Abstract
Although the complete genome of Mycoplasma genitalium has been sequenced, the functional identification of various genes, including those involved in virulence, has not been accomplished. Further compounding these difficulties has been the failure to develop genetic tools in mycoplasmas that permit the assessment of gene and operon function and regulation. To determine whether homologous recombination could be developed as a tool to analyze the function of genes in M. genitalium, a plasmid that replicates in Escherichia coli but not in M. genitalium was constructed to disrupt the cytadherence-related gene mg218 of M. genitalium. The electroporation of this disruption plasmid into wild-type hemadsorption-positive (HA+) M. genitalium cells permitted the isolation of HA− (strain JB1) and partial HA+ (strains JB2 and JB20) transformants. Analysis of the transformants by Southern hybridization indicated that homologous recombination occurred at the mg218 locus by single-crossover events in JB1 and JB2 and by a double-crossover event in JB20. While integration of the disruption construct abolished the expression MG218 in JB1, strains JB2 and JB20 exhibited a truncated MG218 protein (160 kDa), possibly because of in-frame fusion of the disrupted mg218 gene with sequences downstream of the gentamycin-resistance gene present in the disruption construct. Strain JB1, which lacked MG218, displayed a post-translational defect, being unable to maintain the structural integrity of the major adhesin P140 and its operon-related protein P110, in contrast to JB2 and JB20. It appears that MG218 influences the stability of other cytadherence-related proteins in vivo. Thus, targeted gene disruption through homologous recombination will be a powerful and promising tool for investigating the biology and pathogenesis of M. genitalium.
Mycoplasmas represent the smallest self-replicating biological entities and are characterized by their streamlined genomes, cholesterol-dependent membranes, absence of a cell wall, fastidious nutritional requirements, and use of UGA as a tryptophan codon. These fascinating prokaryotes continue to reemerge as important human and animal pathogens (1). Several Mycoplasma species cause acute and chronic diseases in humans, and recent studies have implicated mycoplasmas in AIDS progression, malignant transformation, chronic fatigue syndrome, and acute and fulminant respiratory distress (1, 2). Therefore, mycoplasmas represent exceptional model systems to study the genetic and biochemical basis of disease pathogenesis. However, little is known about pathogenic mechanisms in mycoplasmas, in part because of the lack of satisfactory genetic tools to dissect gene function. To overcome these experimental restrictions and clarify the genetic basis of mycoplasma virulence, the genomes of Mycoplasma genitalium (580 kb) and Mycoplasma pneumoniae (816 kb) were sequenced (3, 4). M. pneumoniae causes primary atypical pneumonia and a spectrum of postrespiratory complications in humans, and M. genitalium, a relatively recently described species, is an important cause of non-gonococcal urethritis and other pathologies (1, 5, 6). Unfortunately, genome sequencing data failed to reveal conventional pathogenic mechanisms, such as two-component regulatory systems, iron acquisition, and oxidative stress responses. In addition, the function of more than 50% of the genes in both M. pneumoniae and M. genitalium could not be predicted, as they lacked significant identity with existing genes in the databases, or they corresponded to sequenced genes of unknown function.
Genetic tools for mycoplasmas have been almost nonexistent, as mycoplasmas appear refractory to common genetic manipulations. Recently, transposons Tn4001 and Tn916, which originate from the Gram-positive bacteria Staphylococcus aureus (7) and Enterococcus faecalis (8), respectively, have demonstrated limited utility in mycoplasmas. The expression of gentamycin- and tetracycline-resistance markers of Tn4001 and Tn916, respectively, has been detected in several Mycoplasma species infecting both animals and humans (9). Also, insertional mutagenesis of transposon Tn4001 has been accomplished in both M. pneumoniae (10, 11) and M. genitalium (11), and complement gene expression in M. pneumoniae has been described (12). Although these successes are considered advancements in dissecting the genetic mechanisms of mycoplasmas, it appears that the transposition events using Tn4001 are nonrandom and appear directed to specific hot spots in the genomes of M. pneumoniae and M. genitalium. Thus, the development of additional genetic tools in mycoplasmas is pivotal to define gene and operon function and regulation.
Targeted disruption of genes through homologous recombination has been an alternate means of generating mutants in bacteria (13, 14). Although this is a powerful approach, occurrence of homologous recombination has been reported only in the nonpathogenic mollicute Acholeplasma laidlawii (15). In this study, we describe the occurrence of homologous recombination in pathogenic mycoplasmas. Furthermore, we show that disruption of mg218 of M. genitalium through homologous recombination results in a nonhemadsorbing phenotype and the instability of other cytadherence-related proteins, indicating that the product of mg218 plays a significant role in protein–protein interactions.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Culture Conditions.
M. genitalium G37 was grown in 100 ml of SP-4 medium (5) at 37°C for 72 h in 150-cm2 tissue culture flasks (Corning). Surface-attached mycoplasmas were washed four times with phosphate-buffered saline (PBS; pH 7.2) and collected by centrifugation at 20,000 × g for 20 min at 4°C. E. coli strain DH5-α harboring the plasmids pISM2061 and pCR2 was grown at 37°C in LB broth or LB agar plates containing 100 μg/ml ampicillin. Plasmid pISM2061 carrying transposon Tn4001 was a kind gift from C. Minion (Iowa State Univ., Ames, IA), and plasmid pCR2 was purchased from Invitrogen.
Electroporation and Transformation.
Transformation of M. genitalium with suicidal plasmid pDP219 containing the gentamycin-resistance gene was performed by electroporation as described before (11).
Isolation of Hemadsorption-Negative (HA−) M. genitalium Transformants.
