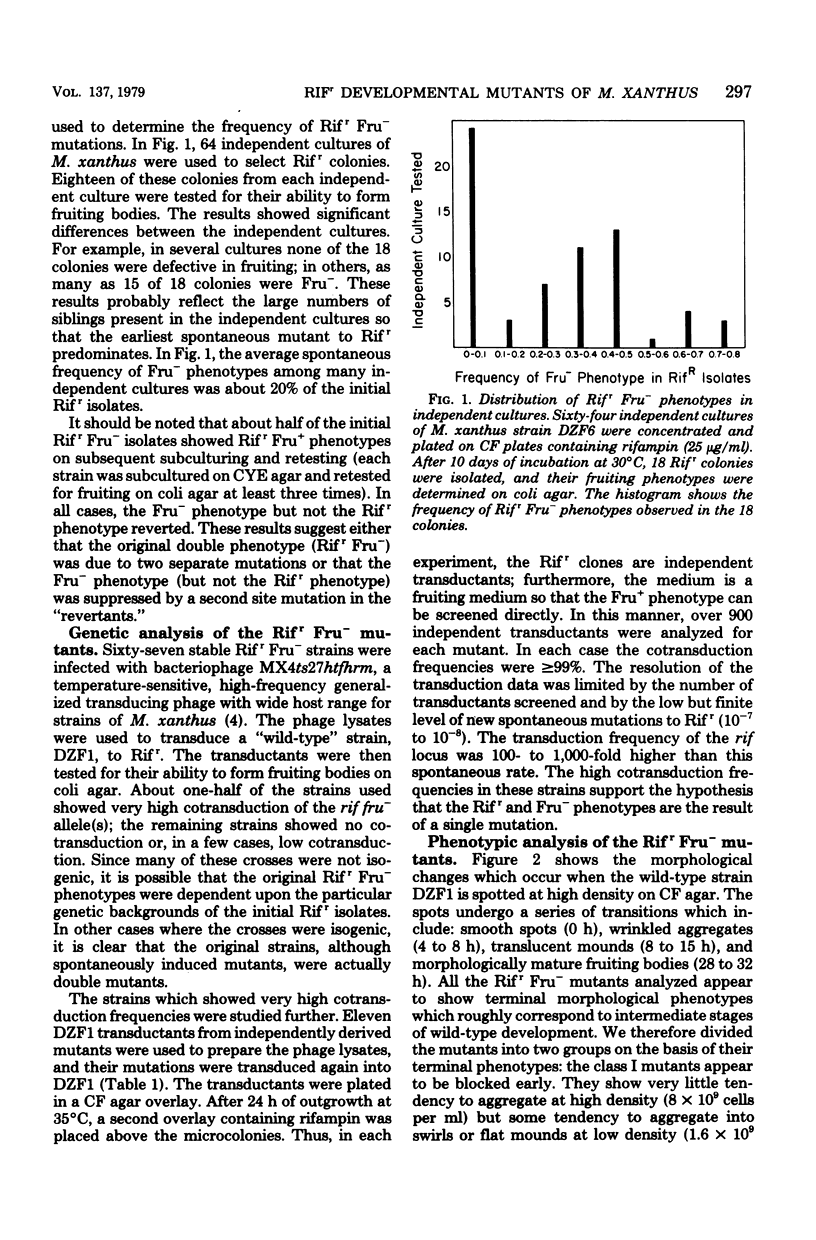
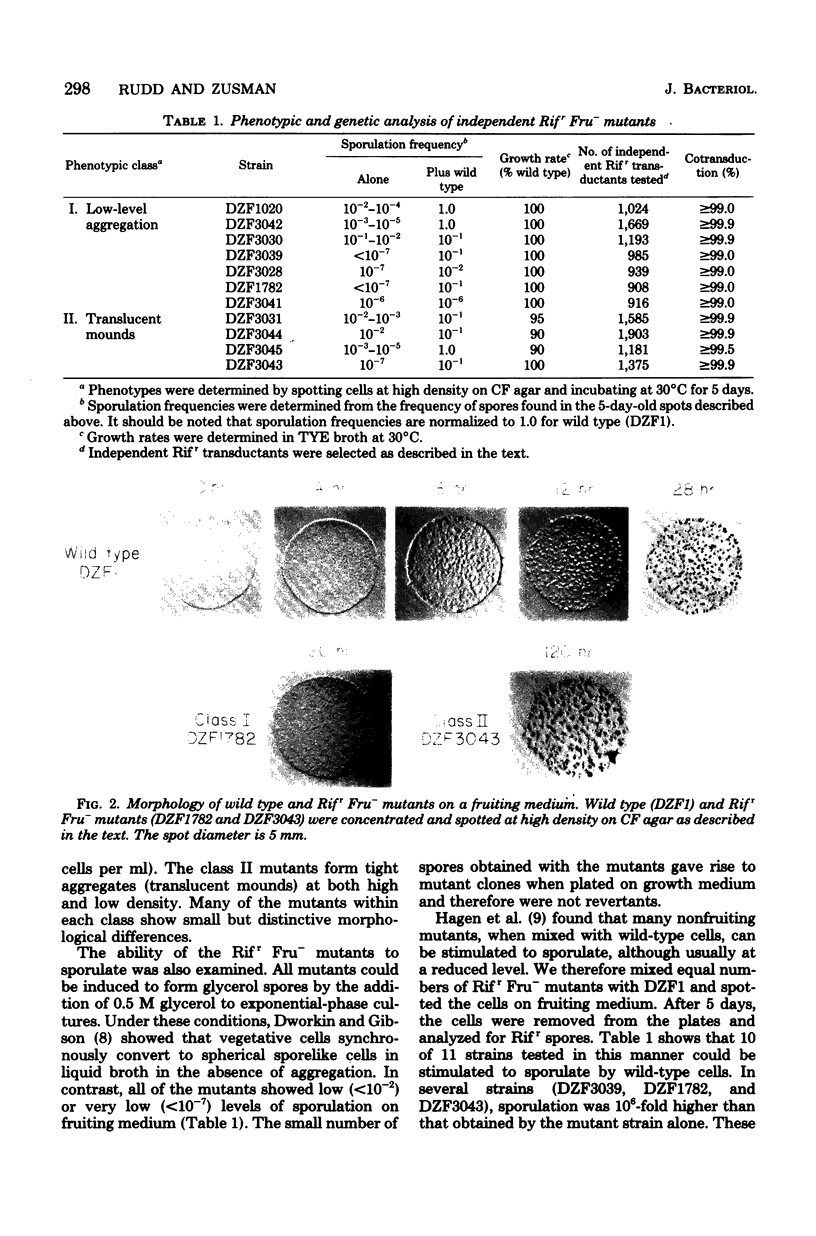
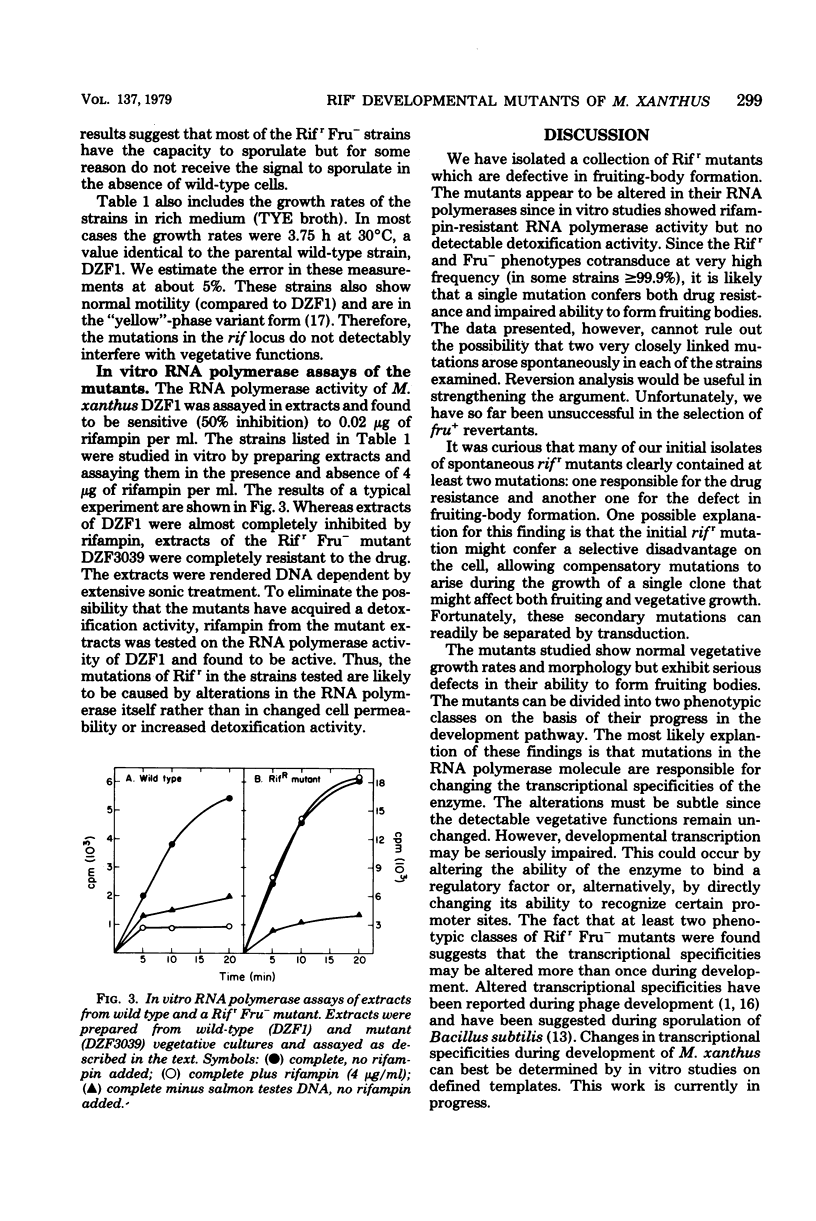
Abstract
Rifampin, an antibiotic which is known to bind to and inhibit RNA polymerase, was used to probe the molecular regulation of development in Myxococcus xanthus. Rifampin-resistant mutants were screened for defects in fruiting-body formation. About 20% of the isolates in the initial screenings showed major defects in developmental aggregation or sporulation. Eleven independent mutants with wild-type growth rates and stable phenotypes were analyzed by transduction. In these strains, the rifampin-resistant and nonfruiting phenotypes showed cotransduction frequencies equal to or greater than 99.0 to 99.9%. The RNA polymerase activities were resistant to rifampin in vitro, indicating that the RNA polymerase is altered in these strains. Although their fruiting phenotypes are heterogeneous, these strains can be divided into two classes based on the level of aggregation. The results suggest that RNA polymerase plays a significant role in the regulation of development in M. xanthus since mutations which cause no apparent changes in vegetative growth result in striking defects in fruiting-body formation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolle A., Epstein R. H., Salser W., Geiduschek E. P. Transcription during bacteriophage T4 development: synthesis and relative stability of early and late RNA. J Mol Biol. 1968 Feb 14;31(3):325–348. doi: 10.1016/0022-2836(68)90413-0. [DOI] [PubMed] [Google Scholar]
