Abstract
Phytochrome is a ubiquitous photoreceptor of plants and is encoded by a small multigene family. We have shown recently that a functional nuclear localization signal may reside within the COOH-terminal region of a major member of the family, phytochrome B (phyB) (Sakamoto, K., and A. Nagatani. 1996. Plant J. 10:859–868). In the present study, a fusion protein consisting of full-length phyB and the green fluorescent protein (GFP) was overexpressed in the phyB mutant of Arabidopsis to examine subcellular localization of phyB in intact tissues. The resulting transgenic lines exhibited pleiotropic phenotypes reported previously for phyB overexpressing plants, suggesting that the fusion protein is biologically active. Immunoblot analysis with anti-phyB and anti-GFP monoclonal antibodies confirmed that the fusion protein accumulated to high levels in these lines. Fluorescence microscopy of the seedlings revealed that the phyB-GFP fusion protein was localized to the nucleus in light grown tissues. Interestingly, the fusion protein formed speckles in the nucleus. Analysis of confocal optical sections confirmed that the speckles were distributed within the nucleus. In contrast, phyB-GFP fluorescence was observed throughout the cell in dark-grown seedlings. Therefore, phyB translocates to specific sites within the nucleus upon photoreceptor activation.
Keywords: green fluorescent protein, nuclear targeting, photomorphogenesis, phytochrome, signal transduction
Light is an important environmental stimulus which plants must perceive and to which they must respond. Plants use light signals to regulate various developmental processes such as seed germination, de-etiolation, and floral induction (Kendrick and Kronenberg, 1994). For this purpose, plants have evolved several different photoreceptors. Among them, phytochrome is the best characterized. Phytochrome is a soluble chromoprotein consisting of an apoprotein of 120 kD and covalently attached linear tetrapyrrole chromophore (Furuya, 1993; Quail et al., 1995). Phytochrome is a ubiquitous photoreceptor in the plant kingdom, the origin of which can be traced back to cyanobacteria (Kehoe and Grossman, 1996; Hughes et al., 1997; Yeh et al., 1997).
Phytochrome undergoes photoreversible conversion between two spectrally distinct forms, a red light absorbing form (Pr) and a far-red light absorbing form (Pfr).1 Only the Pfr form is believed to be biologically active. Red light activates phytochrome by converting it from the Pr to Pfr form. Conversely, far-red light cancels the effects of red light. In this way, phytochrome acts as a molecular switch. The phytochrome protein is comprised of two domains. The NH2-terminal portion, to which the chromophore is attached, confers the spectral properties characteristic of phytochrome. The COOH-terminal portion is involved in dimerization of the molecule and transfer of the signal to downstream components (Quail, 1997).
Phytochrome has been studied intensively since its discovery in 1959 (Sage, 1992). Nevertheless, little was known about the initial step of the phytochrome signal transduction until recently. However, two recent studies hint at how phytochrome transduces the light signal to downstream components. Firstly, a phytochrome-interacting factor, PIF3, has been identified through a yeast two-hybrid screen (Ni et al., 1998). Interestingly, PIF3 is a nuclear-localized basic helix-loop-helix protein. Hence, a direct interaction between phytochrome and a transcriptional regulator might be involved in the signaling pathway within the nucleus. Secondly, phytochrome appears to be a light-regulated serine/threonine kinase (Yeh and Lagarias, 1998), and therefore may transmit light perception via protein phosphorylation.
A wide range of physiological and developmental processes is under the control of phytochrome. Accordingly, phytochromes are expressed in various tissues throughout the life cycle of plants (Nagatani, 1997). The mode of phytochrome action varies substantially (Mancinelli, 1994). For example, some responses are induced by a relatively low fluence of red light, whereas prolonged irradiation with far-red light is required for some responses. The rate of escape from the red/far-red reversibility varies substantially depending on responses. The diversity of phytochrome action can be explained in part by the multiple molecular species. Phytochrome is known to be encoded by a small multigene family (Mathews and Sharrock, 1997). In Arabidopsis, the complete family consists of five members, phytochromes (phy) A–E, that are encoded by respective genes (PHYA-E). Analysis of mutants deficient in phyA and phyB suggests that the modes of their action are different (Furuya and Schaefer, 1996; Shinomura et al., 1996). It has also been shown that phyC (Halliday et al., 1997; Qin et al., 1997), phyD (Aukerman et al., 1997), and phyE (Devlin et al., 1998) are functionally different from phyA and phyB. These findings imply that different molecular species of phytochrome may transduce light signals via distinct mechanisms.
