), defining a second family of arsenite resistance proteins that we call the Acr3p family (Fig. 4B). From our experiments, it is clear that a subgroup of the ABC superfamily forms a third class of arsenite resistance proteins (Fig. 4C).
Figure 4.
Detoxification of arsenicals in prokaryotes and eukaryotes. On the left is the phylogenetic relationships of representative members of the three independently evolved families of arsenite transporters (accession numbers in parentheses). (A) ArsB family: pI258, Staphylococcus aureus plasmid pI258 (M86824); pSX267, Staphylococcus xylosus plasmid pSX267 (M80565); Staphylococcus epidermis (S57521); E. coli (U00039); E. coli plasmid R46 (U38947); E. coli plasmid R773 (J02591); Yersinia enterocolitica (U58366); Mycobacterium tuberculosis (Z96072). (B) Acr3p family: S. cerevisiae Acr3p (Q06598); B. subtilis YqcL (D844321); Archaeglobus fulgidus (AE001071); Synechocystis sp. (D90914). (C) ABC ATPases: Three members of this large superfamily have been shown to confer arsenite resistance, including PgpA of Leishmania tarentolae (P21441); the S. cerevisiae Ycf1p cadmium resistance protein (S51863); and the human MRP1 (2828206). A closely related homologue is AtMRP3 from A. thaliana (U92650). Other members of the ABC superfamily that are probably not arsenite pumps are human cystic fibrosis transmembrane conductance regulator (CFTR) (L49339) and P-glycoprotein (MDR1) (P08183). (D) On the right is a comparison of arsenite transport pathways in E. coli and S. cerevisiae. Although the pathways of resistance are similar in overall design, the prokaryotic proteins are unrelated evolutionarily to their eukaryotic analogues. In both E. coli and yeast, the first step is arsenate detoxification by reduction to arsenite catalyzed by the ArsC or Acr2p enzyme, respectively. Arsenite then is extruded from cells by a plasma membrane carrier protein, ArsB or Acr3p, respectively. Both carriers are proposed to be anion uniporters, coupled to the membrane potential. In S. cerevisiae, there is a second, parallel pathway for removal of arsenite from cytosol in which the ATP-coupled Ycf1p pump catalyzes sequestration of As(GS)3 in the vacuole.
We propose that arsenite detoxification mechanisms are nearly universal in nature and that the pathways will have similar steps even if they are the products of independent evolution (Fig. 4D). The first step is reduction of arsenate to arsenite. Because geological sources of arsenicals are exposed to the oxidizing atmosphere, aqueous arsenical salts, such as those that contaminate drinking water from geochemical sources, are usually in the form of arsenate. In both bacteria and yeast, enzymatic reduction is catalyzed by members of three evolutionarily unrelated families of arsenate reductases. (7, 8, 37, 38). The next step in the pathway is removal of arsenite from the cytosol (Fig. 4D). As discussed above, transport proteins for the detoxification of arsenite also have evolved independently at least three times: ArsB, Acr3p, and Ycf1p. In both bacteria and yeast, active extrusion out of the cell is catalyzed by either ArsB or Acr3p, but, in yeast, Ycf1p provides a second, independent way to remove arsenite from the cytosol by sequestering it in the vacuole as the glutathione conjugate.
Arsenicals and antimonials have a long history of usage as antimicrobial agents (39, 40), and resistance to these metalloids is of clinical importance in treatment of infectious diseases caused by trypanosomatid parasites (40–42). Moreover, arsenite in the form of arsenic trioxide has been shown to be an effective treatment for acute promyelocytic leukemia (43). In a recent study, arsenite treatment produced complete remission of acute promyelocytic leukemia in 11 of 12 patients (44). Three of the eleven patients became unresponsive to treatment with arsenite, suggesting that resistance may have arisen in those individuals. Thus, it is important to determine the routes of arsenic detoxification in higher eukaryotes, including humans. At this point, S. cerevisiae is the only eukaryotic organism for which the pathways of arsenical resistance have been defined, and arsenite-sensitive yeast strains may prove of value in identifying the genes for other eukaryotic transporters.
