Abstract
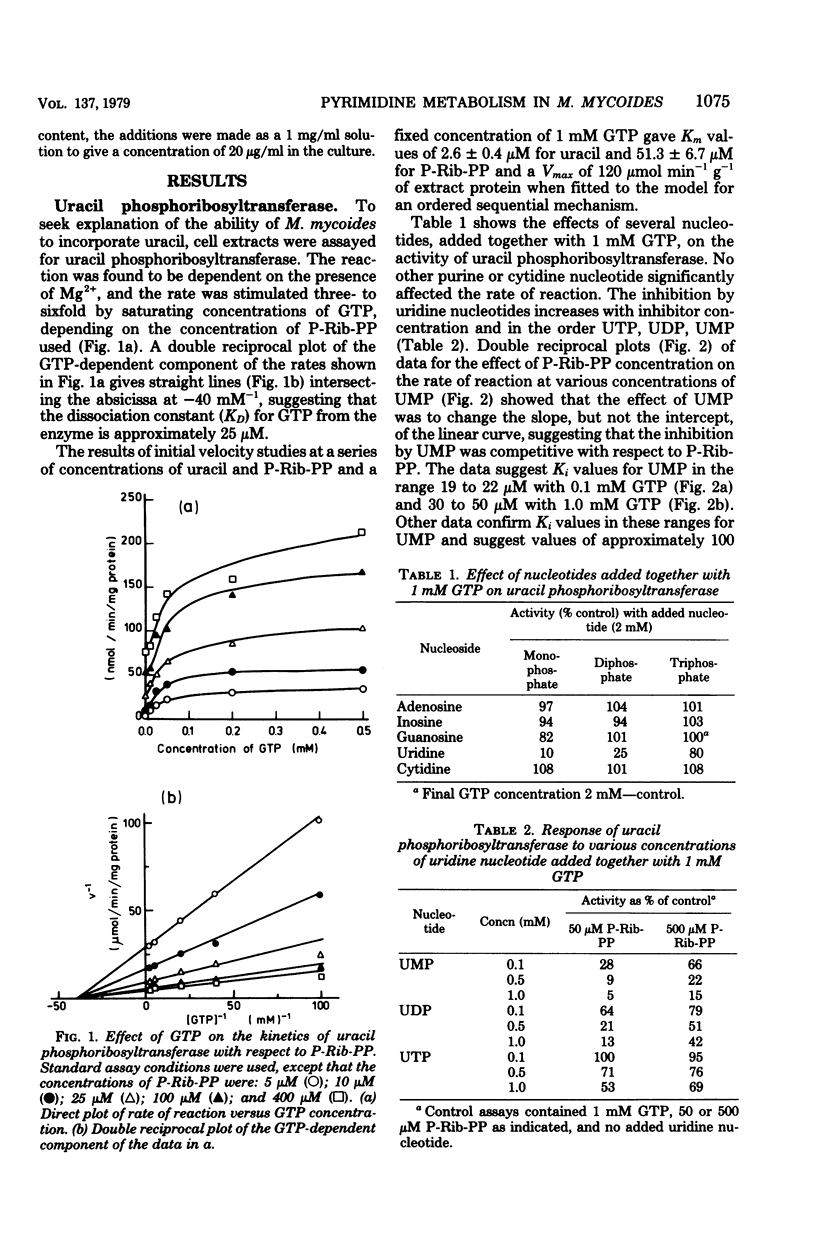
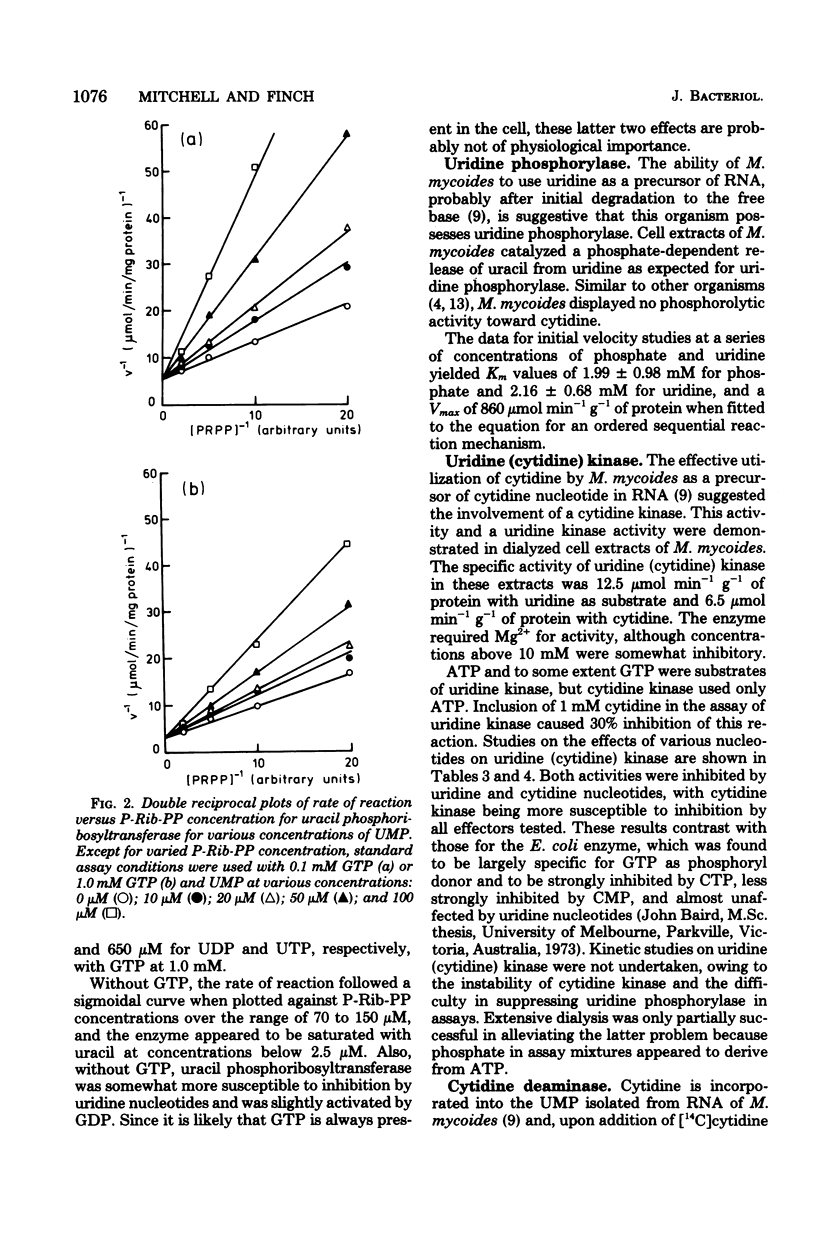
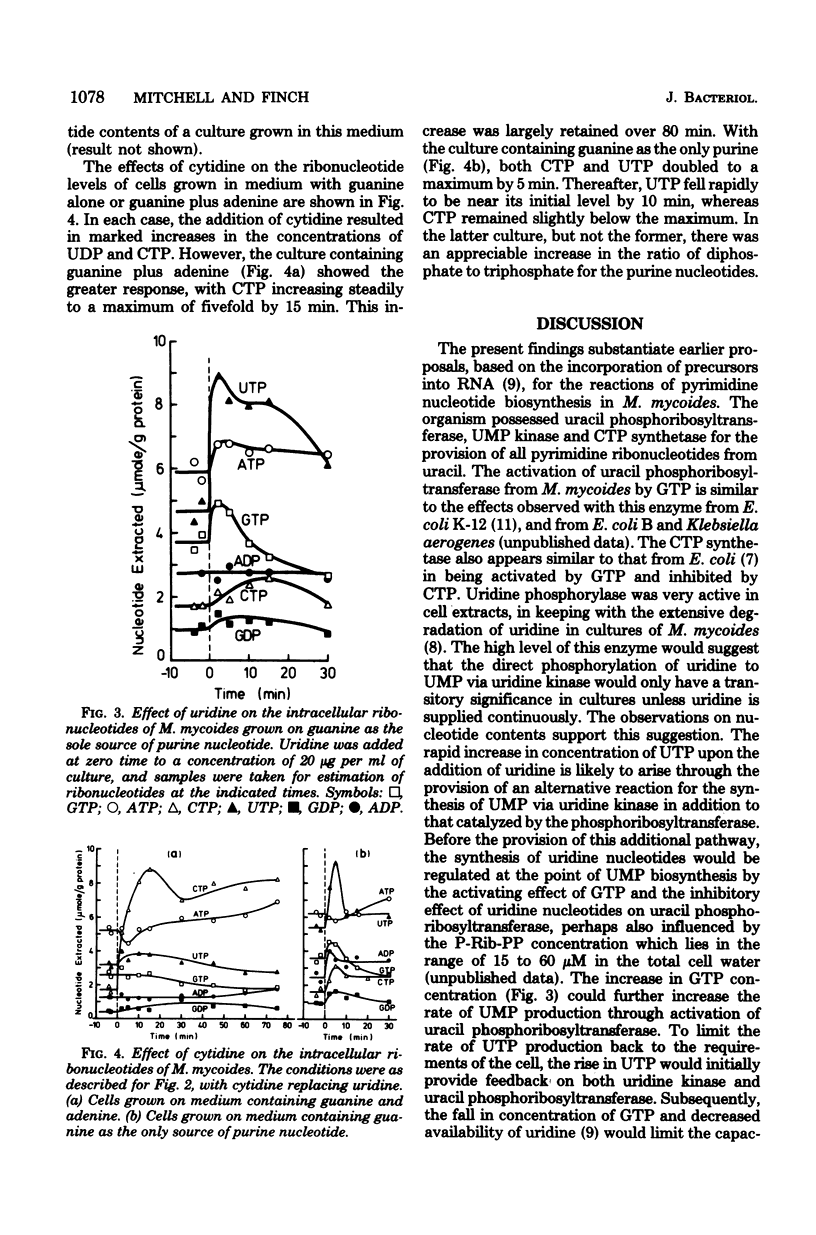
The major pathways of ribonucleotide biosynthesis in Mycoplasma mycoides subsp. mycoides have been proposed from studies on its use of radioactive purines and pyrimidines. To interpret more fully the observed pattern of pyrimidine usage, cell extracts of this organism have been assayed for several enzymes associated with the salvage synthesis of pyrimidine nucleotides. M. mycoides possessed uracil phosphoribosyltransferase, uridine phosphorylase, uridine (cytidine) kinase, uridine 5′-monophosphate kinase, and cytidine 5′-triphosphate synthetase. No activity for phosphorolysis of cytidine was detected, and no in vitro conditions were found to give measurable deamination of cytidine. Of the two potential pathways for incorporation of uridine, our data suggest that this precursor would largely undergo initial phosphorolysis to uracil and ribose-1-phosphate. Conversely, cytidine is phosphorylated directly to cytidine 5′-monophosphate in its major utilization, although conversion of cytidine to uracil, uridine, and uridine nucleotide has been observed in vivo, at least when uracil is provided in the growth medium. Measurements of intracellular nucleotide contents and their changes on additions of pyrimidine precursors have allowed suggestions