Abstract
Chondroitin sulfate A (CSA) is an important receptor for the sequestration of Plasmodium falciparum in the placenta, but the parasite ligand involved in adhesion has not previously been identified. Here we report the identification of a var gene transcribed in association with binding to CSA and present evidence that the P. falciparum erythrocyte membrane protein 1 product of the gene is the parasite ligand mediating CSA binding. Description of this gene and the implication of P. falciparum erythrocyte membrane protein 1 as the parasite ligand paves the way to a more detailed understanding of the pathogenesis of placental infection and potential therapeutic strategies targeting the interaction.
Adherence of Plasmodium falciparum-infected erythrocytes to the endothelium of postcapillary venules assists the parasite in avoiding splenic clearance and promotes sequestration in organs such as the brain and placenta, which is a characteristic of malaria infection (1, 2) that is implicated in the pathogenesis of severe disease. Several molecules have been shown to act as receptors for infected erythrocytes in vitro, including CD36, intercellular adhesion molecule-1 (ICAM-1), thrombospondin, vascular cell adhesion molecule 1, E-selectin, P-selectin, chondroitin sulfate A (CSA), αvβ3-integrin, and platelet endothelial cell adhesion molecule-1/CD31 (3–11). Of these receptors CSA has to date the most clearly established role in clinical disease, namely placental malaria (12).
The glycosaminoglycan polysaccharide CSA has been shown to support adhesion of P. falciparum-infected erythrocytes in static and flow-based assays (9, 13, 14). Although the extent of its distribution is unclear, the presence of CSA on the surface of brain and lung endothelial cells (13) and placental syncytiotrophoblasts (15) makes it potentially relevant as a sequestration receptor in the pathogenesis of severe malarial disease. Studies conducted in Africa (12) (J.G.B. and S.J.R., unpublished data) suggest that adhesion of infected erythrocytes to CSA is important in placental parasitization and malaria during pregnancy, an infection that affects large numbers of women globally and contributes significantly to intrauterine growth retardation, infant mortality, and maternal anemia (16). In the African studies, infected erythrocytes harvested from parasitized placentae were more likely to bind to CSA and at higher levels than infected erythrocytes from the peripheral blood of the same individuals or from nonpregnant individuals. Subsequent work (17) has demonstrated an association between antibodies that block the adhesion of infected erythrocytes to CSA and reduced placental infection in pregnant women. Blocking the interaction between the infected erythrocyte and CSA, for example by an antiadhesive vaccine or a small molecule, is therefore a potentially important strategy for the protection of pregnant women from malaria, but depends on identification of the parasite adhesion ligand.
Adhesion to CD36 and ICAM-1 is mediated by the parasite-derived antigen P. falciparum erythrocyte membrane protein 1 (PfEMP1) (18), a high molecular weight protein (19, 20) associated with knob-like structures at the surface of the infected erythrocyte (21). P. falciparum has been shown to undergo clonal antigenic variation (22), spontaneously switching expression of antigenic and cytoadherence types, concomitantly with size changes in PfEMP1 (23), a mechanism which enables the parasite to evade the antibody response against the exposed protein. PfEMP1 is encoded by an extensive (up to 50 members per genome) and diverse family of var genes (24–26) with a structure characterized by the presence of a variable number of Duffy binding-like (DBL) domains.
Previous work on var gene switching has demonstrated transcription of a particular gene in a CSA adherent line (27), but no evidence has been presented to link this transcriptional event with the expression of a novel PfEMP1, nor to demonstrate that PfEMP1 is directly associated with binding to CSA. To establish the involvement of PfEMP1 in the adhesive interaction between infected erythrocytes and CSA, we have used a P. falciparum isolate selected for high-level binding to CSA to identify a member of the var gene family whose transcription is associated with this phenotype and demonstrated that the expressed var gene encodes a PfEMP1 species that mediates adhesion to CSA.
MATERIALS AND METHODS
Parasite Selection and Phenotyping.