Acknowledgments
We thank S. Ackerman (Wayne State University) for strains, plasmids, advice, and thoughtful discussion. We also thank R. Nakamoto (University of Virginia) and R. Rao (Johns Hopkins University) for suggestions and advice. We are grateful to D. Thiele (University of Michigan) for strains and advice. Thanks are due C. Slayman (Yale University) for Pma1p antibody and N. Davis (Wayne State University) for Pho8 antibody. This work was supported by U.S. Public Health Service Grant GM55425.
ABBREVIATIONS
- MRP
multidrug resistance-associated protein
- GSH
glutathione
- kb
kilobase
References
- 1.Dey S, Rosen B P. In: Drug Transport in Antimicrobial and Anticancer Chemotherapy. Georgopapadakou N H, editor. New York: Dekker; 1995. pp. 103–132. [Google Scholar]
- 2.Rosen B P. J Biol Inorg Chem. 1996;1:273–277. [Google Scholar]
- 3.Xu C, Zhou T, Kuroda M, Rosen B P. J Biochem. 1998;123:16–23. doi: 10.1093/oxfordjournals.jbchem.a021904. [DOI] [PubMed] [Google Scholar]
- 4.Dey S, Papadopoulou B, Haimeur A, Roy G, Grondin K, Dou D, Rosen B P, Ouellette M. Mol Biochem Parasitol. 1994;67:49–57. doi: 10.1016/0166-6851(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Dey S, Rosen B P, Rossman T G. Toxicol Appl Pharmacol. 1996;137:112–119. doi: 10.1006/taap.1996.0062. [DOI] [PubMed] [Google Scholar]
- 6.Cole S P, Sparks K E, Fraser K, Loe D W, Grant C E, Wilson G M, Deeley R G. Cancer Res. 1994;54:5902–5910. [PubMed] [Google Scholar]
- 7.Bobrowicz P, Wysocki R, Owsianik G, Goffeau A, Ulaszewski S. Yeast. 1997;13:819–828. doi: 10.1002/(SICI)1097-0061(199707)13:9<819::AID-YEA142>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 8.Mukhopadhyay R, Rosen B P. FEMS Microbiol Lett. 1998;168:127–136. doi: 10.1111/j.1574-6968.1998.tb13265.x. [DOI] [PubMed] [Google Scholar]
- 9.Wysocki R, Bobrowicz P, Ulaszewski S. J Biol Chem. 1997;272:30061–30066. doi: 10.1074/jbc.272.48.30061. [DOI] [PubMed] [Google Scholar]
- 10.Li Z S, Szczypka M, Lu Y P, Thiele D J, Rea P A. J Biol Chem. 1996;271:6509–6517. doi: 10.1074/jbc.271.11.6509. [DOI] [PubMed] [Google Scholar]
- 11.Szczypka M S, Wemmie J A, Moye-Rowley W S, Thiele D J. J Biol Chem. 1994;269:22853–22857. [PubMed] [Google Scholar]
- 12.Li Z S, Lu Y P, Zhen R G, Szczypka M, Thiele D J, Rea P A. Proc Natl Acad Sci USA. 1997;94:42–47. doi: 10.1073/pnas.94.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tommasini R, Vogt E, Fromenteau M, Hortensteiner S, Matile P, Amrhein N, Martinoia E. Plant J. 1998;13:773–780. doi: 10.1046/j.1365-313x.1998.00076.x. [DOI] [PubMed] [Google Scholar]
- 14.Tommasini R, Evers R, Vogt E, Mornet C, Zaman G J, Schinkel A H, Borst P, Martinoia E. Proc Natl Acad Sci USA. 1996;93:6743–6748. doi: 10.1073/pnas.93.13.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams A, Gottschling D E, Kaiser C, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Plainview, NY.: Cold Spring Harbor Lab. Press; 1998. [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Guthrie C, Fink G R. Methods Enzymol. 1991;194:1–933. [Google Scholar]
- 18.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothstein R J. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 20.Wemmie J A, Szczypka M S, Thiele D J, Moye-Rowley W S. J Biol Chem. 1994;269:32592–32597. [PubMed] [Google Scholar]
