Abstract
We describe a detailed time course of the assembly and disassembly of a STAT3-dependent, glucocorticoid-supplemented enhanceosome for the α2-macroglobulin (α2-M) gene and compare this with a detailed time course of transcription of the gene by run-on analysis. The glucocorticoid receptor (GR) can associate with the enhanceosome without STAT3. Furthermore, the enhanceosome contains c-Jun/c-Fos and OCT-1 constitutively. All of these factors (GR, c-Jun, OCT-1) have transcription activation domains, but STAT3 is required for the observed transcriptional increase. The time course of enhanceosome occupation by GR and tyrosine-phosphorylated STAT3 shows that these transcription factors precede by ∼5-10 min the arrival of RNA polymerase II (Pol II). The enhanceosome remains assembled for ∼90 min in the continued presence of both inducers. When IL-6 and Dex are removed (after 30 min of treatment), the disappearance within an additional 30 min of the established enhanceosome indicates that renewal of STAT3 and GR binding must occur in the continued presence of IL-6+Dex. Compared with the total nuclear tyrosine-phosphorylated STAT3 capable of binding DNA, the chromatin-associated STAT3 resists dephosphorylation and appears to recycle to maintain the enhanceosome. Run-on transcription shows a lag after full enhanceosome occupation that can be largely but not completely explained by the ∼30 min transit time of Pol II across the α2-Mlocus.
Keywords: STAT3, GR, enhanceosome, ChIP, transcription rate
It is well established that most cytokines and many growth factors elicit increased transcription (often transiently) through activation of one of the seven STATs (signal transducers and activators of transcription). As revealed by mouse genetics and by extensive gene expression studies, each STAT has a separate physiologic function and each activates a different set of genes (Horvath 2000; Levy and Darnell 2002). However, STATs 1, 3, 5A, and 5B can each be activated by a wide array of extracellular proteins or peptides. The target genes for any of the STATs following different extracellular ligand stimulation are to some degree constant (regardless of ligand or cell type) and to some degree variable (e.g., in different cell types). Very likely, most of the regulatory sequences in the STAT-responsive genes, as is true for many other genes, associate together with several other transcription factors in enhanceosomes (Grosschedl 1995; Carey 1998; Lomvardas and Thanos 2002). Such clusters could be similar or vary among the genes dependent for activation by a particular STAT allowing for the constant or differing responses to individual STAT activation. However, no details of the assembly and disassembly of a STAT-dependent enhanceosome or the timing of its effect on the steps of transcriptional induction have been described. In fact, no detailed analysis of the time course of enhanceosome assembly and disassembly on chromosomal genes compared with a detailed measurement of transcription by run-on analysis in nuclei has been described.
To make a beginning at describing STAT-dependent enhanceosome function, we (Zhang et al. 1999; Zhang and Darnell 2001) have been analyzing the joint IL-6 and dexamethasone (Dex) induction of the α2-macroglobulin (α2-M) gene in a rat hepatoma cell line (Geiger et al. 1988; Northemann et al. 1988; Hattori et al. 1990; Hocke et al. 1992; Wegenka et al. 1993; Schaefer et al. 1995). This gene was chosen because it is activated by IL-6 through action of STAT3 and because STAT3 has gained attention as a persistently active and necessary factor for human cancer cells to avoid apoptosis (Darnell 2002). Furthermore, at this locus, STAT3 cooperates with GR, induced by Dex, to achieve maximum accumulation of α2-M mRNA in cultured hepatoma cells, affording the chance to study the cooperative interaction of these two transcription factors.
Previous and current transfection experiments (Hattori et al. 1990; Wegenka et al. 1993; Schaefer et al. 1995; Zhang and Darnell 2001) have identified numerous protein-binding sites within the α2-M enhancer/promoter region, two for STAT3, two for AP-1, one for OCT-1, and potentially one for SOX9. However, we find no binding site for GR. Because GR can be demonstrated to bind proteins in the enhanceosome (c-Jun and STAT3), we infer that it is held at the promoter through protein:protein interaction.
We describe the several proteins that are associated with the chromosomal α2-M enhanceosome and analyze the time course of assembly, disassembly, and stability of the enhanceosome. The evidence shows clearly that STAT3 is required for transcriptional activation and that GR alone does not increase transcription but serves, without binding DNA, to boost transcription.
Results
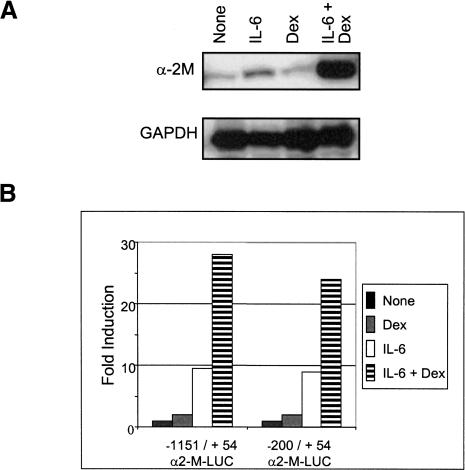
The rat α2-macroglobulin (α2-M) gene is induced in the liver by IL-6 during the so-called acute phase response (caused by injury of various kinds). The induction can be mimicked by IL-6 treatment of hepatoma cells in culture (Lutticken et al. 1995; Heinrich et al. 1998). We have used a rat hepatoma cell line (H-35), which showed an increase in α2-M mRNA accumulation when treated with IL-6 only, which activates STAT3 (Fig. 1A). Treatment with dexamethasone (Dex) alone, which activates the glucocorticoid receptor (GR), produced little or no increase in mRNA level. However, greatly increased accumulation of α2-M mRNA occurred when the IL-6-treated cells received Dex as well (Fig. 1A). That the proteins required for this induced mRNA increase are present in cells prior to stimulation was demonstrated by the approximately equal mRNA accumulation in induced cells treated or not treated with cyclohexamide (data not shown). In transfection experiments with H-35 cells, two upstream reporter constructs containing sequences spanning either -200 to +54 or -1151 to +54 of the promoter, produce equivalently induced luciferase transcription signals, in agreement with earlier experiments (Fig. 1B; Hattori et al. 1990; Heinrich et al. 1998). Again, as with the chromosomal gene, Dex combined with IL-6 enhanced the luciferase signal by several-fold for each construct. That the accumulation of α2-M mRNA from the chromosomal gene is accompanied by increased transcription was verified by run-on transcription analysis (see time course of run-on transcription in Fig. 6, below).
Figure 1.
Synergistic effect of IL-6 and Dex on rat α2-M gene expression. (A) H-35 cells were treated with IL-6, Dex, or IL-6+Dex for 4 h followed by RT-PCR analysis for α2-M and GAPDH mRNA. (B) H-35 cells were transfected with a luciferase reporter construct (either -1151/+54 or -200/+54 of the upstream region of α2-M). Then, 24 h posttransfection, cells were left untreated or treated as indicated for 6 h. The luciferase activity was normalized against the internal β-Gal activity control. Means of three experiments are shown.
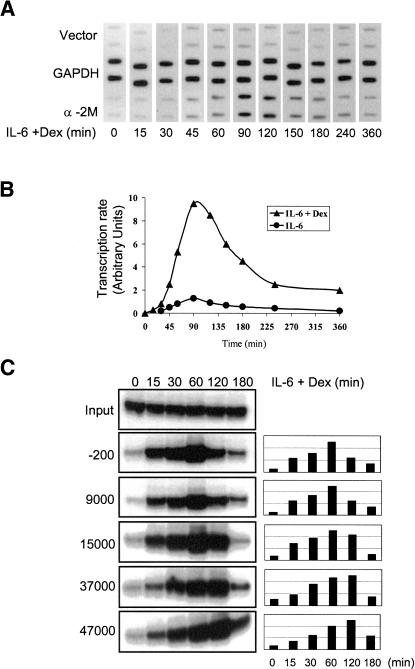
Figure 6.
Transcription rate and RNA Pol II occupancy of α2-M locus. (A) Run-on analysis of H-35 cells treated with IL-6+Dex for the various times indicated. Nuclei were labeled with [α-32P]UTP and labeled RNA hybridized to full-length α2-M cDNA. The autoradiogram of the hybridization filter is shown with duplicate slots on each filter for α2-M and for GAPDH cDNAs as a control. (B) Quantitation of hybridization images carried out by PhosphorImaging. The baseline (empty vector signal) was subtracted, and intensity was scored in arbitrary units. (C) Transit of RNA Pol II across the α2-M locus after induction. Cells treated with IL-6+Dex for indicated periods of time were used in ChIP analysis for RNA Pol II on segments of the α2-M locus. The -200 is the promoter and RNA start site fragment (-200 to +54); other primer coordinates were 9000 (9116-9404), 15,000 (14,867-15,067), 37,000 (37,368-37,678), and 47,000 (47,444-47,698).