P. falciparum parasites were maintained in culture as described (28) and synchronized by sorbitol lysis (29) the day before phenotyping and harvest. FAF-EA8 was derived from ItG2 by selection for adhesion to endothelial cells (23), and the CS2 line was created by panning this line five times for adhesion to Chinese hamster ovary cells and twice to purified CSA (14). Cytoadherence phenotype was determined by assay of infected erythrocytes binding to spots of purified receptors in 2-ml Petri dishes (28), and agglutination assays were performed by standard methods (28).
Reverse Transcription–PCR.
Trophozoite-stage parasites, from the same harvest as the phenotyping, were lysed with 0.15% saponin and poly(A)+ RNA was isolated by using the MicroFast RNA extraction kit (Invitrogen). The RNA was treated with RNase-free DNase (GIBCO/BRL), and first-strand cDNA was synthesized by using a MicroFast cDNA synthesis kit (Invitrogen). Genomic contamination and cDNA quality was checked by amplifying across the intron of the DHPS gene (30). var gene expression was detected by amplification with the degenerate primers UNIEBP5′ and UNIEBP3′ as described (31), and the products were cloned into pGEM-4Z (Promega) and sequenced. Unique CS2-DBL3 primers were derived from the sequence of the initial product (forward 5′-gccggatCCA, GAA, GGT, TTT, AGA, AAA, CAA, ATG and reverse 5′-ggcccccatggTCG, GCG, TTG, TTC, AGT, TTG, TTG, TAT, G) and used to confirm differential reverse transcription–PCR and to PCR screen a yeast artificial chromosome (YAC) library. All PCRs used a standard 50-μl reaction mixture containing 10 mM Tris⋅HCl (pH 8.3), 2.5 mM MgCl2, 50 mM KCl, 200 μM dNTP mix, 1 unit of Taq polymerase (Cetus), and 25 pMol of the individual oligonucleotides. Thermocycling conditions for the degenerate primers were 94°C/1 min, 45°C/1 min, and 67°C/1 min for 38 cycles. For the unique primers the annealing temperature was raised to 50°C. A variety of standard cloning techniques and vectors were used to sequence the whole gene from the YAC clone.
Expression of Fusion Proteins and Raising of Antisera.
Primers carrying restriction sites were designed to create PCR products spanning the domains indicated in Fig. 1c, which could be cloned directly into pGEX expression vectors (Amersham Pharmacia). Transformed Escherichia coli was induced with 2 mM isopropyl β-d-thiogalactoside, pelleted, and sonicated in the presence of 0.5% Triton X-100/PBS and Gex fusion proteins purified from the supernatant by affinity chromatography on glutathione-agarose (Sigma). Rabbits were immunized with 100–200 μg of protein emulsified in Freund’s complete adjuvant followed by a single boost of 50–100 μg in Freund’s incomplete adjuvant and then aqueous boosts at 4-week intervals. All rabbit antisera were collected 10–14 days after each boost and tested by immunoblotting for recognition of the recombinant protein to which they were raised. No cross-recognition was observed.
Figure 1.
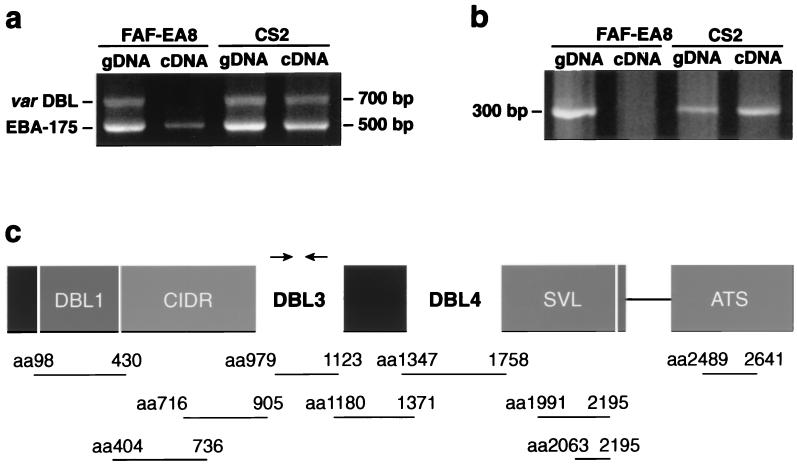
A differentially expressed var gene associated with CSA binding was detected and sequenced. (a) Degenerate universal primers amplified a var gene DBL3 region from genomic DNA of parent and selected line, and from cDNA only in the CSA binding line. The related EBA-175 gene also was amplified under these conditions. (b) The differential expression of the cDNA transcript was confirmed with sequence specific primers. (c) Full-length sequence was obtained for a var-CS2 gene, including three DBL domains. The approximate positions of the primers used to generate a gene-specific probe are marked with arrows. SVL, sequence of variable length. The lines below the gene represent nine different regions that were expressed as fusion proteins and to which rabbit antisera were raised.