The hemadsorption-positive (HA+) phenotype is a characteristic property of wild-type M. genitalium which correlates well with the adherence of this species to eukaryotic cells (16). To isolate HA− and partial HA+ mycoplasma colonies, SP-4 plates were flooded with 2 ml of diluted (1:50) sheep erythrocytes (40% erythrocytes in 60% Alsever’s solution; BioWhittaker). After 1 h of incubation at 37°C, mycoplasma colonies were washed repeatedly with PBS and observed by light microscopy. Individual HA− and partial HA+ colonies were picked and grown in 2 ml of SP4 medium with 100 μg/ml gentamycin.
DNA Manipulations.
Standard recombinant DNA techniques were followed (17), and chromosomal DNA from M. genitalium was isolated as reported earlier (18). PCR was performed by using standard protocols, and a 2-kb PCR fragment encompassing the 5′ portion of mg218 was obtained by using primers MG218A (5′-AAGAATCAAGCAGAAGATAATCCT-3′) and MG218B (5′-GGTTTGTAATTCTTGCAACCGTTT-3′) and M. genitalium genomic DNA as template. This PCR fragment was cloned in the pCR2 vector (Invitrogen) to create plasmid pDP218. Plasmid pDP219 was produced by cloning a 2.5-kb blunt-ended HindIII fragment carrying the gene encoding gentamycin resistance, which was obtained from plasmid pISM2061, at the StuI site located in the insert DNA of plasmid pDP218. PCR was also performed to identify the integration of the disruption construct in transformants by using primers GENTA5 (5′-TATGAAAAAGGTGATAAATAAATG-3′) and GENTA6 (5′-AATTTCTGGTGTTAAAAAAGTTCC-3′). These primers are based on DNA sequences of the 2.5-kb gentamycin-resistance gene fragment, and PCR amplification with these primers in the presence of template is expected to yield an 880-bp DNA fragment. Sequencing of DNA fragments was performed with an automated cycle sequencing system (Applied Biosystems model 373, Center for DNA Technology Core Facility of University of Texas Health Science Center at San Antonio) with fluorescent terminators and by an Amplicycle sequencing kit (Perkin–Elmer) using a PCR machine. Southern hybridization was performed at 68°C, and probes for Southern hybridization were labeled with [α-32P]dCTP by using the random primer method.
RNA and Slot-Blot Analysis.
RNA from M. genitalium strains was isolated as described by us recently (19). For slot-blot analysis, RNA was diluted to different concentrations in 10 μl of diethyl pyrocarbonate (DEPC)-treated water in Eppendorf tubes. To each tube, 20 μl of 100% formamide, 7 μl of 37% formaldehyde, and 2 μl of 20× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate, pH 7.0) were added, and incubation was continued at 68°C for 15 min. Individual samples were mixed with 500 μl of 10× SSC and then blotted on nitrocellulose paper with a slot-blot apparatus (Bio-dot SF Micro filtration apparatus). The blots were baked at 80°C for 2 h and then hybridized with [α-32P]dCTP-labeled probes. A 3.8-kb EcoRI fragment containing 90% of the p140 gene, obtained from plasmid pMG140, was used to probe mRNA transcripts for P140. A 1.6-kb PCR fragment encompassing the 5′ portion of the p110 gene, obtained by using primers MG114F (5′-CCCCATCACCGATTCTAAAAG-3′) and MG114R (5′-GAATAGTTATAGGGAACAGGG-3′) and M. genitalium genomic DNA as template, was used as probe to detect mRNA transcripts for the P110 protein. A 700-bp PCR fragment containing part of the rrnA gene, obtained by using primers GPO1 (5′-ACTCCTACGGGAGGCAGCAGTA-3′) and MGSO (5′-TGCACCATCTGTCACTCTGTTAACCTC-3′) and M. genitalium genomic DNA as template, was used as probe to detect mRNA for 16S rRNA. To generate sense and antisense probes for the p110 gene, the 1.6-kb PCR fragment was initially cloned in pCR2, cleaved as an EcoRI fragment, and cloned at the EcoRI site of pBluescript (SK) (Stratagene) between T3 and T7 promoters. After the sequence for the sense and antisense strand of p110 was verified, [α-32P]CTP-labeled RNA probes were produced by using T3/T7 RNA polymerases obtained from the Riboprobe combination systems kit (Promega) and hybridization was performed (17).
Pulse–Chase Analysis of Protein Synthesis.
Wild-type M. genitalium strain G37 or mg218 mutant strain JB1 was cultured to midlogarithmic phase in SP-4 medium and harvested by centrifugation at 20,000 × g. Mycoplasma pellets were washed with PBS and resuspended in 5 ml of Dulbecco’s modified Eagle’s medium (DMEM) (without cysteine and methionine) containing 10% bovine fetal serum. After the addition of 100 μl of 35S-labeled l-methionine/l-cysteine (1000 Ci/mmol; 37 TBq/mmol, NEN), each suspension was incubated for 3 h at 37°C, centrifuged, and resuspended in 6 ml of SP-4 medium prior to distribution to six microcentrifuge tubes. These tubes were incubated at 37°C, and one tube was removed each h for a period of 5 h. Mycoplasmas in each tube were spun down, resuspended in SDS-loading buffer, and kept at −70°C. Proteins were separated by SDS/polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose membrane, and exposed to x-ray film.
Characterization of Proteins.
SDS/PAGE (7.5% acrylamide) and Western blotting were performed as described (17). Mycoplasma proteins were visualized by staining with Coomassie brilliant blue or exposing to film in the case of radiolabeled extracts. Proteins transferred to nitrocellulose membranes by the Western blot method were probed with polyclonal rabbit antisera raised against M. genitalium proteins or specific peptides. Alkaline phosphatase-conjugated antibodies to rabbit immunoglobulin were used as secondary antibodies, and color development was performed by standard protocols.