These initial experiments justified exploring the α2-M chromosomal gene during transcriptional activation to study the assembly and disassembly of a STAT3-dependent enhanceosome whose maximal activity also depended on the presence of the GR.
Proteins that bind sequences in the α2-M promoter
Through the use of transfection analysis of reporter constructs, the original mutagenesis of the α2-M promoter uncovered a STAT3 site (-165 to -158) and an apparent AP-1 site (-108 to -102; Schaefer et al. 1997). We reported a noncanonical second STAT3 site (a half site, -187 to -179) that acts together with the original site to allow STAT3 dimer:dimer formation and a consequent increase of the signal from a transfected gene (Zhang and Darnell 2001). Two additional AP-1 sites (-147 to -141, AP1H and -194 to -188, AP1H2; Figs. 2A, 3A) were also suggested by sequence analysis. However, by DNA-binding analysis (EMSA), only the canonical AP-1 site and the AP1H site gave evidence of c-Jun binding (data not shown). In vitro footprint analysis (DNase I) with recombinant c-Jun, which forms one of the possible b-zip dimers that bind AP-1 sites (Fig. 2A; Karin et al. 1997), showed clear protection with surrounding hypersensitive sites in the canonical region site (-108 to -102). Both the other possible sites gave only weak footprints. Because the AP1H site did give evidence by EMSA of DNA binding, we performed mutagenesis analysis at this site in addition to the canonical site (see below).
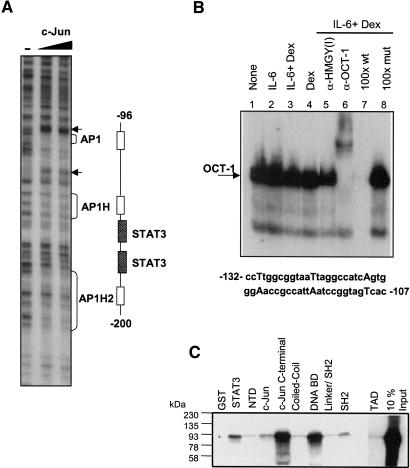
Figure 2.
Proteins in the α2-M enhanceosome. (A) c-Jun binds AP-1 sites. In vitro DNase I footprint analysis of the 5′-end-labeled α2-M DNA fragment using recombinant c-Jun protein showed strong interaction around -110 to -100 with flanking hypersensitive sites (arrows). Weaker interactions occurred around -150 to -140 and -200 to -150 (brackets). (B) EMSA of nuclear extract (NE) from H-35 cells were incubated with an OCT-1 radiolabeled probe. Use of antisera against OCT-1 and 100 M excess of the wild-type probe demonstrated OCT-1 binding to this region of the α2-M promoter. (C) In vitro translated full-length GRα interacted weakly with GST-fused full-length c-Jun and strongly with the c-Jun:C-terminal fusion product. GST-fused STAT3 and various domains of STAT3 fused to GST were interacted with full-length labeled GR. Full-length STAT3 interacted weakly and the STAT3 DNA-binding domain (amino acids 320-495) interacted strongly. Proteins bound to GST on beads were eluted and analyzed by 10% SDS-PAGE and radiography.
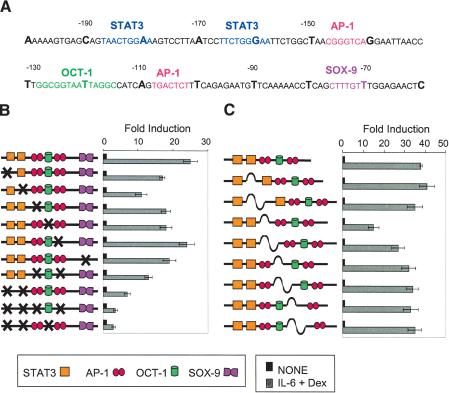
Figure 3.
Transfection analysis of mutagenized rat α2-M promoter. (A) Sequence of the α2-M promoter from -200 to -60 nt. Transcription factor binding sites are colored. (B) Fold induction of the (-1151/+54) α2-M-luciferase reporter containing mutations in different activator binding sites. H-35 cells were transfected with β-Gal vector and different mutants of rat α2-M promoter upstream of the luciferase reporter (-1151/+54). After 24 h, cells were treated with IL-6+Dex for 6 h, and the luciferase signal was scored and normalized against β-Gal. (C) Dependence on helical phasing at the α2-M promoter. Transfection with wild-type and mutant constructs [insertions of one-half of a helical turn (5 bp) or a full helical turn (10 bp)] as in B. The results in B and C are the means of data from three independent experiments.
Using 30-bp segments of the α2-M promoter (illustrated in Fig. 3A) to screen by EMSA for other binding proteins in either induced or uninduced cells, we found two other regions that bound proteins. One segment (-129 to -105) contained a site that resembled the canonical OCT-1 site. Indeed, the EMSA band formed by a protein constitutively bound to this site was supershifted with OCT-1 antiserum (Fig. 2B, lanes 1,6). Likewise, based on preliminary results, a representative of the SOX family of proteins, SOX9, may bind to a site at -76 to -70 (data not shown). There was no difference in the binding to the OCT-1 or potential SOX9 sites in extracts from IL-6-treated cells compared with untreated cells.
No DNA-binding site was found for purified GR either with or without added ligand, and aside from the OCT-1 and AP-1 sites, no binding occurred in the presence of nuclear extracts from Dex-treated cells. Precedent exists in transfection experiments with the casein promoter for cooperation of GR in STAT5-dependent transcription without GR binding to DNA (Stocklin et al. 1996). In this promoter, mutation of a GR-binding site blocked binding but did not remove the extra stimulation that Dex added to prolactin on transcription assayed by transfection.
That an interaction between STAT3 and GR can occur in vitro was detected in pull-down experiments (Fig. 2C). In vitro synthesized, radiolabeled GRα interacted weakly with a GST fusion of full-length STAT3 and more strongly with a GST fusion of residues 320-495 of STAT3, which embraces the DNA-binding domain. The labeled GRα also interacted with the C-terminal half of c-Jun, as has been previously demonstrated (Schaefer et al. 1995). We have shown earlier that c-Jun interacts with STAT3 (residues 130-358). Mutations in the STAT3 coiled-coil domain and in the DNA-binding domain disrupted the STAT3-c-Jun interaction (Zhang et al. 1999). Thus, the GR could participate in α2-M activation by binding to either STAT3 or c-Jun or to both.
Functional analysis of the α2-M promoter by mutagenesis and transfection
We next undertook mutagenesis of binding sites in the α2-M promoter in an attempt to judge from transfection analysis which of the binding sites for putative positive-acting factors discussed above may have relevance in activation of transcription of the chromosomal gene. A panel of mutated constructs that were tested by transfection for response to IL-6+Dex is shown on the left in Figure 3B. The single mutant site with the greatest effect on the transcriptional signal was the canonical STAT3 site, but mutation of this site only lowered the signal by ∼60% (Fig. 3B). Mutations of both STAT sites together decreased the signal by 75%, and ∼40% suppression was observed with mutation of both AP-1 sites. Only a 30% depression of signal was observed with mutation of the OCT-1 or the putative SOX9 sites alone. The most severe suppression of the transcriptional signal (∼90%) occurred with simultaneous mutation of both STAT3 sites plus the two AP-1 sites or the two STAT3 plus the OCT-1 site.
That a functional cooperation could be caused by an AP-1/STAT3 interaction was illustrated further by increasing the spacing by 5 bases between the canonical STAT3 and AP-1 sites, thereby altering the helical phasing between these two binding sites. This insertion suppressed transcription by ∼60% (Fig. 3C). The restitution of the helical phasing (10-bp insertion) at the same site partially rescued the transcription of the reporter. No other insertions tested had a significant effect in the transfection analysis.