- Bretscher A. P., Kaiser D. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J Bacteriol. 1978 Feb;133(2):763–768. doi: 10.1128/jb.133.2.763-768.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess R. R. A new method for the large scale purification of Escherichia coli deoxyribonucleic acid-dependent ribonucleic acid polymerase. J Biol Chem. 1969 Nov 25;244(22):6160–6167. [PubMed] [Google Scholar]
- Campos J. M., Geisselsoder J., Zusman D. R. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978 Feb 25;119(2):167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- Campos J. M., Zusman D. R. Regulation of development in Myxococcus xanthus: effect of 3':5'-cyclic AMP, ADP, and nutrition. Proc Natl Acad Sci U S A. 1975 Feb;72(2):518–522. doi: 10.1073/pnas.72.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DWORKIN M., GIBSON S. M. A SYSTEM FOR STUDYING MICROBIAL MORPHOGENESIS: RAPID FORMATION OF MICROCYSTS IN MYXOCOCCUS XANTHUS. Science. 1964 Oct 9;146(3641):243–244. doi: 10.1126/science.146.3641.243. [DOI] [PubMed] [Google Scholar]
- Doi R. H., Brown L. R., Rodgers G., Hsu Y. Bacillus subtilis mutant altered in spore morphology and in RNA polymerase activity. Proc Natl Acad Sci U S A. 1970 Jun;66(2):404–410. doi: 10.1073/pnas.66.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen D. C., Bretscher A. P., Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978 Jun;64(2):284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Leighton T. J. An RNA polymerase mutation causing temperature-sensitive sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A. 1973 Apr;70(4):1179–1183. doi: 10.1073/pnas.70.