RESULTS
Construct for Gene Disruption.
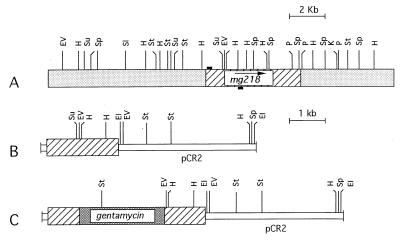
To demonstrate gene disruption through homologous recombination in pathogenic mycoplasmas, we chose mg218 of M. genitalium as the target gene. This is the largest ORF in the M. genitalium genome and encodes a predicted 216-kDa protein that shows high similarity to the cytadherence-related HMW2 protein of M. pneumoniae (11, 20). Hence, we thought that disruption of this gene in M. genitalium may implicate its role in cytadherence, which had not been previously determined. A 2-kb PCR fragment that encompasses the 5′ end of mg218 and is positioned 200 bp downstream of an ATG codon was initially generated by using primers MG218A and MG218B to create the gene disruption construct (Fig. 1A). After cloning this fragment into the pCR2 vector (pDP218; Fig. 1B), which is a pUC-based E. coli vector with genes encoding ampicillin and kanamycin resistance, a 2.5-kb gentamycin-resistance gene was cloned within the insert fragment of pDP218 at the StuI site (Fig. 1C). The resulting construct pDP219 replicates only in E. coli and has no origin of replication for mycoplasmas. Therefore, the selection of gentamycin-resistant transformants can result only from integration of the plasmid into the mycoplasma genome. The gentamycin-resistance gene of the construct was obtained from Tn4001, for which the expression in M. genitalium has already been established (11).
Figure 1.
Schematic representation of the derivation of the mg218 disruption construct. (A) The mg218 locus in the M. genitalium chromosome. Restriction sites are based on the DNA sequences available in National Center for Biotechnology Information databases with accession no. U39699. The hatched box represents the coding region of mg218 and the stippled boxes represent the flanking regions of mg218. The small black boxes represent the location of the primers used for amplifying a portion of mg218 DNA. The arrow indicates the direction of transcription of mg218. (B) Schematic representation of plasmid pDP218. The hatched box represents the 2-kb PCR fragment encompassing the 5′ portion of mg218. The open box represents the pCR2 vector. (C) Schematic representation of plasmid pDP219. The hatched boxes indicate the mg218 DNA. The densely stippled box indicates the gentamycin-resistance gene from Tn4001 inserted at the StuI site of pDP218. The open box represents the pCR2 vector. A detailed description of the construction of plasmids pDP218 and pDP219 appears in Materials and Methods. EI, EcoRI; EV, EcoRV; H, HindIII; K, KpnI; P, PstI; SI, SalI; Sp, SpeI; St, StyI; Su, StuI.
Analysis of Transformants.
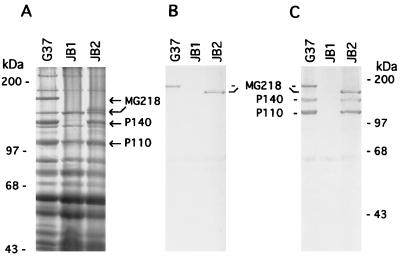
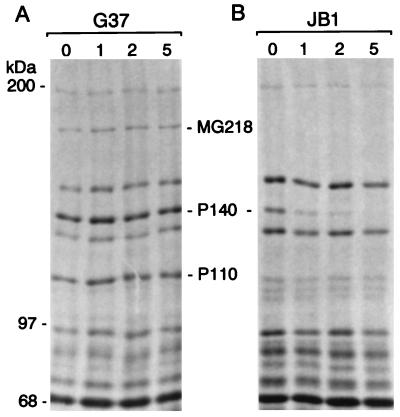
We initially selected transformants after electroporation of the mg218 disruption construct pDP219 into M. genitalium on SP-4 plates containing 50 μg/ml gentamycin. Preliminary analysis of the transformants, however, revealed that transformants selected at this concentration of gentamycin included spontaneous M. genitalium mutants. Therefore, subsequent selections were made on SP-4 plates containing 100 μg/ml gentamycin. Four electroporations yielded a total of 22 transformants (3–7 transformants per electroporation), which all showed the presence of the gentamycin-resistance gene in their genome when analyzed by PCR (data not shown). Because MG218 is a homologue of the cytadherence-related HMW2 protein of M. pneumoniae, we predicted that disruption of mg218 would lead to the generation of HA− mycoplasmas. Thus, the transformants were screened for hemadsorption, and four transformants were found to be totally HA−, whereas the remaining 18 transformants exhibited reduced hemadsorption capabilities (i.e., partial HA+). In addition, all 22 transformants were screened by SDS/PAGE for the absence of MG218. Protein profiles of HA− and partial HA+ transformants were distinctly different from each other and from the profile of wild-type M. genitalium strain G37 (Fig. 2A). With JB1, a representative of the HA− transformants, an approximately 190-kDa protein was completely absent. Further, two major high molecular weight proteins with sizes of 140 kDa and 110 kDa appeared absent or markedly reduced in this category of transformants as compared with wild-type M. genitalium strain G37. On the basis of their sizes, these proteins corresponded to P140 and P110, which are encoded by the p140 cytadherence-related operon of M. genitalium (16). In contrast, protein profiles of JB2, which represents the partial HA+ transformant category, revealed the absence of the 190-kDa protein but the presence of 140-kDa and 110-kDa proteins. In addition, a new protein appeared in this category of transformants at approximately 160 kDa (Fig. 2A).
Figure 2.