To elucidate the signal transduction mechanisms of different phytochromes, it is essential to know the sites of their action within the cell. Since immunochemical analysis has indicated that phyA resides in the cytoplasm in darkness, it has been assumed that phytochrome action takes place in the cytoplasm (Nagatani, 1997). In accordance with this notion, phyA and phyB, and probably other phytochromes, are soluble proteins. In addition, microbeam irradiation experiments in green algae and fern gametophytes have indicated that phytochrome, which mediates various cellular responses in these systems, resides in the cytoplasm (Wada et al., 1993). Although associations of phyA with isolated organelles have been reported repeatedly, the biological relevance of these observations remains obscure (Pratt, 1994).
More recently, we have produced transgenic Arabidopsis expressing fusion proteins consisting of GUS and COOH-terminal fragments of phyB (Sakamoto and Nagatani, 1996). The GUS staining from the fusion proteins is observed in the nucleus, suggesting that a functional nuclear localization signal may reside in the phyB sequence. Furthermore, we have confirmed that a substantial fraction of total cellular phyB is recovered in the isolated nuclei. Interestingly, the level of nuclear phyB is substantially reduced by the dark adaptation of plants. On the basis of these findings, we have proposed that phyB translocates to the nucleus upon photoactivation (Sakamoto and Nagatani, 1996; Nagatani, 1997). However, we could not exclude the possibility that those observations were due to technical artifacts.
In this work, the green fluorescent protein (GFP) of the jelly fish was fused to phyB and expressed in the phyB mutant of Arabidopsis to determine its intracellular localization in vivo. Since GFP is relatively small and tolerates protein fusion, it has been shown to be potentially useful as a fluorescent tag (Chiu et al., 1996). The fluorescence emission of GFP does not require any cofactor or substrate, which enables us to observe its fluorescence without making any pretreatment of the tissue. The resulting transgenic lines exhibited pleiotropic phenotypes reported previously for the phyB overexpressing plants, indicating that the phyB-GFP fusion protein is biologically active. Fluorescent microscopic observation revealed that the fusion protein was localized to the nuclear region in the light. Confocal microscopic analysis confirmed that the fusion protein was indeed inside the nucleus. The effects of light on the nucleocytoplasmic partitioning of phyB were then examined. In dark-grown seedlings, fluorescence was observed throughout the cell. Treatment of the seedlings with continuous red light induced accumulation of phyB-GFP fusion protein in the nucleus. Hence, we suggest that phyB translocates to the nucleus upon light stimulation.
Materials and Methods
Plant Materials
The phyB-5 mutant (Reed et al., 1993) of Arabidopsis thaliana (ecotype, Landsberg er) was used as the host for transformation. Arabidopsis thaliana (ecotype Landsberg er) and the phyB-5 mutant were used as controls for physiological, immunochemical, and microscopic experiments.
Plasmid Construction and Transformation
A full-length PHYB cDNA clone was isolated from an Arabidopsis (ecotype Columbia) cDNA library. Cloned PHYB cDNA was almost identical to a previously reported sequence (accession number X17342, submitted by Dr. R. Sharrock, Montana State University, Bozeman, MT) except that a C to T substitution at the base position 971, which does not cause amino acid difference, was detected. To construct the PHYB-GFP fusion sequence, PHYB translational termination codon (TAG) was replaced with an oligonucleotide sequence (GGAGGTGGAGGTATCGAT) by PCR. This oligonucleotide introduces a unique ClaI restriction site at its 3′ terminus.
The GFP clone (blue-sGFP-TYG-nos KS) (Chiu et al., 1996) was a kind gift from Dr. J. Sheen (Massachusetts General Hospital, Boston, MA). This clone contains a unique ClaI restriction site that shortly precedes the ATG start codon of the GFP gene. The PHYB and GFP clones were ligated at the ClaI restriction site to generate PHYB-GFP translational fusion. As the result, an oligoamino acid sequence (GGGGIDKLDP) was inserted between the phyB and GFP amino acid sequences (Fig. 1 a). This PHYB-GFP chimeric cassette was inserted between the constitutive cauliflower mosaic virus 35S promoter and the Nos terminator of an Agrobacterium transformation vector pBI-Hyg/35S-NosT, which is derived from another transformation vector pBI101-Hm (a gift from Dr. Kenzo Nakamura, Nagoya University, Japan) by removing its uidA gene (Nakamura, M., unpublished observation). The resulting vector was designated pBI-Hyg/35S-PHYB-sGFP-NosT (Fig. 1 a).
Figure 1.