Recruitment of components to the α2-M enhanceosome
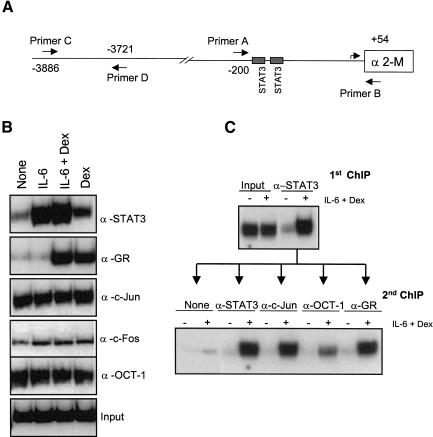
Chromatin immunoprecipitation (ChIP) experiments (Strahl-Bolsinger et al. 1997) have recently achieved great currency in attempts to determine specific DNA:protein and protein:protein associations in enhanceosome regions in the native chromatin of whole cells (Shang et al. 2000; Agalioti et al. 2002; Edelstein et al. 2003). Treating whole cells with formaldehyde fixes protein:DNA complexes as well as associated proteins in enhanceosomes not bound to DNA. In addition, polymerases can be fixed to DNA both at the RNA initiation site and at a distance from it (Cheng and Sharp 2003). Sonication fractures the fixed chromatin, maintaining protein:DNA and protein:protein associations; specific antibody precipitation followed by the release of DNA from cross-linked proteins then allows PCR identification of DNA in the precipitate. Such analysis at the α2-M promoter with antibodies against STAT3, GR, c-Jun, c-Fos, and OCT-1 was carried out using PCR primers that bracket the α2-M promoter and RNA start site (-200 to +54; Fig. 4A). Chromatin accumulation of STAT3 after IL-6 treatment or of GR after Dex treatment by 30 min was clear (Fig. 4B). Joint IL-6 and Dex treatment resulted in significantly more accumulation of both STAT3 and GR (Figs. 4B, 5B). The AP-1 DNA-binding site(s) that was identified (Fig. 2A) could theoretically bind many different proteins, among which are c-Jun:c-Jun dimers or c-Jun:c-Fos heterodimers. Without IL-6 or Dex treatment, c-Jun and c-Fos antisera precipitated the α2-M promoter (Fig. 4B) but not a distant site using primers C and D (data not shown). Given that we have previously shown, using the same cells, that supplemental c-Jun but not c-Fos stimulated the transcription signal in transfection experiments with α2-M reporter constructs, c-Jun dimers might be the most effective α2-M activator (Zhang et al. 1999). However, both c-Fos and c-Jun could be considered constitutively present on this promoter based on chromatin precipitation. OCT-1 was also present without ligand treatment. Therefore, OCT-1 is also a constitutive member of the α2-M enhanceosome.
Figure 4.
Recruitment of proteins to the α2-M promoter. (A) Schematic representation of the α2-M promoter -4000 nt to +400 nt from the transcription-starting site. The α2-M box represents the beginning of the coding region, and two gray rectangles represent STAT3-binding sites. The inverted arrow pairs indicate the primers used in the ChIP assays shown below. (B) Soluble chromatin was prepared from H-35 cells treated with IL-6, Dex, or IL-6+Dex for 30 min and immunoprecipitated (IP) with the indicated antibodies. (C) Double immunoprecipitation ChIP assay. Soluble chromatin was prepared from H-35 cells treated or not with IL-6+Dex for 60 min and immunoprecipitated with antibodies against STAT3 (first IP) as in B. The immunoprecipitates were washed, resuspended, disrupted, and immunoprecipitated a second time with antibodies against STAT3, GR, c-Jun, or OCT-1 (second IP). The final DNA extractions were amplified using the primers A and B that cover the regions of the α2-M enhancer. No signal was obtained with primers C and D (data not shown).
Figure 5.
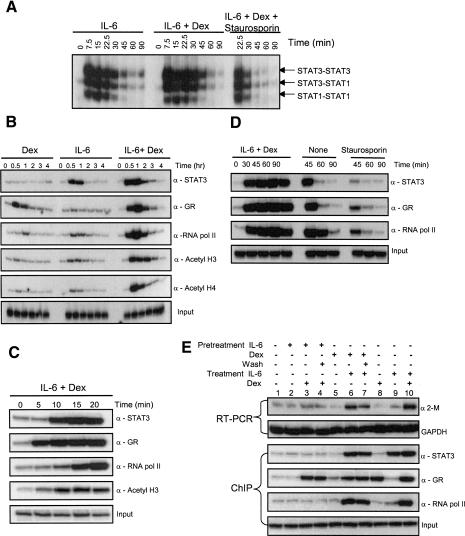
Dynamics of α2-M enhanceosome occupancy. (A) Time course of IL-6-induced STAT3 activation and inactivation. NE from H-35 cells treated with IL-6, IL-6+Dex, or the addition of a kinase inhibitor, staurosporin (500 nM), after 15 min of IL-6+Dex treatment, were used in EMSA experiments with an m67 radiolabeled probe. Arrows mark the STAT3 homodimer, STAT1:3 heterodimer, and STAT1 homodimer. (B,C) The time course of the recruitment of STAT3, GR, RNA Pol II, and acetylation of histones at the α2-M enhanceosome was determined using ChIP. (D) α2-M enhanceosome disassembly. ChIP analysis for presence of STAT3, GR, or RNA Pol II during various times of IL-6+Dex treatment. IL-6 and Dex were removed after 30 min for panel marked NONE and for panels marked Staurosporin, where staurosporin (500 nM) was also added. (E) GR cooperation persists after Dex removal. H-35 cells were pretreated for 15 min with IL-6 or Dex. Medium was removed or washed with serum-free medium. Then, cells were treated for 90min with IL-6 and/or Dex and subjected to either RT-PCR or ChIP analysis.
To test the simultaneous in vivo presence of all the possible components of the α2-M enhanceosome, a primary and secondary ChIP analysis was performed on H-35 cells untreated or treated with IL-6+Dex for 60 min. After a first cycle of immunoprecipitation with STAT3 antibody, the antibody-chromatin complex was disrupted by the addition of a reducing agent to the mixture as previously described (Shang et al. 2000). After the appropriate dilution of the reaction mixture, a second immunoprecipitation was carried out: STAT3 antisera served as a positive control and no antibody as a negative control; c-JUN, OCT-1, and GR antisera (Fig. 4C) were tested for the presence of each of these proteins together with STAT3 on the α2-M promoter. STAT3, c-JUN, OCT-1, and GR antisera precipitated the promoter sequences in the secondary precipitation, demonstrating the presence of each of these proteins bound to the α2-M chromosomal promoter together with STAT3 after IL-6+Dex treatment.
Timing of events at the induced α2-M promoter
Many more STAT molecules are activated by cytokine treatment than actually participate in transcription (Haspel et al. 1996; Haspel 1998). We therefore determined by EMSA the time course of activation of total nuclear STAT3 by IL-6 before determining the time course of chromatin-bound STAT3. Maximal activation was at 7.5 min with <10% remaining at 45 min (Fig. 5A). The three EMSA bands are STAT3 dimers, STAT1 dimers, and STAT1:STAT3 heterodimers. The basis for this rapid turnover of total activated (dimeric) STAT3 is presumably a balance between stopping cytoplasmic activation (e.g., by SHP phosphatases and by action of induced SOCS proteins) and nuclear dephosphorylation (for review, see Starr and Hilton 1999; Levy and Darnell 2002). To estimate the speed of turnover in the absence of any further activation, staurosporin was added after 15 min of IL-6+Dex treatment. The almost complete loss of EMSA signal (i.e., tyrosine-phosphorylated STAT3) between 22.5 and 30 min indicates a half-life of the dimer is no more than 5-10 min, which is in accord with the 7.5-min peak of activation (Fig. 5A).
We next determined by ChIP the time course of STAT3 and GR appearance on chromatin at the promoter site (Fig. 5B,C). Treatment with IL-6 alone showed accumulation of STAT3 by 30 min that peaked between 30 and 60 min and was diminished greatly by 120 min (Fig. 5B). Chromatin precipitation assays with GR antibody in cells treated with Dex alone showed a transient chromatin accumulation at 30 min with a diminution by 60 min and virtual disappearance by 120 min. In cells treated with both IL-6 and Dex, maximal accumulation of both STAT3 and GR was also between 30 and 60 min (Fig. 5B). However, there was much greater accumulation of both STAT3 and of GR, and both remained at a high level through 90 min (Fig. 5D). This persistence on the enhanceosome significantly outlasted the available total nuclear pool of activated STAT3, which had virtually disappeared by 45 min (Fig. 5A). Thus, the STAT3 that becomes bound to chromatin is more resistant to tyrosine dephosphorylation that removes the majority of nuclear DNA-binding STAT3 activity by 45 min. (We return to the stability of the enhanceosome below.)