SDS/PAGE and Western blot profiles of M. genitalium wild-type G37 and mg218 mutants JB1 and JB2 (strains JB2 and JB20 showed similar protein profiles; hence, only JB2 is shown). (A) Total proteins separated by SDS/7.5% PAGE and stained with Coomassie brilliant blue. (B) Western blot probed with anti-MG218 antibodies identifying intact and truncated MG218. (C) Western blot reprobed with anti-P140 and anti-P110 antibodies. MG218, P140, and P110 represent the corresponding M. genitalium proteins. Marker proteins are indicated on the extreme left and right.
To prove that the missing 190-kDa protein in JB1 and JB2 was MG218 of M. genitalium, we performed Western blot analysis using anti-MG218 antibodies generated against an immunogenic synthetic peptide of MG218 as described earlier. Although this antiserum reacted with the 190-kDa protein in the wild-type G37 strain and, surprisingly, with the newly appeared 160-kDa protein in JB2, it showed no reactivity in JB1 (Fig. 2B). This finding indicated that JB1 completely lacked the MG218 protein, whereas JB2 possessed a truncated version of MG218 (i.e., the 160-kDa protein). Western blots previously probed with anti-MG218 were reprobed with anti-P140 and anti-P110 antibodies to confirm the earlier evidence (Fig. 2A) of their apparent absence or marked reduction in JB1. As expected, anti-P140 and anti-P110 antibodies reacted with wild-type G37 and JB2 but not with JB1 (Fig. 2C). While the absence of immunoreactivity at 140 kDa of JB1 is consistent with the protein profile in this region (Fig. 2A), the nonreactivity at the 110-kDa region is somewhat unexpected because a strongly staining protein band appears at this region. Lack of immunoreactivity may be due to improper folding of putative P110-related peptides or the presence of unrelated peptides.
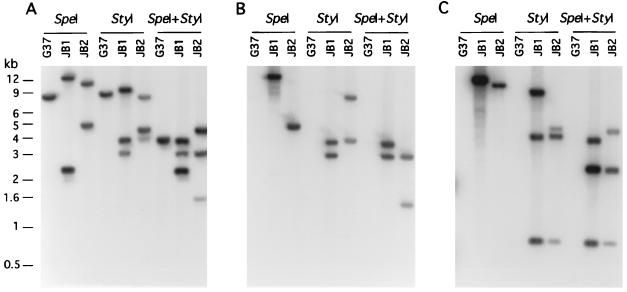
To further demonstrate that the different protein profiles of HA− and partial HA+ transformants directly correlated with the disruption of their mg218 locus, we digested genomic DNA from wild-type G37 and HA− and partial HA+ transformants with SpeI, StyI, and SpeI and StyI together and probed with a 2-kb PCR fragment of mg218 (probe I), originally used for making the disruption construct. While the Southern profiles for all HA− and partial HA+ transformants were distinct from the profile of wild-type G37, consistent with the disruption of the mg218 locus, two different categories of transformants were identified among the partial HA+ transformants, despite their similarity in partial HA+ phenotype and protein profiles. The first category of 11 partial HA+ transformants was identical to JB2 (Fig. 3A), while the second category of 7 partial HA+ transformants was identical to JB20 (Fig. 4A). All four HA− transformants exhibited Southern profiles similar to that of JB1 (Fig. 3A). Thus, strains JB1, JB2, and JB20 were subjected to detailed Southern analysis.
Figure 3.
Southern hybridization profiles of DNA from M. genitalium strains. G37, wild-type; JB1 and JB2, mg218 mutants. StyI, SpeI, and SpeI+StyI represent the restriction enzymes used to cut genomic DNA. DNA fragments were resolved in 0.8% agarose gel, Southern transferred to nitrocellulose membrane, and probed with the following: a 2-kb mg218 gene fragment (A; probe I); a 2.5-kb gentamycin gene fragment (B; probe II); and a 3.9-kb pCRII plasmid DNA (C; probe III). The sizes of the DNA bands are indicated in kilobases (kb).
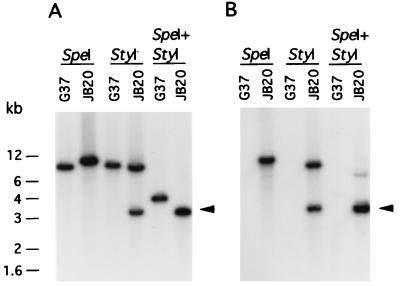
Figure 4.
Southern hybridization profiles of DNA from M. genitalium strains. G37, wild-type; JB20, mg218 mutant. StyI, SpeI, and SpeI+StyI represent the restriction enzymes used to cut genomic DNA. DNA fragments were resolved in 0.8% agarose gel, Southern transferred to nitrocellulose membrane, and probed with the following: a 2-kb mg218 gene fragment (A; probe I); or a 2.5-kb gentamycin gene fragment (B; probe II). There was no positive signal with 3.9-kb pCR2 plasmid DNA (probe III) with JB20 DNA. Arrows indicate the hybridization signals due to similar size fragments of 3.2 kb and 3.3 kb. The sizes of the DNA bands are indicated in kilobases (kb).
Mapping of the Integration Sites.