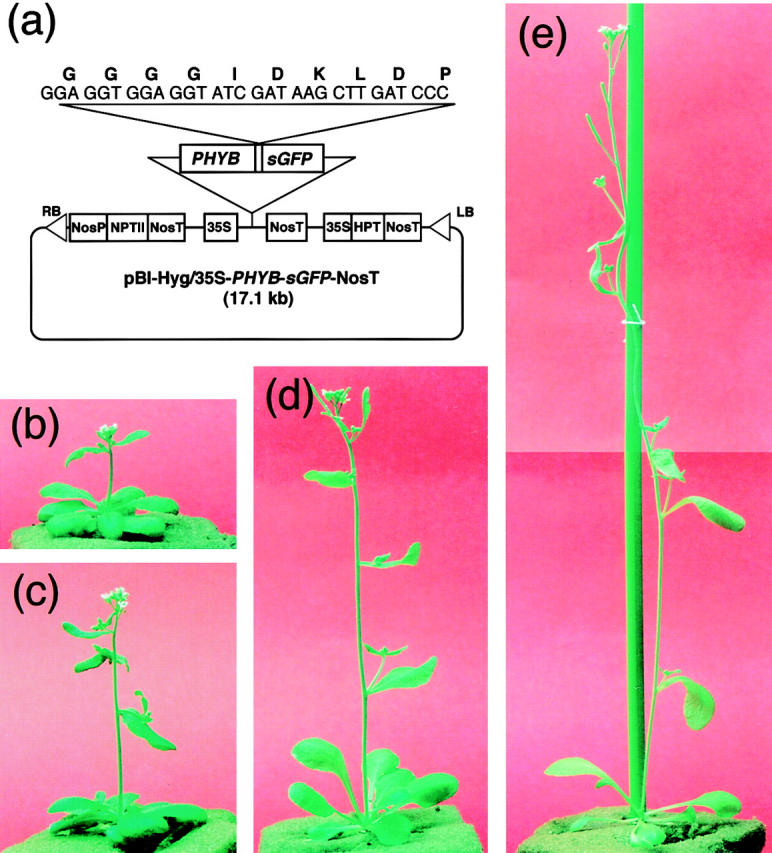
Two independent lines of transgenic Arabidopsis, PBG-5 and PBG-7, which overexpress the phyB-GFP fusion protein. Plants were grown for 4 wk under continuous white light. (a) pBI-Hyg/35S-PHYB-sGFP-NosT used for transformation of Arabidopsis plants. sGFP, synthetic GFP; RB, right border of T-DNA; LB, left border of T-DNA; NosP, nopaline synthase promoter; NosT, nopaline synthase terminator; NPTII, neomycin phosphotransferase II; 35S, cauliflower mosaic virus 35S promoter; HPT, hygromycin phosphotransferase. (b) Picture of the PBG-7 plant. (c) Picture of the PBG-5 plant. (d) Picture of the wild-type plant. (e) Picture of the phyB-5 mutant plant.
Arabidopsis phyB mutant was transformed using Agrobacterium-mediated in planta transformation (Bechtold et al., 1993). Transformed plants were selected on the medium containing 25 mg ml−1 hygromycin B (Boehringer Mannheim) and 166 mg ml−1 claforan (Hoechst). The transgenic lines PBG-5 and PBG-7 were selected from the drug-resistant lines by phyB immunoblotting and GFP epifluorescence microscopy.
Growth Conditions and Light Treatments
For growth of plants, seeds were sown on 0.6% agar plates containing the Murashige-Skoog medium with 2% (wt/vol) sucrose and grown under continuous white light from fluorescent tubes (FLR40SW/M-B; Hitachi). The plants were then transplanted to pots containing vermiculite and grown to maturity under continuous white light from fluorescent tubes. For the immunochemical detection of the fusion protein, rosette leaves were harvested from 3-wk-old plants. For the hypocotyl assay and microscopic observation, seeds were sown on agar plates containing Murashige-Skoog salt mixture without sucrose. The plates were placed at 4°C for 12 h and then irradiated with continuous white light for 12 h at 23°C to induce germination. For the hypocotyl assay, seedlings were grown for 5 d under continuous red light (6.0 W m−2) from red fluorescent tubes (FL20S/R-F; National) or in darkness. For microscopic observation, seedlings were grown for 5 d under continuous white light (15 W m−2) from fluorescent tubes (FLR40SW/M-B; Hitachi) or in darkness.
Immunochemical Experiments
To detect the phyB-GFP fusion protein and the authentic phyB, ∼0.1 g of rosette leaves was glass homogenized in the presence of 0.1 ml of the phytochrome extraction buffer (100 mM Tris-HCl, 2 mM DTT, 5 mM EDTA, pH 8.3) containing proteinase inhibitor cocktails for general use (P2714; Sigma Chemical Co.) and for fungal and yeast extracts (P8215; Sigma Chemical Co.) at the concentrations recommended by the manufacturer. Debris was removed by centrifugation. Proteins were concentrated from the crude homogenate by ammonium sulfate precipitation. The precipitated protein was dissolved in the SDS-PAGE sample buffer and subjected to immunoblot analysis (Sakamoto and Nagatani, 1996). Antibodies used were an anti-phyB mAb, mBA2 (Shinomura et al., 1996), and an anti-GFP mAb (Clontech). Molecular weight markers (prestained SDS molecular weight standard mixture) were from Sigma Chemical Co.
