Abstract
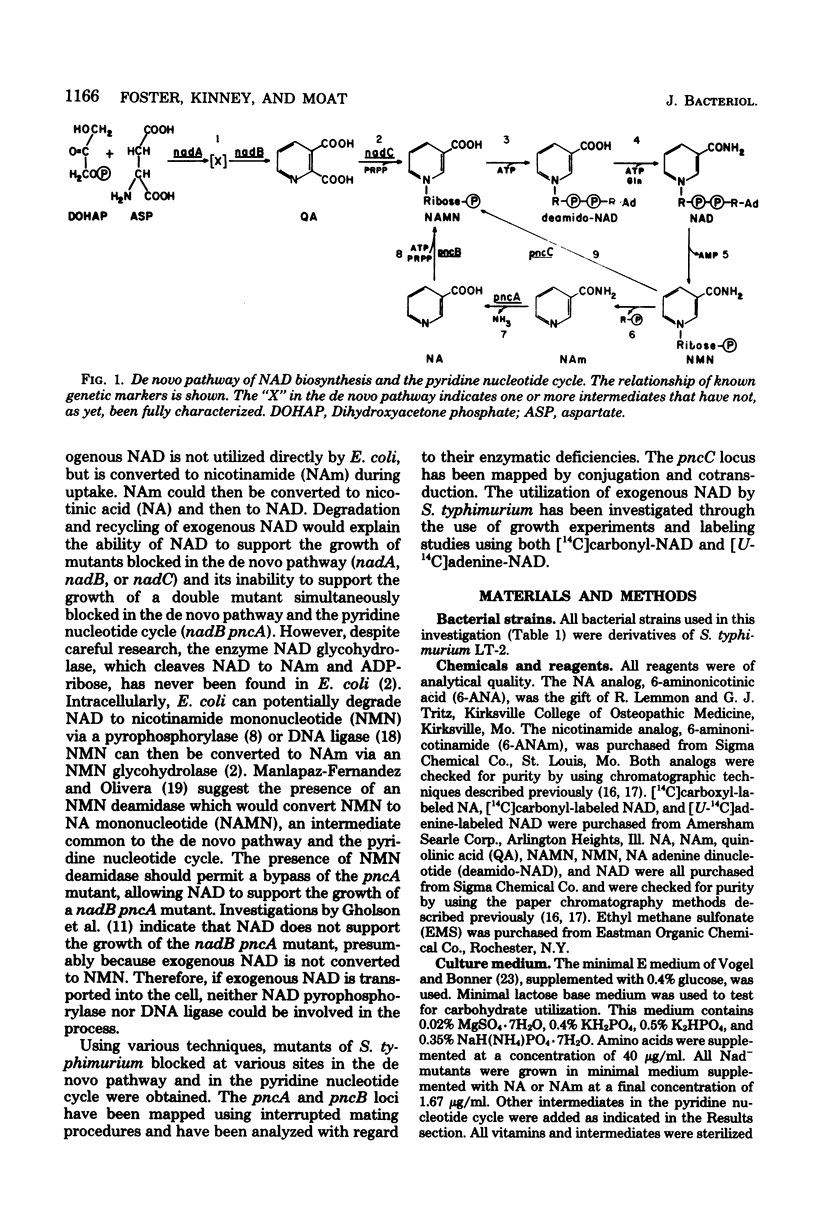
Mutants of Salmonella typhimurium LT-2 deficient in nicotinamidase activity (pncA) or nicotinic acid phosphoribosyltransferase activity (pncB) were isolated as resistant to analogs of nicotinic acid and nicotinamide. Information obtained from interrupted mating experiments placed the pncA gene at 27 units and the pncB gene at 25 units on the S. typhimurium LT-2 linkage map. A major difference in the location of the pncA gene was found between the S. typhimurium and Escherichia coli linkage maps. The pncA gene is located in a region in which there is a major inversion of the gene order in S. typhimurium as compared to that in E. coli. Growth experiments using double mutants blocked in the de novo pathway to nicotinamide adenine dinucleotide (NAD) (nad) and in the pyridine nucleotide cycle (pnc) at either the pncA or pncB locus, or both, have provided evidence for the existence of an alternate recycling pathway in this organism. Mutants lacking this alternate cycle, pncC, have been isolated and mapped via cotransduction at 0 units. Utilization of exogenous NAD was examined through the use of [14C]carbonyl-labeled NAD and [14C]adenine-labeled NAD. The results of these experiments suggest that NAD is degraded to nicotinamide mononucleotide at the cell surface. A portion of this extracellular nicotinamide mononucleotide is then transported across the cell membrane by nicotinamide mononucleotide glycohydrolase and degraded to nicotinamide in the process. The remaining nicotinamide mononucleotide accumulates extracellularly and will support the growth of nadA pncB mutants which cannot utilize the nicotinamide resulting from the major pathway of NAD degradation. A model is presented for the utilization of exogenous NAD by S. typhimurium LT-2.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreoli A. J., Grover T., Gholson R. K., Matney T. S. Evidence for a functional pyridine nucleotide cycle in Escherichia coli. Biochim Biophys Acta. 1969 Dec 30;192(3):539–541. doi: 10.1016/0304-4165(69)90408-5. [DOI] [PubMed] [Google Scholar]
- Andreoli A. J., Okita T. W., Bloom R., Grover T. A. The pyridine nucleotide cycle: presence of a nicotinamide mononucleotide-specific glycohydrolase in Escherichia coli. Biochem Biophys Res Commun. 1972 Oct 6;49(1):264–269. doi: 10.1016/0006-291x(72)90039-3. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baecker P. A., Yung S. G., Rodriguez M., Austin E., Andreoli A. J. Periplasmic localization of nicotinate phosphoribosyltransferase in Escherichia coli. J Bacteriol. 1978 Mar;133(3):1108–1112. doi: 10.1128/jb.133.3.1108-1112.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J. L., Gholson R. K. De novo biosynthesis of nicotinamide adenine dinucleotide in Escherichia coli: excretion of quinolinic acid by mutants lacking quinolinate phosphoribosyl transferase. J Bacteriol. 1972 Jul;111(1):98–102. doi: 10.1128/jb.111.1.98-102.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J. L., Gholson R. K. Studies on the de novo biosynthesis of NAD in Escherichia coli. II. Quantitative method for isolating quinolinic acid from biological materials. Anal Biochem. 1972 Aug;48(2):529–535. doi: 10.1016/0003-2697(72)90108-x. [DOI] [PubMed] [Google Scholar]