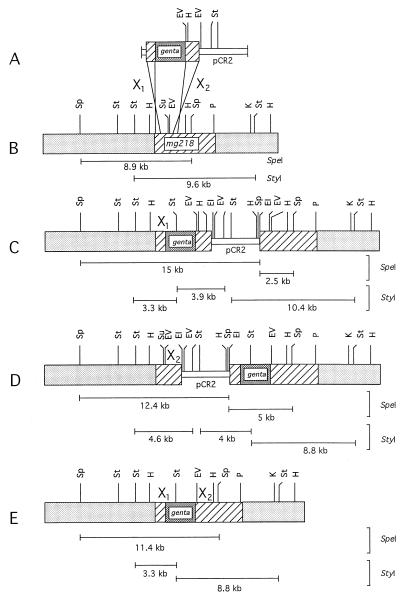
Based on the restriction sites in and around the mg218 locus (see Fig. 1A), as expected probe I hybridized with 8.9-, 9.6-, and 4.1-kb DNA fragments when wild-type G37 DNA was cut with SpeI, StyI, and SpeI + StyI, respectively. However, DNA from transformant JB1 that was cut with SpeI, StyI, and SpeI + StyI showed two bands (15 and 2.5 kb), three bands (10.4, 3.9, and 3.3 kb) and three bands (3.9, 3.3, and 2.5 kb), respectively (Fig. 3A). In contrast, transformant category JB2 showed numbers of bands similar to those of JB1 but different sizes when DNA was cut with SpeI (12.4 and 5 kb), StyI (8.8, 4.6, and 4 kb) and SpeI + StyI (4.6, 3.3, and 1.7 kb) (Fig. 3A). Interestingly, transformant JB20 displayed a Southern profile with one band (11.4 kb) for SpeI, two bands (3.3 and 8.8 kb) for StyI, and two bands (although it appears as a single band, this band is composed of two similar-size DNA fragments of 3.2 and 3.3 kb; see Fig. 5) for SpeI + StyI-cut DNA (Fig. 4A). These results suggested that the mg218 locus in JB1, JB2, and JB20 has been disrupted possibly by three different mechanisms. To further delineate this possibility, a 2.5-kb DNA fragment containing the gentamycin-resistance gene (probe II) and a 3.9-kb DNA of plasmid pCR2 (probe III) were used as probes in Southern analysis. Although probe II hybridized with DNA from JB1, JB2 (Fig. 3B), and JB20 (Fig. 4B), probe III showed positive signals only with DNA from JB1 and JB2 (Fig. 3C) and failed to detect any signal with strain JB20. Also, probe II and probe III showed distinct Southern profiles for JB1 and JB2 (Fig. 3 B and C). On the basis of the sizes of the bands obtained with the different probes and the overlaying of the autoradiographs on each other, we predicted that the mg218 locus in JB1 and JB2 was disrupted by the integration of disruption construct pDP219 at two different sites by single-crossover events. In the case of JB20, the disruption of the mg218 locus appeared to be by a double-crossover event. The mg218 locus in JB1, JB2, and JB20, along with the observed sizes of SpeI and StyI fragments, are schematically depicted in Fig. 5. It should be noted that, despite disruption of the mg218 locus, strains JB2 and JB20 can still produce a 160-kDa truncated MG218. Unlike JB1, strains JB2 and JB20 possess similar size mg218 regions downstream of the gentamycin-resistance gene (Fig. 5 B and C). Therefore, expression of a truncated MG218 in these strains may be due to the fusion of part of the mg218 gene to an ORF or promoter region available in the 2.5-kb DNA that contains the gentamycin-resistance gene.
Figure 5.
Homologous recombination at the mg218 locus in M. genitalium. (A) Schematic representation of the two possible crossover events (X1 and X2) of pDP19 (mg218 disruption construct) with the mg218 locus in the genomic DNA of M. genitalium. (B) mg218 locus and its flanking regions in the wild-type M. genitalium genome. (C) mg218 locus disrupted by single crossover event X1 in the M. genitalium mutant strain JB1. (D) mg218 locus disrupted by single crossover event X2 in the M. genitalium mutant strain JB2. (E) mg218 locus disrupted by double crossover events X1 and X2 in the M. genitalium mutant strain JB20. Hatched boxes, mg218 DNA region; lightly stippled boxes, flanking regions of mg218 locus; densely stippled box, gentamycin-resistance gene; open box, pCR2 plasmid DNA. The different sizes of SpeI and StyI fragments observed in Southern blotting are represented below the mg218 locus of each strain. EI, EcoRI; EV, EcoRV; H, HindIII; K, KpnI; P, PstI; Sp, SpeI; St, StyI; Su, StuI.
Evaluation of P140 and P110 Synthesis.
Although the absence of MG218 in JB1 (Fig. 2) could be attributable directly to the disruption of the mg218 gene, the reasons for the absence or markedly reduced levels of adhesin P140 and the cytadherence-related P110 protein were less clear. Further, the fact that the expression of a truncated MG218 in JB2 and JB20 correlated with complete retention of P140 and P110 proteins indicated that MG218 plays a critical role in the stability of P140 and P110. The situation was somewhat analogous to that of M. pneumoniae, where insertional inactivation at the crl locus, which includes the gene encoding the cytadherence-related HMW2 protein, a homologue of MG218, was reported to affect the synthesis of other proteins implicated in cytadherence (10). The absence of specific proteins in mutant M. pneumoniae strains was associated with a post-translational defect (21). Therefore, we investigated whether mutant M. genitalium strain JB1 exhibited a similar defect with respect to cytadherence-related proteins P140 and P110. This investigation was accomplished by labeling transformant JB1 and wild-type G37 strains with [35S]methionine/[35S]cysteine for 1 h and analyzing the stability of the translated proteins over time by SDS/PAGE (Fig. 6). Whereas P140 and P110 were stable in wild-type G37 even after 5 h (Fig. 6A), P140 in JB1 was stable for only about 1 h after synthesis (Fig. 6B). Concerning P110, only faint multiple bands were detected just below the P110 region in mutant JB1. The protein profiles of the pulse–chase experiments were also subjected to Western blot analysis (data not shown) to confirm the existence or absence of P140- or P110-related proteins in JB1. The band at 140 kDa that showed diminishing appearance with time exhibited a clear reactivity with anti-P140 antibody. However, the 110-kDa region showed no reactivity with anti-P110 antibodies. These results corroborate earlier data suggesting that the absence of MG218 in JB1 results in the loss of stability and conformational integrity of P140 and P110. Further, the half-life of P110 in JB1 was much shorter than that of P140, since no 110-kDa protein was detected even after 1 h of the pulse–chase experiment. To rule out the possibility that the absence of P110 in JB1 was caused by a transcriptional defect, RNA was isolated from G37 and JB1 and probed for mRNA transcripts against P110, P140, and 16S rRNA. The latter served as a housekeeping gene product control. Although slightly reduced levels of P110 mRNA were detected in JB1, the data clearly showed the presence of mRNA for both P110 and P140 in mutant JB1. In the case of P110, the presence of mRNA transcripts was further established by probing mRNA with sense and antisense RNA probes. Only the antisense probe gave positive signals, indicating that the defect was at the translational or post-translational levels.