Microscopic Observation
Arabidopsis seedlings were soaked in 2 μg ml−1 Hoechst No. 33342 (Sigma Chemical Co.) solution made in H2O for visualization of the nucleus in some experiments. Epidermal layers including cortex were peeled from the hypocotyls and placed on glass slides. For the other parts of seedlings, whole organs were placed on glass slides and pressed gently. The specimens were observed using an Olympus BX60 microscope equipped with ×20, ×40, and ×100 objectives, differential interference contrast (DIC) optics, and a 100-W mercury arc light source. Fluorescence was filtered using UV (U-MWU) or FITC (U-MNIBA) filter sets (Olympus).
For confocal microscopy, trichomes were removed from the surface of cotyledons with a razor blade and placed on glass slides. Root tips were placed on glass slides without any pretreatment. The specimens were observed using an inverted laser scan microscope (LSM410 invert; Carl Zeiss Jena) equipped with ×40 and ×63 objectives. The laser scan images were obtained with a combination of 488 nm laser excitation and 515 nm longpass emission filter (LP515; Carl Zeiss Jena). Sequential images from different focus planes were recorded automatically.
Results
phyB-GFP Is Biologically Active in Transgenic Plants
To examine biological activity and intracellular localization of the phyB-GFP fusion protein, the phyB-5 mutant of Arabidopsis was transformed with a vector harboring the 35S::PHYB-GFP construct. The resulting transgenic lines, PBG-5 and PBG-7, exhibited an overall dwarfing of mature plants under continuous white light (Fig. 1, b–e). They flowered a few days later than the wild-type under the conditions tested. Similar phenotypes, which are opposite to those of the phyB-deficient mutants (Reed et al., 1993), have been reported in phyB overaccumulating plants (Wester et al., 1994). Hence, the phyB-GFP fusion protein is likely to be fully functional.
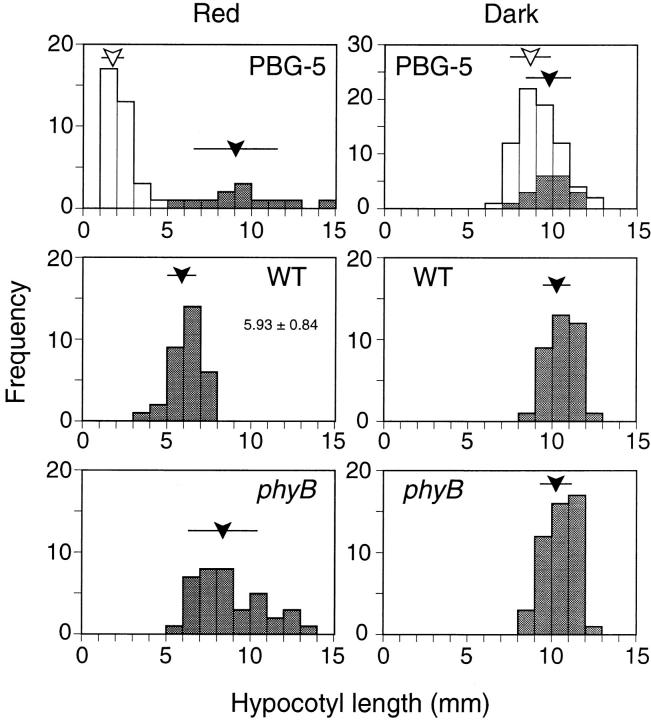
It is known that inhibition of hypocotyl elongation by continuous red light is mediated primarily by phyB (Quail et al., 1995). To confirm the biological activity of phyB-GFP further, heterozygous progeny of the PBG-5 plant was examined for this response. The seedlings were grown under continuous red light for 5 d and hypocotyl lengths were determined. As shown in Fig. 2, a short population segregated from a longer one at about a 3:1 ratio. Hypocotyl lengths in the longer population matched well with those in the parental phyB mutant. In contrast, the shorter seedlings were significantly shorter than the wild-type seedlings, which is consistent with the phyB overexpression phenotypes reported by other groups (Wagner et al., 1991; McCormac et al., 1993). Cosegregation of the short phenotype with the expression of phyB-GFP was then examined. As expected, all the short seedlings exhibited GFP fluorescence whereas no fluorescence was observed in the longer seedlings (Fig. 2).
Figure 2.
Frequency distribution of hypocotyl lengths in PBG-5, phyB mutant, and the wild-type seedlings under continuous red light (left) or in darkness (right). Heterozygous progeny of PBG-5 plant was examined. Individuals that exhibited GFP fluorescence are unshaded. Open and closed arrowheads indicate average hypocotyl lengths of fluorescent and nonfluorescent populations, respectively. Bars indicate the standard deviation.