To test the accumulation of STAT3 and GR on the enhanceosome at times shorter than 30 min, we treated cells with IL-6+Dex and examined the enhanceosome association at 5-min intervals (Fig. 5C). GR, which by itself does not stimulate transcription, was clearly present at 5 min (20%-30% of the 20-min value), whereas only a trace of STAT3 was present at 10 min. By 10 min, GR had increased and STAT3 had appeared (∼50% of the 20-min amount). Histone H3 acetylation paralleled the accumulation of STAT3 and GR.
Given the time course of formation and the duration of enhanceosome assembly, we wished to determine the time course of RNA polymerase II accumulation at the promoter site and also the time course of transcription of the gene by run-on analysis. ChIP analysis of RNA Pol II on the promoter was initially carried out at longer times (Fig. 5B; from 30 min to 4 h). In these experiments, recruitment of Pol II by STAT3 (i.e., after IL-6 treatment) was greater than by GR alone, but recruitment of Pol II in the presence of both GR and STAT3 was substantially increased and was maximal at 60 min. However, we also examined Pol II on the promoter after short intervals of IL-6+Dex treatment (Fig. 5C; 5, 10, 15, and 20 min). Very little Pol II was associated with the promoter until ∼15 min. Thus, there was a distinct lag between the appearance of GR and STAT3, and only after STAT3 appeared did Pol II accumulation begin.
Dynamics of the α2-M enhanceosome
We tested the stability of the α2-M enhanceosome by allowing its assembly and then removing IL-6 and Dex or removing both with the addition of a kinase inhibitor, staurosporin, which quickly stops any additional STAT phosphorylation (Haspel and Darnell 1999). Chromatin association at the promoter of STAT3, GR, and RNA Pol II was then assayed (Fig. 5D). In the continuous treatment sample, maximal accumulation of all three occurred (as noted earlier, Fig. 5B) after 60 min with only a small decrease by 90 min. When both activators were removed after 30 min of treatment (panel labeled NONE), a small decrease was seen at 45 min, but both STAT3 and GR were largely gone by 60 min. However, RNA Pol II was still present at (or near) the promoter at 60 min. The Pol II that was still present at 60 min when the enhanceosome was essentially unoccupied was paused either at the promoter or conceivably had just entered into transcription. In carrying out ChIP analysis, chromatin fragments averaging 500 nt of DNA are produced, and the PCR in our ChIP experiments used primers from -200 to +54. Polymerase molecules that were beginning to transcribe the α2-M gene but had not passed 250-300 nt could be precipitated. Thus, it is clear that the Pol II associated with the -200 to +54 fragment definitely does not immediately “clear” the promoter (on average the Pol II rate is 30 NT/sec; 10 sec of full transcription would remove Pol II from the promoter region; see later discussion notes). By 90 min after IL-6+Dex removal, the RNA Pol II detected with the -200 to +54 primers was greatly diminished, indicating that all recruited polymerases had cleared the promoter between 60 and 90 min.
When staurosporin was added after 30 min (coincident with removal of both IL-6 and Dex), STAT3 was largely gone from the promoter by 45 min while about one-third of the GR remained associated. But over half of the Pol II had departed the promoter by 45 min, and the great majority was gone by 60 min, again emphasizing the importance of STAT3 in Pol II recruitment to the enhanceosome region.
These results (Fig. 5D) show that the maintenance of full (or near full) occupation of the enhanceosome for 90 min in the continuous presence of IL-6+Dex requires renewal of bound STAT3 and GR during this 90-min time period. The staurosporin results suggest a short lifetime for activated STAT3 on the enhanceosome (∼15 min) and a somewhat longer lifetime for GR even in the absence of STAT3. Furthermore, the results (Fig. 5D) support the argument that promoter clearance requires at least 10-15 min.
The experiments of Figure 5B and C indicate that GR can bind the α2-M enhanceosome without STAT3 being present, and we wished to test whether such chromatin-bound GR would, if furnished STAT3, stimulate transcription and mRNA accumulation. Cells were pretreated with either IL-6 or Dex for 15 min or left untreated; the inducers were removed and then after restoration of the opposite inducer were treated for a total of 90 min. Cells were then monitored for mRNA levels by reverse transcriptase PCR (RT-PCR) or for chromatin-bound STAT3, GR, or RNA Pol II. In control cells treated continuously with IL-6, Dex, or both, mRNA accumulation was strong after joint treatment, weakly increased after IL-6 alone, but not after Dex alone (Fig. 5E, lanes 8-10). In cells pretreated with IL-6, and the medium then replaced with fresh medium lacking IL-6, there was little or no α2-M mRNA increase stimulated by Dex (Fig. 5E, lanes 2-4). If the pretreatment was with Dex, a lasting effect was seen when Dex was removed and cells were treated with IL-6. In the Dex-pretreated cells, IL-6, now in the absence of Dex, elevated mRNA to a level similar to (slightly less than) combined IL-6+Dex treatment. ChIP assays showed that whereas IL-6 removal caused almost complete lack of active STAT3 by 90 min, there was still GR present in the initially Dex-treated cells that had subsequently been IL-6 treated. These experiments definitely support the idea that GR can remain nuclear and potentially chromatin associated for some time after Dex removal and that STAT3 can capture the functioning of this residual GR. This interaction of the joint partners stabilizes the nuclear STAT3 (Fig. 5E) and leads to transcriptional increase. This experiment is further evidence not only of the dominance of STAT3 in initiating transcription, but also of the importance of synergism with GR for maximal transcriptional increase.
Timing of transcription by run-on analysis
To relate the time course of appearance of inducible transcription factors on the α2-M enhanceosome to actual transcription, as measured by nuclear run-on analysis, we examined the time course of transcriptional increase after either IL-6 treatment alone or IL-6+Dex treatment (Fig. 6B). Using the entire 4.5-kb cDNA complementary to the α2-M mRNA to hybridize labeled “run-on” nuclear RNA, an extensive series of run-on analyses clearly showed that the transcription rate was detectably elevated by 45 min but required between 90 and 120 min to achieve a maximal signal (Fig. 6A,B), which thereafter declined. However, as noted before (Fig. 5B,D), the maximal appearance of STAT3 and GR on the -200 to +54 chromatin segment had definitely occurred by 30-60 min and even in continued presence of IL-6+Dex had decreased greatly by 120 min.
Although the transcription of the α2-M sequences was raised by IL-6 alone by only about twofold above background (Fig. 6B), the time course of elevation was very similar to the maximum elevation owing to combined IL-6+Dex treatment (Fig. 6A; no signal above background was obtained with Dex alone). In either case, there was a distinct delay between the maximal accumulation of activators and Pol II on the -200 to +54 promoter region compared with maximal transcription scored by run-on.
Transcription across the ∼50-kb α2-M locus
The maximal run-on signal from an induced gene requires the maximal number of initiated polymerases on a locus. The time to reach a maximal signal is controlled in part by the time required to transit the whole gene. The rat α2-M locus is ∼50 kb (42 introns with similar spacing between exons; rat database access: NW_0436 or gi: 26007627), which would require a Pol II transit time of ∼16-40 min based on several earlier estimates of the Pol II synthesis rate (20-50 NT/sec; Miyazaki et al. 1976; Sehgal et al. 1976; Jackson et al. 2000). To further explore the delay between maximal enhanceosomal occupancy and maximal transcription signal, we followed the transit of RNA polymerase across the locus by ChIP (Fig. 6C). ChIP analysis with Pol II antibody at various times following induction was carried out using primers located at the promoter site and approximately at 9, 15, 37, and 47 kb from the start site. The intensity of signals (RNA Pol II presence) at the various segments of the transcription unit at each time point after induction was then determined (Fig. 6C). The presence of polymerase at the promoter (-200 to +54; labeled -200 in Fig. 6C) was significant (25%-30% of maximal) by 15 min and was maximal at 60 min with a sharp decline by 120 min.
The experiment shows a wave of polymerase movement across the α2-M locus. At the earliest time (15 min), the Pol II was already associated with the promoter and the 9- and 15-kb sites but very little beyond this segment. By 30 min, polymerases were present at all sites across the locus, but neither the enhanceosome nor the gene segments were fully loaded. By 60 min, the polymerase was maximally loaded on gene segments but still gave a high signal on the -200 to +54 segment (as seen in Fig. 5). Such a distribution of polymerases should produce a maximal transcription signal by this time or shortly after. By 120 min, there was a decline of Pol II at the enhancer and a distinct shift in distribution of the remaining Pol II toward the more distant sites on the gene. This overall picture suggests that the strongest transcription should occur at 60 min or shortly thereafter and decline by 120 min, which was seen (Fig. 6A,B). This experiment shows that on a 50-kb locus, there is displacement (delay) of maximal rate of transcription to a later time than maximal loading of Pol II on the enhancer that must be caused by the transit time of the polymerase.