Figure 6.
Pulse–chase analysis of M. genitalium proteins. (A) Total protein profiles of wild-type strain G37 at 0 h, 1 h, 2 h, and 5 h. (B) Total protein profiles of mg218 mutant strain JB1 at 0 h, 1 h, 2 h, and 5 h. MG218, P140, and P110 represent the corresponding M. genitalium proteins. The pulse with [35S]methionine/[35S]cysteine was for 3 h, and individual samples were removed at 1-h intervals for up to 5 h. Then, mycoplasma proteins were separated by SDS/5% PAGE, and the gels were dried prior to autoradiography.
DISCUSSION
In the absence of suitable genetic tools to target mutations in mycoplasma genes, the characterization of spontaneously arising mycoplasma mutants has provided initial insights into gene function and virulence. In particular, the analysis of spontaneously arising HA− mutants of M. pneumoniae and M. genitalium, which exhibited marked defects in adherence to various mammalian target cells, helped to partially resolve mechanisms of cytadherence (16, 22, 23). For example, M. genitalium cytadherence is mediated by the P140 adhesin protein, which shares extensive immunological crossreactivity and DNA sequence homologies with the major M. pneumoniae cytadhesin P1 (24, 25). Further, spontaneous cytadherence-negative mutants of M. genitalium either lacked P140 or exhibited defects in its processing, thus reinforcing the importance of P140 as a major adhesin (16). The gene encoding P140 is located in the p140 operon, which contains two other ORFs encoding predicted proteins of 29 kDa and 114 kDa (i.e., P110) situated upstream and downstream of P140 but with unknown functions (26). A similar organization has been described for the p1 operon of M. pneumoniae (27). In addition to the P140 cytadherence-associated operon, other major adhesins or putative cytadherence-related genes in M. genitalium and M. pneumoniae have been identified and assigned to distinct cytadherence-associated operons (28, 29). To gain further insights into the functional role of genes in mycoplasma pathogenesis, we and others used transposon mutagenesis (10, 11). These studies identified additional genes and loci associated with cytadherence, thus revealing an unexpected level of genetic and biochemical complexity. Interestingly, these cytadherence-negative mutants of M. genitalium and M. pneumoniae also lost their ability to attach avidly to glass and plastic surfaces, possibly due to changes in membrane electrostatic potentials (30).
Although the initial use of transposons appeared promising, this approach did not yield the desired results, since the most effective transposon, Tn4001, failed to insert randomly into the genomes of M. pneumoniae and M. genitalium. Thus, alternative and improved methods of gene disruption in mycoplasmas were required. The results presented here clearly show that mg218 was disrupted in M. genitalium through homologous recombination by single- and double-crossover events. This is an unequivocal demonstration of homologous recombination in pathogenic mycoplasmas. In addition to the disruption of mg218, our unpublished results with similar disruption constructs indicate that homologous recombination occurs at several other loci in the M. genitalium genome, including genes encoding excinuclease and P140-related repeat regions. Thus, homologous recombination-mediated gene disruption using related constructs should be useful in analyzing the function of numerous genes in M. genitalium as well as other mycoplasmas infecting humans and animals. In bacteria, the RecA protein plays a major role in mediating homologous recombination (9), and its presence has been demonstrated in a wide variety of Mycoplasma species, including M. genitalium (4) and M. pneumoniae (3).
An interesting finding in this study is the expression of a truncated MG218 (160 kDa) in JB2 and JB20, despite disruption of the mg218 locus. As shown in Fig. 5 B and C, the mg218 locus in these two strains, as opposed to JB1 (Fig. 5A), exhibits similar downstream regions that follow the gentamycin-resistance gene. This part of the mg218 gene (4.4 kb) has the capacity to encode an approximately 160-kDa protein, which includes the peptide region (amino acids 419–429) used for immunization of rabbits. The fact that the truncated proteins in transformants JB2 and JB20 could be detected by these antibodies supports the hypothesis that an in-frame fusion is formed with sequences downstream of the gentamycin-resistance gene, resulting in the expression of the truncated proteins. Sequences following the gentamycin-resistance gene in the 2.5-kb DNA fragment constitute part of IS256, and it is not known whether this region serves as a promoter for the expression of truncated MG218 proteins. Although the mechanisms of expression of truncated MG218 in the strains JB2 and JB20 remain unclear, the fact that P140 and P110 were not rapidly degraded in these strains, in contrast to JB1, reinforces the importance of MG218 in maintaining cytadherence of M. genitalium. The partial HA+ phenotype of JB2 and JB20 may arise from suboptimal conformational integrity and reduced functional binding of the cytadherence-implicated proteins in the presence of truncated MG218. Further, the partial HA+ phenotype may be due to the instability of the integrated plasmid in these strains or to the unstable expression of truncated MG218 within a population of mycoplasma colonies. Instability or reversion should lead to the reappearance of full MG218, and the protein extracts from these strains should contain full and truncated MG218 when analyzed by Western blotting. However, we have never encountered such an occurrence even after multiple passages of these strains.