The seedling phenotype was examined in darkness as well. As is the case with the phyB overexpressing plants (Wagner et al., 1991), no clear segregation of shorter seedlings was observed in the PBG-5 heterozygous progeny (Fig. 2). The average hypocotyl lengths in the fluorescent and nonfluorescent populations were indistinguishable. Hence, phyB-GFP was suggested to be not only biologically but also photochemically active.
Immunoblot Analysis Confirms Accumulation of phyB-GFP
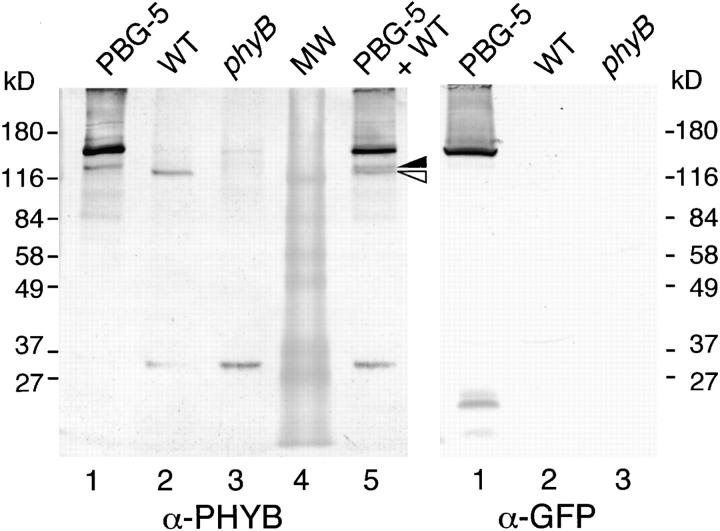
To examine the accumulation of phyB-GFP fusion protein in the transgenic plants, immunoblot analysis was performed. Proteins were extracted from rosette leaves of the PBG-5 plants and probed with anti-phyB and anti-GFP antibodies (Fig. 3). The anti-phyB mAb detected a major band of ∼143 kD in the PBG-5 extracts (Fig. 3, left). The size was consistent with the expected mass of the phyB-GFP fusion protein. A band at the same size was detected with the anti-GFP antibody (Fig. 3, right), confirming that the band represented the phyB-GFP fusion protein. The higher intensity of the phyB-GFP band compared with that of the authentic phyB indicated that the phyB-GFP was overaccumulated in the transgenic plants. A similar result was obtained for the other transgenic line, PBG-7 (data not shown).
Figure 3.
Immunoblot analysis of phyB-GFP fusion protein in the PBG-5 rosette leaves. Extracts from rosette leaves were probed with anti-phyB (left) or anti-GFP mAb (right). Lane 1, PBG-5; lane 2, the wild-type; lane 3, the phyB mutant; lane 4, molecular weight markers; lane 5, 1:1 mixture of the extracts from PBG-5 and the wild-type plants. Closed triangle, a proteolytic phyB-GFP fragment; open triangle, authentic phyB. Each lane contains either 25 (left) or 112 (right) μg total protein.
In addition to the major 143-kD band, a weak band of ∼123 kD was detected in the PBG-5 plants (Fig. 3). The intensity of the band was comparable to that of authentic phyB. To confirm that the fragment is larger than the authentic phyB (117 kD on the blot), extracts from the PBG-5 and the wild-type plants were mixed and probed with the anti-phyB antibody. As expected, the two bands were separated on the blot. Since the fragment was not detected with anti-GFP antibody, it is speculated that proteolysis of phyB-GFP within the GFP portion yielded this fragment. In accordance with this, minor bands around 20 kD were detected on the anti-GFP blot. In the absence of the protease inhibitor cocktails, fragmentation was much more severe (data not shown). Hence, the 123-kD fragment is likely to be produced by the residual proteolytic activity in the extract during the extraction procedure.