Immunoblotting of PfEMP1.
For the detection of PfEMP1, Triton X-100-insoluble/SDS-soluble proteins were extracted, separated on 6% SDS/PAGE gels, and immunoblotted as reported (21). Filters were probed with either affinity-purified anti-acidic terminal sequence (ATS) or whole rabbit antisera to the other domains at 1:100 or 1:200 dilutions. Extracts from noninfected erythrocytes and rabbit serum collected before immunization were used as controls of nonspecific reaction. Fusion proteins were separated on 10% SDS/PAGE gels, blotted, and probed under similar conditions. Anti-glutathione S-transferase antisera were used to confirm the presence of the fusion protein on the filters.
Immunofluorescence Microscopy.
Smears of trophozoite stage parasites were fixed in 90% acetone and 10% methanol and incubated with a range of dilutions of recombinant domain-specific var-CS2 antisera or control preimmunization sera for 60 min at room temperature. They were then washed, incubated with FITC-conjugated goat anti-rabbit Ig antibodies, mounted in an antiquenching agent, and visualized by using a laser confocal microscope. For live fluorescence a similar process was carried out but unfixed erythrocytes that were incubated and washed in microfuge tubes and visualized as a wet preparation were used. Infected erythrocytes could be easily identified by light microcopy without further counterstaining. The DBL3 and DBL4 antisera were used at titers above 1:1,000 to give CS2-specific reactivity.
Adhesion Inhibition Assays.
Assays were performed on CS2 P. falciparum trophozoite-infected erythrocytes at a parasitemia of 3–8%, using previously described methods (32). CSA from porcine rib cartilage (Sigma-Aldrich) was immobilized overnight onto plastic Petri dishes in 10-μl spots, at a concentration of 20 μg/ml, and blocked with 1% BSA. Infected erythrocytes, at a final hematocrit of 0.25–0.5%, were incubated with a 50% dilution of the test sera in RPMI-Hepes, pH 6.8, for 45 min at 37°C, on a rotating wheel, and 30-μl aliquots then were spotted over the receptor. After incubation at 37°C for 15 min the Petri dishes were washed five times with RPMI-Hepes to remove unbound cells, fixed with 2% glutaraldehyde, and stained with 10% Giemsa for microscopic examination.
RESULTS
Parasites Selected for CSA Binding Show Comodulation of Adherence Phenotype and Variant Antigenic Type.
To identify the mechanism of CSA adhesion, the cloned parasite line FAF-EA8 (23) was selected by panning for binding to CSA, creating the CS2 line. Cytoadherence assays performed on synchronized late trophozoite-stage parasites showed a distinct shift in adherence phenotype after selection (Table 1). FAF-EA8-infected erythrocytes adhered at high levels to ICAM-1 and CD36 but at only low levels to CSA, whereas CS2-infected erythrocytes showed virtually no binding to ICAM-1 and CD36, but markedly up-regulated binding to CSA. Agglutination assays with two rabbit antisera raised against FAF-EA8 trophozoites (R1341 and R1360) and two against CS2 (R1945 and R1944) demonstrated that a distinct antigenic shift also had taken place (Table 1). The comodulation of adherence and antigenicity strongly suggests that the variant antigen also determines the CSA binding phenotype.