as to the operation of regulatory mechanisms on pyrimidine nucleotide biosynthesis in M. mycoides in vivo. With uracil alone or uracil plus uridine as precursors of pyrimidine ribonucleotides, the regulation of uracil phosphoribosyltransferase and cytidine 5′-triphosphate synthetase is probably most important in determining the rate of pyrimidine nucleotide synthesis. When cytidine supplements uracil in the growth medium, control of cytidine kinase activity would also be important in this regard.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagnara A. S., Finch L. R. Quantitative extraction and estimation of intracellular nucleoside triphosphates of Escherichia coli. Anal Biochem. 1972 Jan;45(1):24–34. doi: 10.1016/0003-2697(72)90004-8. [DOI] [PubMed] [Google Scholar]
- Beck C. F., Ingraham J. L., Neuhard J., Thomassen E. Metabolism of pyrimidines and pyrimidine nucleosides by Salmonella typhimurium. J Bacteriol. 1972 Apr;110(1):219–228. doi: 10.1128/jb.110.1.219-228.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doskocil J., Sorm F. The mode of action of 5-aza-2'-deoxycytidine in Escherichia coli. Eur J Biochem. 1970 Mar 1;13(1):180–187. doi: 10.1111/j.1432-1033.1970.tb00916.x. [DOI] [PubMed] [Google Scholar]