The observation that both P140 and P110 proteins, which belong to the cytadherence-associated p140 operon, were unstable in transformant JB1, which lacks MG218, was surprising. Although a related phenomenon occurred in M. pneumoniae when hmw2 was disrupted (10, 11), a significant difference exists between M. genitalium and M. pneumoniae in terms of the proteins and operons involved. For example, the expression of ORFs associated with the p1 operon of M. pneumoniae, which is equivalent to the p140 operon of M. genitalium, was unaffected by mutations in the hmw2 or crl locus of mutant M. pneumoniae strains. Instead, the proteins belonging to the hmw gene cluster, HMW1 and HMW3, were reported absent or produced at low levels (10, 11, 20). HMW1 and HMW3 play critical roles in the cytadherence of M. pneumoniae and are localized along the leading filamentous extension of the cell (31) and in the terminal button of the tip structure of M. pneumoniae (32), respectively. The M. genitalium homologues of HMW1 and HMW3 encode 130- and 69-kDa proteins, respectively, based on similarities in DNA sequences. No protein alterations in this region were detected in the SDS/PAGE profiles of JB1. Thus, it appears that distinctions exist in the overall regulation of cytadherence of M. genitalium and M. pneumoniae, despite the remarkable similarities in their adhesins and adherence-related operons.
Does MG218 have a direct regulatory role on the expression of the p140 operon of M. genitalium? Unfortunately, our results do not favor such a hypothesis, since the pulse–chase analysis of proteins in JB1 demonstrated that P140 was indeed synthesized and appeared relatively stable for several hours. At the same time the absence of P110 may be due to a decreased half-life of the synthesized polypeptide because the slot-blot analysis clearly detected mRNA transcripts for both P140 and P110. In addition, because the gene encoding P110 is located just downstream of p140 in the same operon, the fate of P110 might be predicted to be similar to P140. It is logical to assume that these proteins were lost post-translationally, similar to HMW1 and HMW3 in the hmw2 mutant strain of M. pneumoniae (21). Therefore, it appears that MG218 has no direct transcriptional or translational impact on the p140 operon. Nonetheless, how the loss of MG218 affects the proteins of the p140 operon remains intriguing. Computer analysis of MG218 reveals that it is a basic protein (calculated pI 8.6) with several leucine zippers and coiled-coil structures, characteristic of binding proteins. In addition, MG218 exhibits sequence similarities with myosin heavy chain, as reported for other adhesins of M. pneumoniae and M. genitalium, possibly indicating that this protein may be involved in cytoskeleton-related structures and functions (33). Krause et al. (20) have observed similar features for HMW2 of M. pneumoniae and suggested that it might play a chaperone-like or an intermediary role between chaperones and proteins. This hypothesis was supported by the findings that HMW1 and HMW3 have “P-Xaa-L/R-Xaa-SS” motifs for proteolytic cleavage at their C termini (21). In the absence of HMW2, the HMW1 and HMW3 proteins are highly prone to proteolysis, since cleavage at the proteolytic motif of HMW1 and HMW3 appears to be the most important step for their complete lysis. In other words, the presence of HMW2 prevents the degradation of these proteins by interacting with them. Although experimental evidence is still needed to define the proteolytic mechanisms of HMW1 and HMW3 in M. pneumoniae, the identifiable mycoplasma proteolytic motifs resemble sequences in the Caulobacter crescentus McpA chemoreceptor protein, where the motif is important for regulating its degradation during cell division (34). Interestingly, the examination of the C termini of the P140, P110, HMW1-like, and HMW3-like proteins of M. genitalium revealed the presence of amino acid sequences PVLNASS, PVSVGSS, PDRTSNSSLLDEDLFESS, and PTRLSS, respectively. These sequences are considered proteolytic cleavage motifs similar to those described for HMW1 and HMW3 of M. pneumoniae (21) and for McpA of C. crescentus (34).
If the hypothesis of proteolytic degradation holds true for M. genitalium, it is possible that the HMW1-like and HMW3-like proteins of M. genitalium, along with P140 and P110, will exhibit proteolytic degradation in JB1. This possibility may be investigated by generating antibodies against the putative HMW1-like and HMW3-like proteins of M. genitalium and performing comparative immunoblotting between wild-type and JB1 strains. At this juncture studies on the interaction of MG218 with P140 and P110 as well as with the HMW1-like and HMW3-like proteins may clarify the role of MG218 in M. genitalium cytadherence.
In conclusion, this study has provided a technology for gene disruption in M. genitalium. The creation and delivery of the disruption construct are relatively straightforward, and mutants resulting from this method are stable in the presence of antibiotics for many passages. The advantage of this technology over existing mutagenesis methodologies is that it can be targeted to specific genes and operons, permitting the development of research strategies to relate structure–function activities and virulence in M. genitalium.
Acknowledgments
We thank Dr. S. Dallo for her help in producing anti-MG218 antibodies. This work was supported in part by Grant AI 41010 from the National Institute of Allergy and Infectious Diseases.