Table 1.
Phenotypic characteristics of CSA-selected line CS2 and parent line FAF-EA8
| Cytoadherence*
|
Agglutination
|
||||||
|---|---|---|---|---|---|---|---|
| CD36 | ICAM-1 | CSA | R1341 | R1360 | R1945 | R1944 | |
| FAF-EA8 | 1666 ± 298 | 426 ± 88 | 47.9 ± 15 | +++ | +++ | – | – |
| CS2 | 13 ± 4 | 3 ± 1.3 | 3,918 ± 186 | – | – | +++ | ++ |
+++ = agglutinates of 20–100 infected erythrocytes; ++ = agglutinates of 5–20 infected erythrocytes.
Infected erythrocytes binding per mm2. Mean of triplicate counts ± SE.
A Specific var Gene Is Transcribed in CS2.
Poly(A)+ RNA was isolated from trophozoite-stage cultures and cDNA-synthesized to investigate var gene expression. To ensure consistency, the parasites were harvested from the same batch of parasites that were tested in cytoadherence assays. The cDNA was amplified by reverse transcription–PCR with degenerate oligonucleotide primers targeting conserved blocks of DBL domains. The amplification of FAF-EA8 and CS2 cDNA (Fig. 1a) produced a common band of 500 bp, which was cloned and found to be erythrocyte-binding antigen 175 (eba-175), a merozoite surface antigen involved in erythrocyte recognition that contains DBL domains (33). A second band of 700 bp was detected only in the CS2 line, and the sequenced clones were homologous to a var gene DBL3 domain. Specific primers designed to a nonconserved region of this sequence confirmed the differential expression of a var gene in the CSA binding line (Fig. 1b). It should be noted that although the “universal” primers provided clear evidence of specific gene transcription in CS2, they also demonstrated their limitations, as transcription of other var genes in FAF-EA8 could be detected with alternative primer sets (data not shown).
A B8(ItG) YAC clone (34) containing the var-CS2 gene was identified by using specific CS2-DBL3 primers, and the presence of a single var gene was confirmed by restriction digests. The gene sequenced from the YAC (Fig. 1c) was characteristic of the var family. The 6.7-kb exon 1 possesses a head region comprising a semiconserved DBL1 domain and a cysteine-rich inter-domain region (CIDR), two downstream DBL domains (numbered 3 and 4 after the first published sequence, ref. 24), and a C-terminal sequence of variable length, terminating in a transmembrane consensus sequence. A 1.4-kb putative cytoplasmic domain (ATS) then follows a 729-bp intron. cDNA clones later were generated from different regions along the gene, including the intron splice junction, by using both specific and degenerate primers. All cDNA fragments sequenced corresponded with the YAC sequence, confirming this to be the sole expressed var gene (data not shown). Little is known about specific motifs that bind to CSA and a search of the var-CS2 gene sequence did not identify any of the known heparin/heparan sulfate-binding consensus sequences or homologous sequences of galactose-binding proteins (35).
Mapping of the gene by pulsed field gel electrophoresis (data not shown) localized the gene to the same subtelomeric fragment of chromosome 6 in both the active (CS2) and inactive (FAF-EA8) state.
Transcription of var-CS2 Results in Expression of a Different Form of PfEMP1 from the Parent Line.