- Hammer-Jespersen K., Munch-Petersen A., Schwartz M., Nygaard P. Induction of enzymes involed in the catabolism of deoxyribonucleosides and ribonucleosides in Escherichia coli K 12. Eur J Biochem. 1971 Apr 30;19(4):533–538. doi: 10.1111/j.1432-1033.1971.tb01345.x. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Levine E. M. Mycoplasma contamination of animal cell cultures: a simple, rapid detection method. Exp Cell Res. 1972 Sep;74(1):99–109. doi: 10.1016/0014-4827(72)90484-3. [DOI] [PubMed] [Google Scholar]
- Long C. W., Pardee A. B. Cytidine triphosphate synthetase of Escherichia coli B. I. Purification and kinetics. J Biol Chem. 1967 Oct 25;242(20):4715–4721. [PubMed] [Google Scholar]
- Mitchell A., Finch L. R. Pathways of nucleotide biosynthesis in Mycoplasma mycoides subsp. mycoides. J Bacteriol. 1977 Jun;130(3):1047–1054. doi: 10.1128/jb.130.3.1047-1054.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A., Sin I. L., Finch L. R. Enzymes of purine metabolism in Mycoplasma mycoides subsp. mycoides. J Bacteriol. 1978 Jun;134(3):706–712. doi: 10.1128/jb.134.3.706-712.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy A., Finch L. R. Uridine-5'-monophosphate pyrophosphorylase activity from Escherichia coli. FEBS Lett. 1969 Nov 12;5(3):211–213. doi: 10.1016/0014-5793(69)80334-0. [DOI] [PubMed] [Google Scholar]