phyB-GFP Localizes to the Nucleus in the Light
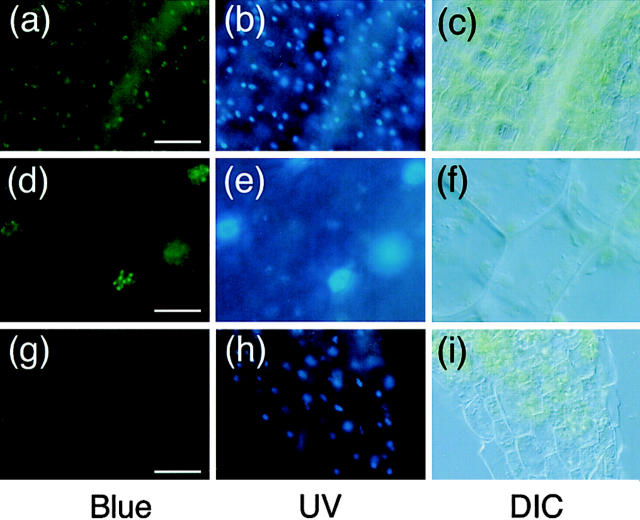
Intracellular localization of the phyB-GFP fusion protein in the PBG-5 seedlings was examined. Epidermal layers including cortex were peeled from the light-grown seedlings and observed under a fluorescence microscope. At lower magnification, bright green spots of GFP fluorescence were observed (Fig. 4, a–c). Positions of the spots matched well with those of the nuclei revealed by the Hoechst staining. Similar fluorescence images were obtained for another transgenic line, PBG-7 (data not shown). Interestingly, observation at higher magnification revealed that the phyB-GFP fluorescence was speckled within the nuclear region (Fig. 4, d–f). The apparent size of each speckle appeared to be <1 μm. Although speckles were observed in all of the nuclei, the number per nucleus varied. In most cases, one nucleus contained 5–10 speckles. The intracellular localization of phyB-GFP was then examined in other parts of the seedling. As shown in Fig. 5, nuclear fluorescence was confirmed in leaf (Fig. 5, a–c), root (Fig. 5, d–f), and root hair cells (Fig. 5, g–i). Furthermore, the speckles were observed in all of the cell types examined.
Figure 4.
Fluorescence microscopic observation of hypocotyl peel from light-grown PBG-5 and the wild-type seedlings. Samples were stained with Hoechst No. 33342 and viewed under epifluorescence optics with blue (left) or UV (middle) excitation. DIC images in the same view are shown (right). (a–c) PBG-5 hypocotyl cells, ×20 objective. Bar, 50 μm. (d–f) PBG-5 hypocotyl cells, ×100 objective. Bar, 10 μm. (g–i) Wild-type hypocotyl cells, ×20 objective. Bar, 50 μm.
Figure 5.
Fluorescence microscopic images of different parts of light-grown PBG-5 seedlings. Samples were stained with Hoechst No. 33342 and viewed under epifluorescence optics with blue (left) and UV (middle) excitation. DIC images of the same sample are shown (right). (a–c) PBG-5 leaf epidermis, ×100 objective. Bar, 10 μm. (d–f) PBG-5 root cells, ×40 objective. Bar, 25 μm. (g–i) PBG-root hair, ×100 objective. Bar, 10 μm.
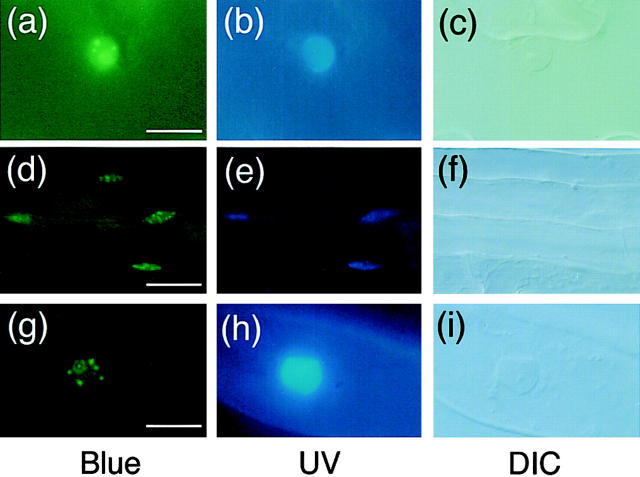
To determine the spatial distribution pattern of the speckles within the nucleus, optical sectioning of the cell with a confocal microscope was performed (Fig. 6). For this purpose, trichomes were chosen for observation because of the large size of their nuclei. As shown in Fig. 6, the speckles appeared to be distributed more or less evenly in the nucleus. In this particular case, at least 24 spots were recognized. This is probably due to the large size of the trichome nucleus. The images clearly demonstrated that the size of each speckle varied substantially even within one nucleus.
Figure 6.
Confocal optical sectioning of the trichome nucleus in PBG-5. Trichomes were removed from light-grown PBG-5 cotyledons and observed on an inverted laser scan microscope (LSM410 invert; Carl Zeiss Jena) with a combination of 488 nm laser excitation and 515 nm longpass emission filter. (a) DIC image, ×63 objective. Bar, 5 μm. (b–i) Serial sections at 2-μm intervals, ×63 objective.
phyB-GFP Is Distributed throughout the Cell in Darkness
A previous study suggested that the nuclear localization of phyB is light dependent (Sakamoto and Nagatani, 1996). In accordance with this, weak fluorescence was observed throughout the cell in PBG-5 dark-grown seedlings (Fig. 7, a–e). Since the intensity of fluorescence was low, it was difficult to determine the intracellular localization in detail. However, higher intensity in the peripheries of the cells indicated that phyB-GFP was distributed in the cytoplasm. Those cells were highly vacuolated and the cytoplasm was observed mostly in the peripheral region, as observed by DIC microscopy (Fig. 7 b). In some cells, fluorescence was observed not only in the peripheries but also in the nuclear region (Fig. 7, a–e). However, it was difficult to conclude that the phyB-GFP exists inside the nucleus even by confocal observation (data not shown).