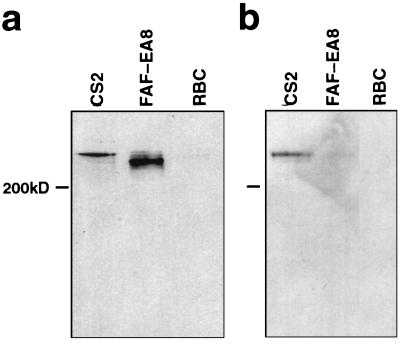
To investigate the biological properties of the var-CS2 product, several recombinant glutathione S-transferase-fusion protein fragments were produced corresponding to the major domains of the gene (Fig. 1c) and rabbit antibodies were raised against them. Membrane proteins of FAF-EA8 and CS2 trophozoite-infected erythrocytes were extracted, and Western blots were performed. The blots then were probed with the rabbit antisera. Antiserum to a conserved region of the ATS reacted with a high molecular weight band with the migration characteristics of PfEMP1 (Fig. 2a). A single band was visible in both FAF-EA8 and CSA, but the phenotypic switch had been accompanied by a marked increase in the size of the molecule expressed by CS2. The anti-DBL4 serum reacted specifically with the CS2-PfEMP1 band under nonreducing electrophoresis conditions (Fig. 2b), confirming the expression of a specific form of PfEMP1 in the line selected for CSA binding. The other antisera tested labeled CS2, but lacked clear specificity in this assay and also labeled FAF-EA8 and the unrelated 3D7 line (data not shown).
Figure 2.
Antibody to recombinant fusion proteins demonstrates that a different form of PfEMP1 is expressed in the CSA selected line. (a) Western blot from a reducing PAGE probed with the conserved anti-ATS affinity purified serum showing size differences in PfEMP1. (b) Western blot from a nonreducing PAGE probed with anti-DBL4 serum differentiates between CS2 and FAF-EA8 PfEMP1.
The anti-DBL3 and DBL4 antisera specifically labeled CS2 in immunofluorescence assays, and not FAF-EA8 or 3D7. They produced a punctate pattern of fluorescence at the surface of trophozoite-infected erythrocytes, similar to our previous report for PfEMP1 (21). The same pattern was seen by using anti-DBL3 and DBL4 antisera on both acetone-fixed and live infected erythrocytes or the ATS antisera on fixed, but not live, infected erythrocytes.
Antibodies to Recombinant var-CS2 Proteins Inhibit Adhesion of CS2-Infected Erythrocytes to CSA.
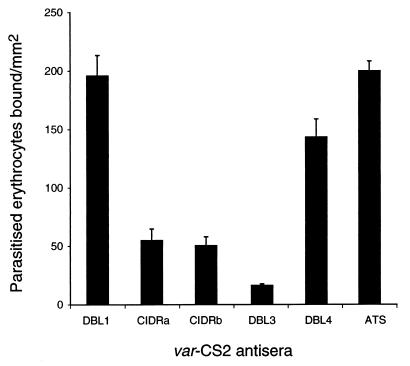
Antibodies raised in rabbits to recombinant var-CS2 proteins were tested for their ability to inhibit the adhesion of CS2-infected erythrocytes to CSA. Antisera against DBL3 almost completely inhibited adhesion to CSA and antisera against two regions of the CIDR (CIDRa: amino acids 404–736 and CIDRb: amino acids 716–905) also had a strong inhibitory effect (Fig. 3), when compared with DBL1, DBL4, ATS, or sera collected before vaccination. When the same antisera were tested for inhibition of the binding of FAF-EA8 and 3D7 to CD36, only the anti-CIDR sera significantly inhibited binding (data not shown). None of the rabbit antisera were reactive in agglutination assays against CS2.
Figure 3.
Antisera to the recombinant var-CS2 CIDR and DBL3 domains significantly inhibit the binding of CS2 to CSA, when compared with DBL1, DBL4, or ATS. The bars represent the mean (±SEM) numbers of CS2-parasitized erythrocytes binding per mm2 to immobilized CSA, determined by triplicate assays performed on four separate occasions.
DISCUSSION
In this study we show that an expressed var gene of a CSA binding parasite line encodes a unique PfEMP1 species that mediates adhesion to CSA. P. falciparum-infected erythrocytes have been shown to adhere, in a highly specific manner, to immobilized purified CSA and to the CSA chains on thrombomodulin (36, 37), under both static (9) and physiological flow (14) conditions, but previously, the parasite ligand involved in the interaction had not been identified. CSA is known to be present on the surface of placental syncytiotrophoblasts (15) where it has been implicated as the receptor for parasite sequestration in the placenta, an essential process in the pathogenesis of maternal malaria (12), and the identification of PfEMP1 as a primary ligand is an important step toward a better understanding of this disease.