- Neuhard J., Ingraham J. Mutants of Salmonella typhimurium requiring cytidine for growth. J Bacteriol. 1968 Jun;95(6):2431–2433. doi: 10.1128/jb.95.6.2431-2433.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhard J. Pyrimidine nucleotide metabolism and pathways of thymidine triphosphate biosynthesis in Salmonella typhimurium. J Bacteriol. 1968 Nov;96(5):1519–1527. doi: 10.1128/jb.96.5.1519-1527.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan G. A., Neuhard J. Pyrimidine metabolism in microorganisms. Bacteriol Rev. 1970 Sep;34(3):278–343. doi: 10.1128/br.34.3.278-343.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RODWELL A. W. Nutrition and metabolism of Mycoplasma mycoides var. mycoides. Ann N Y Acad Sci. 1960 Jan 15;79:499–507. doi: 10.1111/j.1749-6632.1960.tb42716.x. [DOI] [PubMed] [Google Scholar]
- Rodwell A. W., Peterson J. E., Rodwell E. S. Striated fibers of the rho form of Mycoplasma: in vitro reassembly, composition, and structure. J Bacteriol. 1975 Jun;122(3):1216–1229. doi: 10.1128/jb.122.3.1216-1229.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrecker A., Jacobsen D. W., Kirchner J. Separation of ribonucleosides from deoxyribonucleosides and arabinonucleosides by thin-layer chromatography. Anal Biochem. 1968 Dec;26(3):474–477. doi: 10.1016/0003-2697(68)90217-0. [DOI] [PubMed] [Google Scholar]
- Sin I. L., Finch L. R. Adenine phosphoribosyltransferase in Mycoplasma mycoides and Escherichia coli. J Bacteriol. 1972 Oct;112(1):439–444. doi: 10.1128/jb.112.1.439-444.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]