Discussion
For some time it has been known that IL-6+Dex led to accumulation of α2-M mRNA (Hocke et al. 1992). The present experiments establish that transcriptional induction of the chromosomal α2-M gene in response to IL-6+Dex treatment of rat hepatoma cells is regulated through a STAT3-dependent enhanceosome. Prior to induction of transcription, c-Jun (and probably c-Fos) and OCT-1, all factors capable of positive transcriptional activation, are present on the chromatin. Furthermore, GR can be recruited to join the constitutive factors without stimulating transcription. However, after IL-6 stimulation and STAT3 appearance, there is an increase in transcription (see model in Fig. 7).
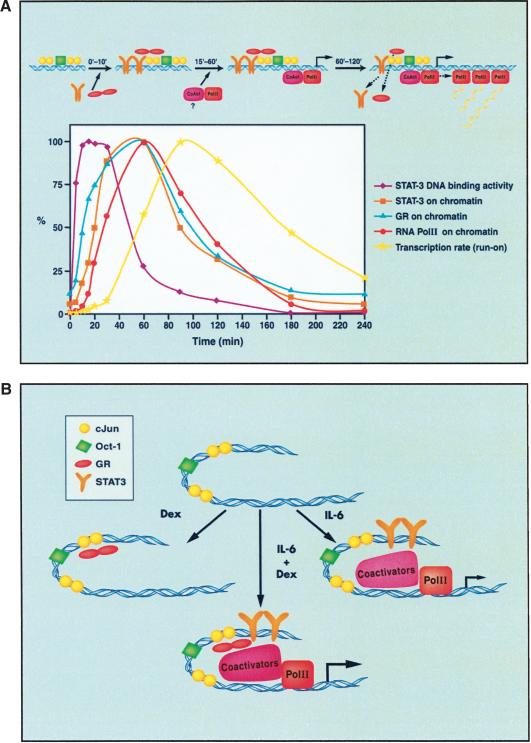
Figure 7.
Model of α2-M enhanceosome assembly and disassembly. (A) Intensity signal scans of STAT3, GR, and RNA Pol II in the ChIP analyses in Figure 5 were taken. For the transcription rate, the data of Figure 6 were used. The maximal signals were scored as 100%, and the results of all samples were then plotted. (B) The sequential formation of complexes leading to the activation of the α2-M gene. OCT-1 and c-Jun bind constitutively the α2-M promoter. Dex-activated GR is recruited to the complex, possibly by interacting with c-Jun. IL-6 treatment recruits STAT3 to the complex, and STAT3 alone is competent to recruit Pol II and stimulate low levels of transcription. The combined treatment of IL-6+Dex results in the recruitment of both STAT3 and GR for a more effective recruitment of RNA Pol II, achieving maximal transcription.
Treatment of cells with IL-6+Dex induces the following sequence of events (Fig. 7). Within 5 min, GR enters the nucleus and associates with the enhancer/promoter region, quite possibly through association with c-Jun (see Fig. 2C; Zhang et al. 1999) or some other enhanceosomal constituent, because there is no GR-binding site in the enhanceosomal DNA. Tyrosine-phosphorylated STAT3 capable of binding DNA rapidly accumulates in the nucleus (peaking by 7.5 min) and by 10 min is clearly present on the α2-M enhanceosome (Fig. 5). The amount of Pol II recruited to the enhanceosome is definitely increased by dual IL-6+Dex treatment (Fig. 5B) compared with either inducer alone. Acetylation of histone H3 in the enhanceosome is stimulated slightly by 5 min and strongly by 10 min, implying that GR and/or STAT3 attract histone acetyl transferases (HATs). One likely candidate is CBP/p300, although other HATs could be involved (Paulson et al. 1999).
The time course of recruitment of pol II by STAT3 and GR is of particular importance with respect to the time of transcriptional induction. RNA Pol II appears in the enhanceosome at least 5-10 min later than GR and STAT3, implying a lag time is required for GR/STAT3/coactivator interaction to capture Pol II. At present, we know nothing further about cofactors other than that signified by the induction of histone acetylation. Perhaps the most widely used coactivator in transcription initiation is the multiprotein complex, “mediator” (Malik and Roeder 2000; Myers and Kornberg 2000; Rachez and Freedman 2001). This complex has been known for some time to associate with steroid receptors (Fondell et al. 1996). Recently a mediator-STAT2 interaction has been described (Lau et al. 2003), and we have found an enhancement of STAT1-dependent in vitro transcription by the mediator (Zakharova et al. 2003). Therefore, it seems very likely that the mediator complex might be involved in α2-M transcriptional initiation. (Available mediator antisera have not enabled us to show such association with this gene by ChIP.) At any rate, there is a definite lag between the appearance on the enhanceosome of GR and STAT3 before RNA Pol II arrives.
We continued the kinetic analysis of transcriptional induction up to 4 h (Fig. 5) and repeatedly found only a small (<2×) increase of GR, STAT3, and Pol II in the enhanceosome between 30 and 60 min and a decrease thereafter. Thus, the period of maximal potential for initiating transcription has been reached by 60 min but is near maximal from 30 to 90 min (Fig. 5). There is then a spontaneous decrease in enhanceosomal factor binding that is virtually complete between 2 and 3 h despite continued presence of inducers in the medium.
The stability of the induced enhanceosome was tested by removing the IL-6 and Dex and/or by using a kinase inhibitor, staurosporin, to halt further STAT3 activation. From the sharp decline of enhanceosome occupancy upon IL-6+Dex removal (Fig. 5D), it is obvious that in the continued presence of IL-6+Dex, the enhanceosomal binding of STAT3 and GR must be renewed every 15-30 min during the 30-90-min interval, while the enhanceosome is occupied at near maximal levels. It is also clear (Fig. 5D) that in the absence of STAT3 and GR, polymerase remains on the promoter/enhancer region for a time (∼15 min), implying some delay before transcription actually begins.
Several additional points of interest come from these time-course experiments. First, consider the pool of available, activated nuclear STAT3. This pool was maximal at 7.5 min, remained high for 30 min, declined greatly by 45 min, and was essentially background at 60 min (Fig. 5A). The maximum enhanceosomal occupation by STAT3 at 60 min and its high maintenance through 90 min (Fig. 5D) in the face of the diminished total nuclear STAT3 pool suggest that (1) the DNA-bound STAT3 resists dephosphorylation and (2) the chromatin-associated molecules of active STAT3 may cycle on and off the promoter at least once or twice before being dephosphorylated. Although there are ample mechanisms known that limit the time of STAT activation (Levy and Darnell 2002), even though a cytokine is still present in the medium, less is known about events that limit GR nuclear (or enhanceosome) lifetime. In one well-studied system, another steroid receptor, that for estrogen, bound to chromatin of an induced gene in a cyclic fashion—appearance, disappearance, and reappearance—in the continued presence of estrogen (Shang et al. 2000). In still another case, androgen receptor bound to chromatin in the prostate-specific antigene (PSA) gene is present in two separate sites between 45 min and 3 h and then is no longer bound. In both those cases, the gene activation demands steroid receptor binding of DNA (Shang et al. 2002). These cases are distinctly different from the case of the α2-M gene, in which no DNA binding of GR is involved and STAT3 has a limited lifetime, leading to disassembly of the α2-M enhanceosome. One other comment on the α2-M enhanceosome is pertinent. At least four proteins in this structure—c-Jun, OCT-1, STAT3, and GR—have transcriptional activation domains (TADs). Three—c-Jun, OCT-1, and GR—can all be present without activating transcription of the α2-M gene. Does that mean that when STAT3 arrives it is only the STAT3 TAD that recruits other factors? That seems less likely than the possibility that a final rearrangement of the four proteins allows presentation of other TADs (at least those in GR). Of course, it remains possible that some negative-acting factor present before either IL-6 or Dex stimulation is only removed after the STAT3 moves in.