- 21.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 22.Dey S, Ouellette M, Lightbody J, Papadopoulou B, Rosen B P. Proc Natl Acad Sci USA. 1996;93:2192–2197. doi: 10.1073/pnas.93.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan R S, Pedersen P L. Anal Biochem. 1985;150:97–104. doi: 10.1016/0003-2697(85)90445-2. [DOI] [PubMed] [Google Scholar]
- 24.Sorin A, Rosas G, Rao R. J Biol Chem. 1997;272:9895–9901. doi: 10.1074/jbc.272.15.9895. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Reay P F, Asher C J. Anal Biochem. 1977;78:557–560. doi: 10.1016/0003-2697(77)90117-8. [DOI] [PubMed] [Google Scholar]
- 27.Ouellette M, Borst P. Res Microbiol. 1991;142:737–746. doi: 10.1016/0923-2508(91)90089-s. [DOI] [PubMed] [Google Scholar]
- 28.Mobley H L, Rosen B P. Proc Natl Acad Sci USA. 1982;79:6119–6122. doi: 10.1073/pnas.79.20.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasilyeva E A, Minkov I B, Fitin A F, Vinogradov A D. Biochem J. 1982;202:15–23. doi: 10.1042/bj2020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klionsky D J, Emr S D. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delnomdedieu M, Basti M M, Otvos J D, Thomas D J. Chem Biol Interact. 1994;90:139–155. doi: 10.1016/0009-2797(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 32.Leier I, Jedlitschky G, Buchholz U, Cole S P, Deeley R G, Keppler D. J Biol Chem. 1994;269:27807–27810. [PubMed] [Google Scholar]
- 33.Luker G D, Rao V V, Crankshaw C L, Dahlheimer J, Piwnica-Worms D. Biochemistry. 1997;36:14218–14227. doi: 10.1021/bi971931z. [DOI] [PubMed] [Google Scholar]
- 34.Sato T, Kobayashi Y. J Bacteriol. 1998;180:1655–1661. doi: 10.1128/jb.180.7.1655-1661.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneko T, Tabata S. Plant Cell Physiol. 1997;38:1171–1176. doi: 10.1093/oxfordjournals.pcp.a029103. [DOI] [PubMed] [Google Scholar]
- 36.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. Nature (London) 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 37.Ji G, Garber E A, Armes L G, Chen C M, Fuchs J A, Silver S. Biochemistry. 1994;33:7294–7299. doi: 10.1021/bi00189a034. [DOI] [PubMed] [Google Scholar]
- 38.Gladysheva T B, Oden K L, Rosen B P. Biochemistry. 1994;33:7288–7293. doi: 10.1021/bi00189a033. [DOI] [PubMed] [Google Scholar]
- 39.Fairlamb A H, Henderson G B, Cerami A. Proc Natl Acad Sci USA. 1989;86:2607–2611. doi: 10.1073/pnas.86.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borst P, Ouellette M. Annu Rev Microbiol. 1995;49:427–460. doi: 10.1146/annurev.mi.49.100195.002235. [DOI] [PubMed] [Google Scholar]
- 41.Fairlamb A H. Parasitology. 1989;99:93–112. doi: 10.1017/s003118200008344x. [DOI] [PubMed] [Google Scholar]
- 42.Grogl M, Thomason T N, Franke E D. Am J Trop Med Hyg. 1992;47:117–126. doi: 10.4269/ajtmh.1992.47.117. [DOI] [PubMed] [Google Scholar]
- 43.Chen Z, Wang Z Y, Chen S J. Pharmacol Ther. 1997;76:141–149. doi: 10.1016/s0163-7258(97)00090-9. [DOI] [PubMed] [Google Scholar]
- 44.Soignet S L, Maslak P, Wang Z G, Jhanwar S, Calleja E, Dardashti L J, Corso D, DeBlasio A, Gabrilove J, et al. N Engl J Med. 1998;339:1341–1348. doi: 10.1056/NEJM199811053391901. [DOI] [PubMed] [Google Scholar]
- 45.Bowman S, Ackerman S H, Griffiths D E, Tzagoloff A. J Biol Chem. 1991;266:7517–7523. [PubMed] [Google Scholar]