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn T., Losick R., Sonenshein A. L. Rifampin resistance mutation of Bacillus subtilis altering the electrophoretic mobility of the beta subunit of ribonucleic acid polymerase. J Bacteriol. 1975 Jun;122(3):1387–1390. doi: 10.1128/jb.122.3.1387-1390.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabussay D., Zillig W. A rifampicin resistent rna-polymerase from E. coli altered in the beta-subunit. FEBS Lett. 1969 Oct 21;5(2):104–106. doi: 10.1016/0014-5793(69)80305-4. [DOI] [PubMed] [Google Scholar]
- Sonenshein A. L., Losick R. RNA polymerase mutants blocked in sporulation. Nature. 1970 Aug 29;227(5261):906–909. doi: 10.1038/227906a0. [DOI] [PubMed] [Google Scholar]
- Talkington C., Pero J. Restriction fragment analysis of the temporal program of bacteriophage SPO1 transcription and its control by phage-modified RNA polymerases. Virology. 1977 Dec;83(2):365–379. doi: 10.1016/0042-6822(77)90181-7. [DOI] [PubMed] [Google Scholar]
- Wireman J. W., Dworkin M. Morphogenesis and developmental interactions in myxobacteria. Science. 1975 Aug 15;189(4202):516–523. doi: 10.1126/science.806967. [DOI] [PubMed] [Google Scholar]
- Zusman D. R., Krotoski D. M., Cumsky M. Chromosome replication in Myxococcus xanthus. J Bacteriol. 1978 Jan;133(1):122–129. doi: 10.1128/jb.133.1.122-129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]