A general comment here on enhanceosomal function in the chromosomal context seems appropriate. In the best studied cases of enhanceosomes (Grosschedl 1995; Thanos and Maniatis 1995; Agalioti et al. 2002), conclusions about cooperative functions between neighboring proteins were drawn by mutating DNA-binding sites in segments of DNA containing binding sites and testing for transcriptional function by transfection. These studies were complemented recently by either in vitro assembly of enhanceosome components or with limited ChIP identification of protein accumulation in cells. These experiments are consonant with an “integrated” function of enhanceosomal protein:protein contacts but are not conclusive. It should be recognized that none of these studies proves a functional interaction between any two proteins within an enhanceosome, let alone the mechanistic basis for any positive interaction that does occur. Only experiments in which such putative interactions on a chromosomal gene are somehow interrupted (e.g., RNAi removal of a participating member of the enhanceosome or introduction of characterized dominant-negative-acting mutants of a participating member) will signal true cooperation between enhanceosomal proteins in specific chromosomal gene activation. Then the question will arise, are the interactions between two proteins the same in all enhanceosomes? Stated another way, how many enhanceosomes can be built from a limited repertoire of participants?
We wished to complement the formation and stability studies of the enhanceosome with measurement of actual induced transcription rates throughout the lifetime of the enhanceosome. This has not been done in any other study. Some variation of run-on transcription analysis is required to measure the actual transcription rate. As we noted earlier, the run-on technique scores total occupancy on a locus of synthesizing RNA polymerases. Thus, the frequency of initiation plus the time to completely transcribe the locus determines the time of maximal run-on signal.
In the α2-M gene response, the time of maximal transcriptional signal lagged the maximal enhanceosomal loading by between 30 and 60 min (Fig. 6). What is the origin of this delay? Is there a pause between enhanceosome recruitment of Pol II and transcriptional action? Alternatively, is all the delay caused by transit time of Pol II across the α2-M locus? RNA Pol II synthesis rates have been determined by a variety of early experiments to be 20-50 NT/sec (Miyazaki et al. 1976; Sehgal et al. 1976; Jackson et al. 2000). Therefore, for the 50-kb α2-M locus, a transit time of ∼16-40 min is estimated. If we take the longer rate, then the maximal transcription signal would be between 70 and 100 min (30-60 + 40) in accord with what is observed (Fig. 6A). However, for the faster synthesis rate, the maximum transcription signal should be between 45 and 75 min (30-60 + 16). Thus, we conclude that the long transit time of the polymerase contributes to the delay between the enhanceosomal loading and the maximal transcription rate but there also may be a pause after polymerase loading on the enhanceosome before rapid transcription across the locus occurs.
This conclusion is strongly supported by another ChIP experiment. Removal of IL-6+Dex (Fig. 5D) was followed by a more rapid departure from the promoter fragment by GR and STAT3 than by Pol II, suggesting a pause after Pol II recruitment before the enzyme leaves the start site. Such a delay has several logical explanations. The polymerase itself must undergo C-terminal serine phosphorylations before transcription (see Cheng and Sharp 2003 and references therein), and because there is substantial evidence in favor of association of transcriptional machinery with RNA processing machinery (Maniatis and Reed 2002), it may take many minutes for a fully equipped Pol II complex to be sent on its way.
Many unanswered questions remain. For example, what is the function of the c-Jun/c-Fos or OCT-1 in the α2-M enhanceosome? (Or, for that matter, what is the function of any constitutive member of an enhanceosome complex prior to its activation?) Many transcription factors including c-Jun/c-Fos and OCT-1 enter the nucleus as soon they are translated (Brivanlou and Darnell 2002) but do not activate genes with which they may be associated until a later signal leads to their phosphorylation (see references on c-Fos gene itself, Treisman 1995). If these ubiquitous proteins associate with the DNA just after each replication, they may serve to institute a chromatin state that can be quickly activated. In fact, they may cause a low level of expression constitutively (see Fig. 1). Whether c-Jun, c-Fos, or OCT-1 has any such “structural role” or even a direct positive activation role in transcription of the α2-M gene can be tested best through introduction of a dominant-negative protein that cannot bind DNA or that has no transcriptional capacity.
The present results, of course, concern a single STAT3-dependent enhanceosome. How does the composition of this assembly of proteins compare with that on other STAT3-stimulated genes? Particularly, is there a difference between STAT3-dependent enhanceosomes that require glucocorticoid receptor for full activity and those that do not? With the present information, availability of the whole genome sequence and the already identified STAT3-induced genes, it should be possible to explore the composition of other STAT3-dependent enhanceosomes directly.
Materials and methods
Reagents
Human IL-6 and the recombinant soluble form of the human IL-6 receptor were purchased from R&D Systems and were used at a concentration of 80 ng/mL (IL-6) and 100 ng/mL (IL-6-R), respectively. Dexamethasone was purchased from Sigma and used at a concentration of 100 nM. Protease inhibitor cocktail and Staurosporin were purchased from Calbiochem.
Antibodies
Anti-STAT3 was purchased from Cell Signaling Technology; anti-GR from Affinity Bioreagents Inc. (PA1-512); anti-OCT-1, anti-c-Jun, anti-c-Fos, and anti-RNA Pol II (C-21 sc-900 and N-20 sc-899) from Santa Cruz Biotechnology; and anti-acetyl-H4 (#06-866) and anti-acetyl-H3 (#06-599) from Upstate Biotechnology. Salmon Sperm DNA/Protein A agarose was purchased from Upstate Biotechnology.
Tissue culture and cell extracts
The rat H-35 hepatoblastoma cell line was grown at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; GIBCO-BRL), supplemented with 100 μg/mL penicillin-streptomycin mix (GIBCO-BRL), 5% fetal bovine serum (Hyclone), and 20% horse serum (Bio-Whittaker). Before experimental treatment, cells were serum starved (1% serum FBS medium) overnight, and all treatments were carried out with the low serum medium. Nuclear extracts (NE) were prepared from untreated or treated H-35 cells as described previously (Shuai et al. 1992).
Plasmids
The (-1151/+54) HindIII/HindIII α2-M luciferase reporter and the α2-M cDNA were a gift from George H. Fey (University of Erlangen-Nuremberg, Erlangen, Germany). The (-200/+54) α2-M luciferase reporter vector was a gift from Daniel Nathans (Johns Hopkins University School of Medicine, Baltimore, MD; Schaefer et al. 1995). The full-length cDNA of GRα was a gift from Ron Evans (The Salk Institute, La Jolla, CA). pCMVβ-Gal construct was purchased from Invitrogen.
RNA isolation and analysis
Total RNA was isolated from cells untreated or treated as indicated in each figure with TRizol (GIBCO-BRL). After DNAse I (GIBCO-BRL) treatment, total RNA was reverse transcribed (SuperScript, GIBCO-BRL) and amplified by PCR using primers specific for α2-M and GAPDH.
Site-directed mutagenesis
All point mutations in the luciferase reporter construct were prepared by the PCR-based mutagenesis method (CMCR QuikChange Site Direct Mutagenesis, Stratagene). The different mutations introduced into the α2-M gene promoter fragment-luciferase are summarized: Core, 5′-AGTCCTTAATCCG GATGGGAATTCTGGC-3′; Homology, 5′-AGCAGTAACTTTCCAGTCCTTAATCTT-3′; AP-1 H, 5′-GGCTAACGGGGA CGGAATTAACCTTGGCG-3′; AP-1, 5′-TTAGGCCATCAGG TTCTCTTTCAGAGAA-3′; Sox-9, 5′-ACCTCAGCTTTACC TGGAGAACTCGTC-3′
To construct pOCT-1-α2-M luciferase, two rounds of mutagenesis were performed using the following oligonucleotides: OCT-1 I, 5′-CCTTGGCGGTACGTAGGCCATCAGTGACTCTTC-3′; OCT-1 II, 5′-CCTTGGCGGCGCGTAGGCCATCAGTGAC-3′.
The α2-M gene promoter fragment-luciferase spacer mutants, in which a half-helical turn (5 bp) or a full-helical turn (10 bp) were introduced between different pairs of activator-binding sites, were prepared using the PCR base mutagenesis method (CMCR QuikChange Site Direct Mutagenesis, Stratagene). The different spacers introduced into the α2-M gene promoter fragment-luciferase are summarized: +5 STAT3 (h)-STAT3 (c), 5′-GCA GTAA CTGG AA AGCTT ACTCCTTAATCCTTCTGGGAA-3′; +10 STAT3 (h)-STAT3 (c), 5′-GTAACTGGAAAGC TAGCTATACTCCTTAATCCTTCTGGGAA-3′; +5 STAT3 (c)-AP-1, 5′-CTTCTGGGAATTCTGGCCTACGTAACGGGTCAGG-3′; +10 STAT3 (c)-AP-1, 5′-CTTCTGGGAATTCTGGCCTATGGCACGTAACGGGTCAGG-3′; +5 AP-1-OCT-1, 5′-ACGGGTCAGGAATTAAGCTTACCTTGGCGGTAATTAGGCC-3′; +10 AP-1-OCT-1, 5′-CGGGTCAGGAATTAAC TATGGCTTACCTTGGCGGTAATTAGGC-3′; +5 OCT-1-AP-1 (h), 5′-TGGCGGTAATTAGGCCAAGCTTTCAGTGACTCTTTC-3′; +10 OCT-1-AP-1 (h), 5′-TGGCGGTAATTAGGCCAAGCATATATTTCAGTGACTCTTTC-3′. All mutations in reporter constructs were verified by nucleotide sequencing.