Figure 7.
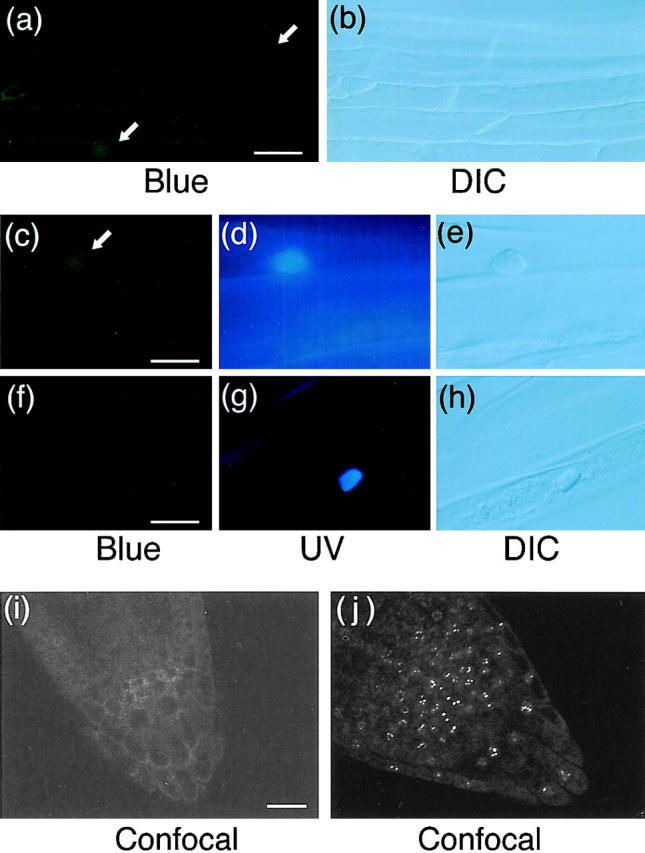
Fluorescence microscopic images of hypocotyl and root tip cells in dark-grown PBG-5 and wild-type seedlings. Hypocotyl specimens were stained with Hoechst No. 33342 and viewed under epifluorescence optics with blue (a, c, and f) and UV (d and g) excitation or under DIC optics (b, e, and h). Root tip specimens were observed on an inverted laser scan microscope (LSM410 invert; Carl Zeiss Jena) with a combination of 488 nm laser excitation and 515 nm longpass emission filter (i and j). Arrows indicate fluorescence detected in the nuclear regions. (a and b) Dark-grown PBG-5 hypocotyl cells, ×40 objective. Bar, 25 μm. (c–e) Dark-grown PBG-5 hypocotyl cells, ×100 objective. Bar, 10 μm. (f–h) Dark-grown wild-type hypocotyl cells, ×100 objective. Bar, 10 μm. (i) Dark-grown PBG-5 root tip cells, ×40 objective. Bar, 20 μm. (j) Light-grown PBG-5 root tip cells, ×40 objective.
Intracellular distribution of phyB-GFP in the light and darkness was compared in root tip cells with a confocal microscope. As shown in Fig. 7 j, the speckles of fluorescence were observed in light-grown seedlings. In contrast, relatively uniform fluorescence was observed in the peripheries of the cells in dark-grown seedlings (Fig. 7 i), which provided further evidence that phyB-GFP was distributed outside the nucleus and throughout the cell in darkness.
Red Light Induces Nuclear Accumulation of phyB-GFP
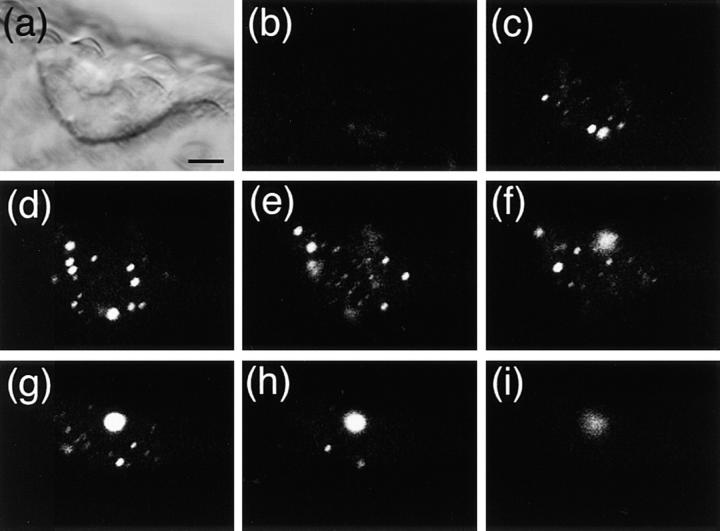
The time course of nuclear accumulation of phyB-GFP during the dark to light transition was followed. The PBG-5 dark-grown seedlings were transferred under continuous red light. As shown in Fig. 8, nuclear fluorescence was not clear at time 0 (Fig. 8, a and b). After 2 h in red light, the intensity of the nuclear GFP signal was increased (Fig. 8, c and d). However, fluorescence remained detectable in the periphery of the cells. Speckles in the nucleus were rarely observed at this time point, although a few tiny spots were detected in some cases. After 4 h in red light, many small speckles were observed (Fig. 8, e and f). Fluorescence in the cell periphery was greatly reduced. After 6 h in red light, the speckles became larger but the number per nucleus was reduced (Fig. 8, g and h). Hence, translocation of phyB-GFP to the nucleus appeared to be completed within 4–6 h in hypocotyl cells under continuous red light. In the course of these experiments, we noticed that the translocation took longer in root cells, although the reason for this was not clear (data not shown).