Selection of the P. falciparum line FAF-EA8 for binding to CSA resulted in dramatically increased adhesion to this receptor and caused a change in variant antigenic type. The comodulation of adherence and antigenic type is a characteristic that suggested PfEMP1 as a potential parasite ligand (22, 23), but further investigation was necessary to confirm the central role of PfEMP1 in this adhesion phenotype. A specific var gene subsequently was shown to be transcribed in the CSA binding line CS2, but not in the parent FAF-EA8, and it then was demonstrated that the switch to this gene correlates with the expression of a var-CS2-specific variety of PfEMP1.
The association of the CS2-PfEMP1 with CSA binding was confirmed by demonstrating that adherence of CS2-infected erythrocytes to CSA is significantly reduced in the presence of antibodies raised against recombinant var-CS2 domains. The evidence that PfEMP1 mediates adhesion was enhanced by the finding that inhibition of CSA binding is not a general effect of antibodies against CS2-PfEMP1, but is restricted to antibodies targeting a particular region of the protein. The direct connection between CSA binding phenotype, var gene transcription, PfEMP1 expression, and PfEMP1-specific disruption of the CSA binding interaction provides compelling evidence that PfEMP1 is the primary parasite ligand in CSA adhesion.
Although proving the involvement of PfEMP1, inhibition of CSA binding by antibodies against specific recombinant domains is not conclusive proof of the location of the binding site, as it is possible that the results for individual antisera reflect physical hindrance of a site in an adjacent domain. Additionally, the lack of native conformation in the fusion-protein strategy may have restricted the activity of antibodies to other domains with potential binding ability.
The inhibitory effect of the CS2-DBL3 antibody was specific to CSA binding, but the anti-CIDR sera also was found to inhibit CD36 binding of other parasite lines. This finding was not entirely surprising, because CD36 adhesion is thought to be mediated by a binding site located in the CIDR (38), and the presence of well-conserved regions in this area of the protein may have led not only to the crossreactivity observed in immunoassay but also cross-interference in binding activity around this domain. The possibility of a CSA binding site in the proximity of the CIDR, which would potentially interfere with the ability of a parasite to bind to CD36, is an attractive speculation, given that field data suggest this exclusive relationship may exist in placental malaria (12). Further investigation of this region is clearly warranted.
Our identification of a dominant var gene transcribed in the CSA selected line is consistent with the findings of Scherf and colleagues (27), who selected the FCR3 line for CD36, ICAM-1, and CSA binding and found in each case a single var gene transcribed in the mature parasite. Those researchers, however, did not expand the studies to associate transcription with PfEMP1 expression. Also consistent with previous work (24, 27) was a lack of evidence of chromosomal rearrangement associated with the switch to the var-CSA. Recent data have shown that multiple genes may be transcribed during the parasite ring-stage before some form of silencing takes place as the parasite matures and directs the expression of a specific var gene (27, 39). Throughout our study particular attention was given to examining parasites harvested at the mature trophozoite stage, when the adhesive phenotype is displayed, thus avoiding confusion from this mechanism when investigating transcription of the functional var gene.
We have found that the binding of parasitized erythrocytes to CSA is mediated by PfEMP1 and that this protein is the product of a dominant var gene. The indication that the binding interaction centers on a specific region of the protein will direct future studies to defining the CSA binding motif and investigating the degree of conservation of the site of interaction among different CSA-binding parasites. The proof that PfEMP1 mediates CSA adhesion will allow a closer investigation of the pathogenesis of placental malaria and the consideration of using functional targets as the basis of malaria therapy.
Acknowledgments
We thank Tim Byrne and Anne Thaus for excellent technical assistance and the Blood Bank of the Red Cross of Victoria for generously providing serum and red cells. S.J.R. and J.G.B. were recipients of National Health and Medical Research Council of Australia Medical Postgraduate Scholarships. The Division of Infection and Immunity is supported by the National Health and Medical Research Council of Australia.