Transfection experiments
Transient transfection was done on 24-well plates with 2.5 × 105 cells (60%-70% confluence) per well using 0.1 μg of reporter construct, 0.2μg of β-galactosidase expression vector, and 0.7 μg of carrier DNA (GIBCO-BRL), as indicated in the figure legends. Cells were transfected with Lipofectamine Plus Reagent (GIBCO-BRL) as instructed by the manufacturer. Then, 24 h posttransfection, cells were treated with IL-6-IL-6R, Dex, IL-6-IL-6R + Dex, or left untreated. luciferase assays were performed according to the manufacturer's directions (luciferase Reporter Assay System; Promega), and β-galactosidase (β-Gal) assays were done as previously described (Ausubel 1994). All results shown are luciferase activities normalized against the internal control β-Gal activity in the corresponding untreated sample. Each sample of an experiment was performed in duplicate and repeated in three different experiments with similar results. luciferase activity values were normalized to transfection efficiency monitored by the cotransfected β-Gal expression vector.
DNase I footprinting
The DNase I footprinting assay was performed according to the manufacturer's directions (Promega) with some modifications. An aliquot of 0.55-1.5 ng of recombinant c-Jun (Promega) was incubated with 20,000-30,000 cpm of α2-M DNA fragment (-270 to +40 bp) 5′-end-labeled probe in 50 μL of binding buffer containing 25 mM Tris-HCl (pH 8.0), 50 mM KCl, 6.25 mM MgCl2, 0.5 mM EDTA, 10% glycerol, and 0.5 mM DTT on ice for 10 min. Next, 50 μL of room temperature (RT) Ca2+/Mg2+ solution (5 mM CaCl2 and 10 mM MgCl2) was added to the mixture and incubated at RT for 1 min. DNase I (GIBCO-BRL) was diluted in 50 mM CaCl2 and 20 mM HEPES (pH 7.9). Then 0.05 U of DNase I was incubated with the mixture for 1 min. The reaction was stopped by 90 μL of Stop Solution (200 mM NaCl, 30 mM EDTA, 1% SDS, and 100 μg/mL yeast RNA). DNA was recovered by phenol:chloroform:isoamyl alcohol and ethanol precipitation, resuspended in Loading Solution, and applied to an 8% polyacrylamide sequencing gel (Amresco).
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts (2-10 μg of protein) from H-35 cells untreated and treated with IL-6, Dex, or IL-6+Dex were incubated with 1 ng of 32P-labeled probe (in a final volume of 12μL) at RT for 15 min. The protein-DNA complex was analyzed by EMSA as previously described (Fried and Crothers 1981).
The oligonucleotide sequences used are as follows: m67 oligo, 5′-CATTTCCCGTAAATCGTCGA-3′ (Wagner et al. 1990) and Oct-1 binding site, 5′-CCTTGGCGGTAATTAGGCCATCAGTG-3′. Klenow polymerase (GIBCO-BRL) was used to radiolabel the probe. For competition assay, unlabeled probes were used at 100 M excess to the radiolabeled probes. For supershift analysis, the nuclear extracts were preincubated with the respective antibody at 4°C for 30 min before EMSA procedures. The reaction products were fractionated on a 4% nondenaturing polyacrylamide gel (29:1, acrylamide-bis) in 0.25% Tris borate/EDTA at 4°C.
GST pull-down assay
Glutathione S-transferase (GST) fusion constructs containing various STAT3 and c-Jun fragments were generated by PCR using primers containing 5′-BamHI sites and 3′-NotI sites. Amplified products were digested with appropriate enzymes and cloned into pGEX-5X-1 (Pharmacia). They include the STAT3 (residues 1-770), N-terminal domain (STAT3 residues 1-150), coiled-coil domain (STAT3 residues 134-319), DNA-binding domain (STAT3 residues 320-495), linker/SH2 domains (STAT3 residues 496-688), SH2 domain (STAT3 residues 584-688), transactivation domain (TAD, STAT3 residues 689-770), c-Jun (residues 1-334), and c-Jun C-term (residues 105-334). Expression of GST fusion proteins was carried out by induction of Escherichia coli containing the fusion vector at 30°C with 1 mM isopropyl-D-thiogalactopyranoside (IPTG). Following lysis by sonication, GST proteins were purified on glutathione-Sepharose beads (Pharmacia) and washed extensively with phosphate-buffered saline. For in vitro translation of GRα, the full-length cDNA was used in program-coupled transcription and translation reactions (TNT; Promega) in the presence of 35S-labeled methionine (DuPont/NEN) according to the manufacturer's directions. GST-protein association assays with translation products were carried out as previously described (Zhang et al. 1996, 1999). After being washed, the resulting complexes were eluted in sodium dodecyl sulfate (SDS) gel-loading buffer and separated by 10% SDS-polyacrylamide gel electrophoresis (PAGE).
ChIP assay
ChIP was performed using the Chromatin Immunoprecipitation Assay Kit (Upstate Biotechnology). A total of 1 × 107 rat H-35 cells under different treatments for the appropriate times (as described in the corresponding figures) were used for each ChIP. Using PCR, the purified DNA precipitated was analyzed for the presence of the following rat α2-M promoter fragments: -3886/-3721 (primers C and D); -200/+54 (primers A and B). Titration of PCR cycles was performed to ensure that experiments were performed in the linear range of amplification.
ChIP reimmunoprecipitation
Complexes were eluted from the primary immunoprecipitation by incubation with 10 mM DTT at 37°C for 30 min and diluted 1:50 in buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl at pH 8.1) followed by reimmunoprecipitation with second antibodies. ChIP reimmunoprecipitations of supernatants were performed essentially as were the primary IPs.
Determination of transcription rates
Run-on assays were performed as previously described (Pine et al. 1990). H-35 cells (1 × 107) were untreated or treated with IL-6, Dex, or IL-6+Dex for the indicated times. For slot blot analysis, 10 μg of linearized α2-M-pUC18, GAPDG-pUC18, or pUC18 was applied to nitrocellulose membranes (Protan, Schleicher & Schuell) and then hybridized with the run-on transcripts (1 × 106 dpm per sample). An additional RNase A (Roche Molecular Biochemicals) wash step was performed (2× SSC RNase A, 2.5 μg/mL) at 37°C for 30 min. Finally, the membrane was rinsed with 2× SSC. “Run-on” signals were then visualized by autoradiography and quantitated with the Molecular Dynamics PhosphorImager.
Acknowledgments
This work was supported by NIH grants AI32489 and AI32440 to J.E.D. L.L. was supported by the Pew Latin American Fellows Program. M.A.H. was a Cancer Research Institute Post Doctoral Fellow. We thank Marc Fucillo for technical assistance and Lois Cousseau for manuscript preparation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1135003.
References
- Agalioti T., Chen, G., and Thanos, D. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111: 381-392. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. 1994. Current protocols in molecular biology. Wiley, New York.
- Brivanlou A.H. and Darnell Jr., J.E. 2002. Signal transduction and the control of gene expression. Science 295: 813-818. [DOI] [PubMed] [Google Scholar]
- Carey M. 1998. The enhanceosome and transcriptional synergy. Cell 92: 5-8. [DOI] [PubMed] [Google Scholar]
- Cheng C. and Sharp, P.A. 2003. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and γ-actin genes. Mol. Cell. Biol. 23: 1961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell Jr J.E. 2002. Transcription factors as targets for cancer therapy. Nat. Rev. Cancer 2: 740-749. [DOI] [PubMed] [Google Scholar]
- Edelstein L.C., Lagos, L., Simmons, M., Tirumalai, H., and Gelinas, C. 2003. NF-κ B-dependent assembly of an enhanceosome-like complex on the promoter region of apoptosis inhibitor Bfl-1/A1. Mol. Cell. Biol. 23: 2749-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell J.D., Ge, H., and Roeder, R.G. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. 93: 8329-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M. and Crothers, D.M. 1981. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 9: 6505-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T., Andus, T., Bauer, J., Northoff, H., Ganter, U., Hirano, T., Kishimoto, T., and Heinrich, P.C. 1988. Cell-free-synthesized interleukin-6 (BSF-2/IFN-β 2) exhibits hepatocyte-stimulating activity. Eur. J. Biochem. 175: 181-186. [DOI] [PubMed] [Google Scholar]
- Grosschedl R. 1995. Higher-order nucleoprotein complexes in transcription: Analogies with site-specific recombination. Curr. Opin. Cell Biol. 7: 362-370. [DOI] [PubMed] [Google Scholar]
- Haspel R.L. 1998. “Inactivation of the JAK-STAT pathway.” Ph.D. thesis. The Rockefeller University, New York.