- Cobb J. R., Pearcy S. C., Gholson R. K. Metabolism of 6-aminonicotinic acid in Escherichia coli. J Bacteriol. 1977 Sep;131(3):789–794. doi: 10.1128/jb.131.3.789-794.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmen W., Webb B., Preiss J. The deamido-diphosphopyridine nucleotide and diphosphopyridine nucleotide pyrophosphorylases of Escherichia coli and yeast. Arch Biochem Biophys. 1967 May;120(2):440–450. doi: 10.1016/0003-9861(67)90262-7. [DOI] [PubMed] [Google Scholar]
- Dickinson E. S., Sundaram T. K. Chromosomal location of a gene defining nicotinamide deamidase in Escherichia coli. J Bacteriol. 1970 Mar;101(3):1090–1091. doi: 10.1128/jb.101.3.1090-1091.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W., Moat A. G. Mapping and characterization of the nad genes in Salmonella typhimurium LT-2. J Bacteriol. 1978 Feb;133(2):775–779. doi: 10.1128/jb.133.2.775-779.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholson R. K., Tritz G. J., Matney T. S., Andreoli A. J. Mode of nicotinamide adenine dinucleotide utilization by Escherichia coli. J Bacteriol. 1969 Sep;99(3):895–896. doi: 10.1128/jb.99.3.895-896.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinathan K. P., Sirsi M., Vaidyanathan C. S. Nicotinamide-adenine dinucleotide glycohydrolase of Mycobacterium tuberculosis H37Rv. Biochem J. 1964 May;91(2):277–282. doi: 10.1042/bj0910277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARTMAN P. E., LOPER J. C., SERMAN D. Fine structure mapping by complete transduction between histidine-requiring Salmonella mutants. J Gen Microbiol. 1960 Apr;22:323–353. doi: 10.1099/00221287-22-2-323. [DOI] [PubMed] [Google Scholar]
- IMSANDE J. A COMPARATIVE STUDY OF THE REGULATION OF PYRIDINE NUCLEOTIDE FORMATION. Biochim Biophys Acta. 1964 Mar 16;82:445–453. doi: 10.1016/0304-4165(64)90436-2. [DOI] [PubMed] [Google Scholar]
- IMSANDE J. Pathway of diphosphopyridine nucleotide biosynthesis in Escherichia coli. J Biol Chem. 1961 May;236:1494–1497. [PubMed] [Google Scholar]
- Kasărov L. B., Moat A. G. Convenient method for enzymic synthesis of 14 C-nicotinamide riboside. Anal Biochem. 1972 Mar;46(1):181–186. doi: 10.1016/0003-2697(72)90410-1. [DOI] [PubMed] [Google Scholar]
- Kasărov L. B., Moat A. G. Metabolism of nicotinamide adenine dinucleotide in human and bovine strainsof Mycobacterium tuberculosis. J Bacteriol. 1972 May;110(2):600–603. doi: 10.1128/jb.110.2.600-603.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman I. R. DNA ligase: structure, mechanism, and function. Science. 1974 Nov 29;186(4166):790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- Manlapaz-Fernandez P., Olivera B. M. Pyridine nucleotide metabolism in Escherichia coli. IV. Turnover. J Biol Chem. 1973 Jul 25;248(14):5150–5155. [PubMed] [Google Scholar]
- Pardee A. B., Benz E. J., Jr, St Peter D. A., Krieger J. N., Meuth M., Trieshmann H. W., Jr Hyperproduction and purification of nicotinamide deamidase, a microconstitutive enzyme of Escherichia coli. J Biol Chem. 1971 Nov 25;246(22):6792–6796. [PubMed] [Google Scholar]
- Sanderson K. E., Hartman P. E. Linkage map of Salmonella typhimurium, edition V. Microbiol Rev. 1978 Jun;42(2):471–519. doi: 10.1128/mr.42.2.471-519.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E. Linkage map of Salmonella typhimurium, edition IV. Bacteriol Rev. 1972 Dec;36(4):558–586. doi: 10.1128/br.36.4.558-586.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]