ABBREVIATIONS
- CSA
chondroitin sulfate A
- ICAM-1
intercellular adhesion molecule-1
- PfEMP1
P. falciparum erythrocyte membrane protein 1
- DBL
Duffy binding-like
- YAC
yeast artificial chromosome
- ATS
acidic terminal sequence
- CIDR
cysteine-rich inter-domain region
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF134154).
References
- 1.Aikawa M, Iseki M, Barnwell J W, Taylor D, Oo M M, Howard R J. Am J Trop Med Hyg. 1990;43:30–37. doi: 10.4269/ajtmh.1990.43.30. [DOI] [PubMed] [Google Scholar]
- 2.Bulmer J N, Rasheed F N, Francis N, Morrison L, Greenwood B M. Histopathology. 1993;22:211–218. doi: 10.1111/j.1365-2559.1993.tb00110.x. [DOI] [PubMed] [Google Scholar]
- 3.Barnwell J W, Asch A S, Nachman R L, Yamaya M, Aikawa M. J Clin Invest. 1989;84:765–772. doi: 10.1172/JCI114234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ockenhouse C F, Tandon N N, Magowan C, Jamieson G A, Chulay J D. Science. 1989;243:1469–1471. doi: 10.1126/science.2467377. [DOI] [PubMed] [Google Scholar]
- 5.Berendt A R, Simmons D L, Tansey J, Newbold C I, Marsh K. Nature (London) 1989;341:57–59. doi: 10.1038/341057a0. [DOI] [PubMed] [Google Scholar]
- 6.Ockenhouse C F, Tegoshi T, Maeno Y, Benjamin C, Ho M, Kan K E, Thway Y, Win K, Aikawa M, Lobb R R. J Exp Med. 1992;176:1183–1189. doi: 10.1084/jem.176.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts D D, Sherwood J A, Spitalnik S L, Panton L J, Howard R J, Dixit V M, Frazier W A, Miller L H, Ginsburg V. Nature (London) 1985;318:64–66. doi: 10.1038/318064a0. [DOI] [PubMed] [Google Scholar]
- 8.Ho M, Schollaardt T, Niu X, Looareesuwan S, Patel K D, Kubes P. Blood. 1998;91:4803–4809. [PubMed] [Google Scholar]
- 9.Rogerson S J, Chaiyaroj S C, Ng K, Reeder J C, Brown G V. J Exp Med. 1995;182:15–20. doi: 10.1084/jem.182.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siano J P, Grady K K, Millet P, Wick T M. Am J Trop Med Hyg. 1998;59:77–79. doi: 10.4269/ajtmh.1998.59.77. [DOI] [PubMed] [Google Scholar]
- 11.Treutiger C J, Heddini A, Fernandez V, Muller W A, Wahlgren M. Nat Med. 1997;2:1405–1408. doi: 10.1038/nm1297-1405. [DOI] [PubMed] [Google Scholar]
- 12.Fried M, Duffy P E. Science. 1996;272:1502–1504. doi: 10.1126/science.272.5267.1502. [DOI] [PubMed] [Google Scholar]
- 13.Robert C, Pouvelle B, Meyer P, Muanza K, Fukioka H, Aikawa M, Scherf A, Gysin J. Res Immunol. 1995;146:383–393. doi: 10.1016/0923-2494(96)81042-x. [DOI] [PubMed] [Google Scholar]
- 14.Cooke B M, Rogerson S J, Brown G V, Coppel R L. Blood. 1996;88:4040–4044. [PubMed] [Google Scholar]
- 15.Parmley R T, Takagi M, Denys F R. Anat Rec. 1984;210:477–484. doi: 10.1002/ar.1092100308. [DOI] [PubMed] [Google Scholar]
- 16.Brabin B J. Bull WHO. 1983;61:1005–1016. [PMC free article] [PubMed] [Google Scholar]
- 17.Fried M, Nosten F, Brockman A, Brabin B J, Duffy P E. Nature (London) 1998;395:851–852. doi: 10.1038/27570. [DOI] [PubMed] [Google Scholar]