ABBREVIATIONS
- HA+ and HA−
hemadsorption-positive and -negative
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Baseman J B, Tully J G. Emerg Infect Dis. 1997;3:21–32. doi: 10.3201/eid0301.970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lo S-C. In: Mycoplasma: Molecular Biology and Pathogenesis. Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Washington, DC: Am. Soc. Microbiol.; 1992. pp. 525–545. [Google Scholar]
- 3.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B, Herrmann R. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 5.Tully J, Taylor-Robinson D, Rose D L, Cole R M, Bove J M. Int J Syst Bacteriol. 1983;33:387–396. [Google Scholar]
- 6.Krause D C, Taylor-Robinson D. In: Mycoplasmas: Molecular Biology and Pathogenesis. Maniloff R N, McElhaney R N, Finch L R, Baseman J B, editors. Washington, DC: Am. Soc. Microbiol.; 1992. pp. 417–444. [Google Scholar]
- 7.Lyon B R, May J W, Skurray R A. Mol Gen Genet. 1984;193:554–556. doi: 10.1007/BF00382099. [DOI] [PubMed] [Google Scholar]
- 8.Franke A E, Clewell D B. J Bacteriol. 1981;145:494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dybvig K, Voelker L L. Annu Rev Microbiol. 1996;50:25–57. doi: 10.1146/annurev.micro.50.1.25. [DOI] [PubMed] [Google Scholar]
- 10.Hedreyda C T, Krause D C. Infect Immun. 1995;63:3479–3483. doi: 10.1128/iai.63.9.3479-3483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy S P, Rasmussen W G, Baseman J B. FEMS Immunol Med Microbiol. 1996;15:199–211. doi: 10.1111/j.1574-695X.1996.tb00086.x. [DOI] [PubMed] [Google Scholar]
- 12.Hahn T-W, Krebes K A, Krause D C. Mol Microbiol. 1996;19:1085–1093. doi: 10.1046/j.1365-2958.1996.455985.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Dhandayuthapani S, Deretic V. Proc Natl Acad Sci USA. 1996;93:13212–13216. doi: 10.1073/pnas.93.23.13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Kasper D L, Ausubel F M, Rosner B, Michel J L. Proc Natl Acad Sci USA. 1997;94:13251–13256. doi: 10.1073/pnas.94.24.13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dybvig K, Woodard A. Plasmid. 1992;28:262–266. doi: 10.1016/0147-619x(92)90058-i. [DOI] [PubMed] [Google Scholar]
- 16.Mernaugh G R, Dallo S F, Holt S C, Baseman J B. Clin Infect Dis. 1993;17:S69–S78. doi: 10.1093/clinids/17.supplement_1.s69. [DOI] [PubMed] [Google Scholar]
- 17.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1989. [Google Scholar]
- 18.Dallo S F, Chavoya A, Su C-J, Baseman J B. Infect Immun. 1989;57:1059–1065. doi: 10.1128/iai.57.4.1059-1065.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhandayuthapani S, Rasmussen W G, Baseman J B. Gene. 1998;215:213–222. doi: 10.1016/s0378-1119(98)00260-1. [DOI] [PubMed] [Google Scholar]
- 20.Krause D C, Proft T, Hedreyda C T, Hilbert H, Plagens H, Herrmann R. J Bacteriol. 1997;179:2668–2677. doi: 10.1128/jb.179.8.2668-2677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popham P L, Hahn T-W, Krebes K A, Krause D C. Proc Natl Acad Sci USA. 1997;94:13979–13984. doi: 10.1073/pnas.94.25.13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krause D C, Leith D K, Wilson R M, Baseman J B. Infect Immun. 1982;35:809–817. doi: 10.1128/iai.35.3.809-817.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baseman J B, Morrison-Plummer J, Drouillard D, Puelo-Scheppke B, Tryon V V, Holt S C. Isr J Med Sci. 1987;23:474–479. [PubMed] [Google Scholar]
- 24.Morrison-Plummer J, Lazzell A, Baseman J B. Infect Immun. 1987;55:49–56. doi: 10.1128/iai.55.1.49-56.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu P-C, Schaper U, Collier A M, Clyde W A, Jr, Horikawa M, Huang Y-S, Barile M F. Infect Immun. 1987;55:1126–1131. doi: 10.1128/iai.55.5.1126-1131.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inamine J M, Loechel S, Collier A M, Braile M F, Hu P-C. Gene. 1989;82:259–267. doi: 10.1016/0378-1119(89)90051-6. [DOI] [PubMed] [Google Scholar]
- 27.Inamine J M, Loechel S, Hu P-C. Gene. 1988;73:175–183. doi: 10.1016/0378-1119(88)90323-x. [DOI] [PubMed] [Google Scholar]
- 28.Reddy S P, Rasmussen W G, Baseman J B. J Bacteriol. 1995;177:5943–5951. doi: 10.1128/jb.177.20.5943-5951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Layh-Schmitt G, Himmelreich R, Leibfried U. FEMS Microbiol Lett. 1997;152:101–108. doi: 10.1111/j.1574-6968.1997.tb10415.x. [DOI] [PubMed] [Google Scholar]
- 30.Razin S. In: The Mycoplasmas. Razin S, Barile M F, editors. Vol. 4. New York: Academic; 1985. pp. 161–201. [Google Scholar]
- 31.Stevens M K, Krause D C. J Bacteriol. 1991;173:1041–1050. doi: 10.1128/jb.173.3.1041-1050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens M K, Krause D C. J Bacteriol. 1992;174:4265–4274. doi: 10.1128/jb.174.13.4265-4274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baseman J B, Reddy S P, Dallo S F. Am J Respir Crit Care Med. 1996;154:S137–S144. doi: 10.1164/ajrccm/154.4_Pt_2.S137. [DOI] [PubMed] [Google Scholar]
- 34.Alley M R K, Maddock J R, Shapiro L. Science. 1993;259:1754–1757. doi: 10.1126/science.8456303. [DOI] [PubMed] [Google Scholar]