- Haspel R.L. and Darnell Jr., J.E. 1999. A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc. Natl. Acad. Sci. 96: 10188-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haspel R.L., Salditt-Georgieff, M., and Darnell Jr., J.E. 1996. The rapid inactivation of nuclear tyrosine phosphorylated Stat1 depends upon a protein tyrosine phosphatase. EMBO J. 15: 6262-6268. [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Abraham, L.J., Northemann, W., and Fey, G.H. 1990. Acute-phase reaction induces a specific complex between hepatic nuclear proteins and the interleukin 6 response element of the rat α 2-macroglobulin gene. Proc. Natl. Acad. Sci. 87: 2364-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich P.C., Horn, F., Graeve, L., Dittrich, E., Kerr, I., Muller-Newen, G., Grotzinger, J., and Wollmer, A. 1998. Interleukin-6 and related cytokines: Effect on the acute phase reaction. Z. Ernahrungswiss. 37 Suppl 1: 43-49. [PubMed] [Google Scholar]
- Hocke G.M., Barry, D., and Fey, G.H. 1992. Synergistic action of interleukin-6 and glucocorticoids is mediated by the interleukin-6 response element of the rat α 2 macroglobulin gene. Mol. Cell. Biol. 12: 2282-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath C.M. 2000. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem. Sci. 25: 496-502. [DOI] [PubMed] [Google Scholar]
- Jackson D.A., Pombo, A., and Iborra, F. 2000. The balance sheet for transcription: An anlaysis of nuclear RNA metabolism in mammalian cells. FASEB J. 14: 242-254. [PubMed] [Google Scholar]
- Karin M., Liu, Z., and Zandi, E. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9: 240-246. [DOI] [PubMed] [Google Scholar]
- Lau J.F., Nusinzon, I., Barkov, D., Freedman, L.P., and Horvath, C.M. 2003. Role of metazoan Mediator proteins in interferon-responsive transcription. Mol. Cell. Biol. 23: 620-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.E. and Darnell Jr., J.E. 2002. Stats: Transcriptional control and biological impact. Nat. Rev. Mol. Cell. Biol. 3: 651-662. [DOI] [PubMed] [Google Scholar]
- Lomvardas S. and Thanos, D. 2002. Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110: 261-271. [DOI] [PubMed] [Google Scholar]
- Lutticken C., Coffer, P., Yuan, J., Schwartz, C., Caldenhoven, E., Schindler, C., Kruijer, W., Heinrich, P.C., and Horn, F. 1995. Interleukin-6-induced serine phosphorylation of transcription factor APRF: Evidence for a role in interleukin-6 target gene induction. FEBS Lett. 360: 137-143. [DOI] [PubMed] [Google Scholar]
- Malik S. and Roeder, R.G. 2000. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 25: 277-283. [DOI] [PubMed] [Google Scholar]
- Maniatis T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416: 499-506. [DOI] [PubMed] [Google Scholar]
- Miyazaki M., Hosoki, K., and Yamamoto, K. 1976. Renin inhibition by synthetic phosphatidyl and phosphorylethanolamines. Japan Circ. J. 40: 901-910. [DOI] [PubMed] [Google Scholar]
- Myers L.C. and Kornberg, R.D. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69: 729-749. [DOI] [PubMed] [Google Scholar]
- Northemann W., Shiels, B.R., Braciak, T.A., Hanson, R.W., Heinrich, P.C., and Fey, G.H. 1988. Structure and acute-phase regulation of the rat α 2-macroglobulin gene. Biochemistry 27: 9194-9203. [DOI] [PubMed] [Google Scholar]
- Paulson M., Pisharody, S., Pan, L., Guadagno, S., Mui, A.L., and Levy, D.E. 1999. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 274: 25343-25349. [DOI] [PubMed] [Google Scholar]
- Pine R., Decker, T., Kessler, D.S., Levy, D.E., and Darnell Jr., J.E. 1990. Purification and cloning of interferon-stimulated gene factor 2(ISGF2): ISGF2(IRF-1) can bind to the promoters of both β interferon- and interferon-stimulated genes but is not a primary transcriptional activator of either. Mol. Cell. Biol. 10: 2448-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachez C. and Freedman, L.P. 2001. Mediator complexes and transcription. Curr. Opin. Cell Biol. 13: 274-280. [DOI] [PubMed] [Google Scholar]
- Schaefer T.S., Sanders, L.K., and Nathans, D. 1995. Cooperative transcriptional activity of Jun and Stat3β, a short form of Stat3. Proc. Natl. Acad. Sci. 92: 9097-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer T.S., Sanders, L.K., Park, O.K., and Nathans, D. 1997. Functional differences between Stat3α and Stat3β. Mol. Cell. Biol. 17: 5307-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal P.G., Derman, E., Molloy, G.R., Tamm, I., and Darnell Jr., J.E. 1976. 5,6-Dichloro-b-D-ribofuranosylbenzimidazole inhibits initiation of nuclear heterogeneous RNA chains in HeLa cells. Science 94: 431-433. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu, X., DiRenzo, J., Lazar, M.A., and Brown, M. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103: 843-852. [DOI] [PubMed] [Google Scholar]
- Shang Y., Myers, M., and Brown, M. 2002. Formation of the androgen receptor transcription complex. Mol. Cell 9: 601-610. [DOI] [PubMed] [Google Scholar]
- Shuai K., Schindler, C., Prezioso, V.R., and Darnell Jr., J.E. 1992. Activation of transcription by IFN-γ: Tyrosine phosphorylation of a 91-kD DNA binding protein. Science 258: 1808-1812. [DOI] [PubMed] [Google Scholar]
- Starr R. and Hilton, D.J. 1999. Negative regulation of the JAK/STAT pathway. Bioessays 21: 47-52. [DOI] [PubMed] [Google Scholar]
- Stocklin E., Wissler, M., Gouilleux, F., and Groner, B. 1996. Functional interactions between Stat5 and the glucocorticoid receptor. Nature 383: 726-728. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht, A., Luo, K., and Grunstein, M. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 11: 83-93. [DOI] [PubMed] [Google Scholar]
- Thanos D. and Maniatis, T. 1995. Virus indication of human IFN β gene expression requires the assembly of an enhanceosome. Cell 83: 1091-1101. [DOI] [PubMed] [Google Scholar]
- Treisman R. 1995. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 14: 4905-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner B.J., Hayes, T.E., Hoban, C.J., and Cochran, B.H. 1990. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 9: 4477-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegenka U.M., Buschmann, J., Lutticken, C., Heinrich, P.C., and Horn, F. 1993. Acute-phase response factor, a nuclear factor binding to acute-phase response elements, is rapidly activated by interleukin-6 at the posttranslational level. Mol. Cell. Biol. 13: 276-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharova N., Lymar, E.S., Yang, E., Malik, S., Zhang, J.J., Roeder, R.G., and Darnell Jr., J.E. 2003. Distinct transcriptional activation functions of STAT1α and β on DNA and chromatin templates. J. Biol. Chem. 278 (in press). [DOI] [PubMed]
- Zhang X. and Darnell Jr., J.E. 2001. Functional importance of Stat3 tetramerization in activation of the α 2-macroglobulin gene. J. Biol. Chem. 276: 33576-33581. [DOI] [PubMed] [Google Scholar]
- Zhang J.J., Vinkemeier, U., Gu, W., Chakravarti, D., Horvath, C.M., and Darnell Jr., J.E. 1996. Two contact regions between Stat1 and CBP/p300 in interferon γ signaling. Proc. Natl. Acad. Sci. 93: 15092-15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wrzeszczynska, M.H., Horvath, C.M., and Darnell Jr., J.E. 1999. Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol. Cell. Biol. 19: 7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]