- 18.Baruch D I, Gormley J A, Ma C, Howard R J, Pasloske B L. Proc Natl Acad Sci USA. 1996;93:3497–3502. doi: 10.1073/pnas.93.8.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leech J H, Barnwell J W, Miller L H, Howard R J. J Exp Med. 1984;159:1567–1575. doi: 10.1084/jem.159.6.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magowan C, Wollish W, Anderson L, Leech J. J Exp Med. 1988;168:1307–1320. doi: 10.1084/jem.168.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crabb B S, Cooke B M, Reeder J C, Waller R F, Caruana S R, Davern K M, Wickham M E, Brown G V, Coppel R L, Cowman A F. Cell. 1997;89:287–296. doi: 10.1016/s0092-8674(00)80207-x. [DOI] [PubMed] [Google Scholar]
- 22.Biggs B A, Goozé L, Wycherley K, Wollish W, Southwell B, Leech J H, Brown G V. Proc Natl Acad Sci USA. 1991;88:9171–9174. doi: 10.1073/pnas.88.20.9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biggs B A, Anders R F, Dillon H E, Davern K M, Martin M, Petersen C, Brown G V. J Immunol. 1992;149:2047–2054. [PubMed] [Google Scholar]
- 24.Su X-Z, Heatwole V M, Wertheimer S P, Guinet F, Herrfeldt J A, Peterson D S, Ravetch J A, Wellems T E. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 25.Baruch D I, Pasloske B L, Singh H B, Bi X, Ma X C, Feldman M, Taraschi T F, Howard R J. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 26.Smith J D, Chitnis C E, Craig A G, Roberts D J, Hudson-Taylor D E, Peterson D S, Pinches R, Newbold C I, Miller L H. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, Gysin J, Lanzer M. EMBO J. 1998;17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeder J C, Rogerson S J, Al-Yaman F, Anders R F, Coppel R L, Novakovic S, Alpers M P, Brown G V. Am J Trop Med Hyg. 1994;51:45–55. doi: 10.4269/ajtmh.1994.51.45. [DOI] [PubMed] [Google Scholar]
- 29.Lambros C, Vanderberg J P. J Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- 30.Triglia T, Cowman A F. Proc Natl Acad Sci USA. 1994;91:7149–7153. doi: 10.1073/pnas.91.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson D S, Miller L H, Wellems T E. Proc Natl Acad Sci USA. 1995;92:7100–7104. doi: 10.1073/pnas.92.15.7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beeson J G, Chai W, Rogerson S J, Lawson A M, Brown G V. Infect Immun. 1998;66:3397–3402. doi: 10.1128/iai.66.7.3397-3402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sim B K L, Chitnis C E, Wasniowska K, Hadley T J, Miller L H. Science. 1994;264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- 34.Triglia T, Kemp D J. Mol Biochem Parasitol. 1991;44:207–211. doi: 10.1016/0166-6851(91)90006-r. [DOI] [PubMed] [Google Scholar]
- 35.Holt G D. Glycobiology. 1991;1:329–336. doi: 10.1093/glycob/1.4.329. [DOI] [PubMed] [Google Scholar]
- 36.Rogerson S J, Novakovic S, Cooke B M, Brown G V. Exp Parasitol. 1997;86:8–18. doi: 10.1006/expr.1996.4142. [DOI] [PubMed] [Google Scholar]
- 37.Gysin J, Pouvelle B, Le Tonqueze M, Edelman L, Boffa M-C. Mol Biochem Parasitol. 1997;88:267–271. doi: 10.1016/s0166-6851(97)00082-0. [DOI] [PubMed] [Google Scholar]
- 38.Baruch D I, Ma X C, Singh H B, Bi X, Pasloske B L, Howard R J. Blood. 1997;90:3766–3775. [PubMed] [Google Scholar]
- 39.Chen Q, Fernandez V, Sundstrom A, Schlichtherle M, Datta S, Hagblom P, Wahlgren M. Nature (London) 1998;394:392–395. doi: 10.1038/28660. [DOI] [PubMed] [Google Scholar]