Figure 8.
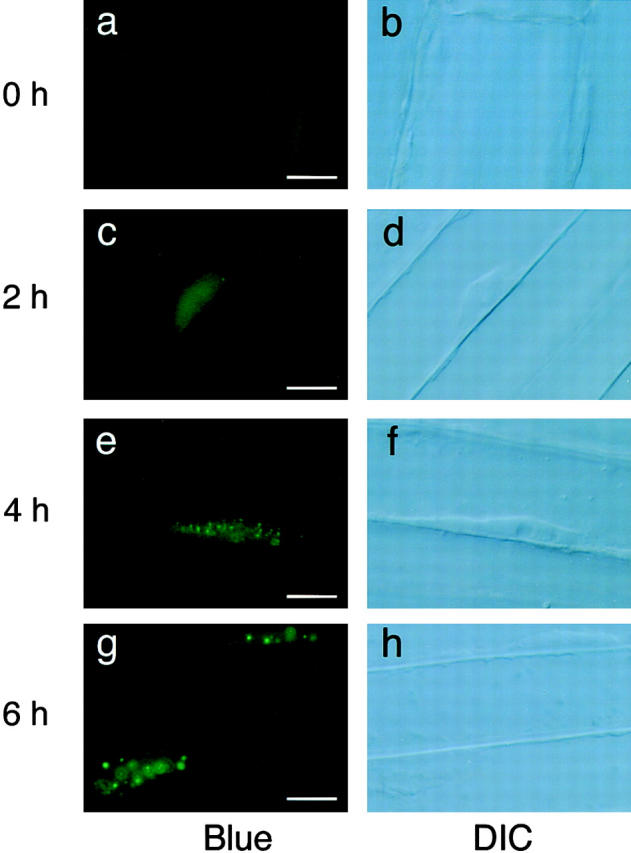
Fluorescence microscopic images of hypocotyl cells in PBG-5 dark-grown seedlings placed under continuous red light for different duration. Hypocotyl cells were viewed under epifluorescence optics with blue excitation (left) and DIC optics (right) with a ×100 objective. Bar, 10 μm. (a and b) Dark-grown seedling. (c and d) 2 h in red light. (e and f) 4 h in red light. (g and h) 6 h in red light.
Discussion
phyB-GFP Is Biologically Active
It is known that phytochromes overexpressed in transgenic plants are biologically active. Transgenic Arabidopsis expressing exogenous phyB exhibits increased sensitivity to red light (Wagner et al., 1991; McCormac et al., 1993). In this study, we have demonstrated that the plants expressing phyB-GFP show similar light-dependent phenotypes (Figs. 1 and 2). Since the expression level of phyB-GFP was comparable to those reported for the phyB overexpressing plants (Wagner et al., 1991), it is concluded that phyB-GFP is as active as authentic phyB. Furthermore, the fusion protein was expressed in the phyB-deficient background in the present study, confirming that the presence of endogenous phyB is not required for correct functioning of the phyB-GFP fusion protein.
In the PBG-5 seedlings, a proteolytic fragment of the fusion protein was detected (Fig. 3). However, its level was as low as the authentic phyB. Furthermore, the fragmentation might have occurred during the extraction. Even if the fragment existed in vivo, it would not contribute to the fluorescence. The immunoblot analysis suggests that the fragment resulted from proteolysis within the GFP portion, which would cause the loss of fluorescence. Likewise, it is unlikely that the fragment alone caused the phyB overexpression phenotypes, although it might have contributed to the phenotype to some extent.
phyB-GFP Is Localized to the Nucleus
Although associations of phyA with various organelles have been reported, only a small portion of the total cellular phyA was recovered in those cases (Pratt, 1994). It is also known that the Pfr form of phyA tends to associate with particulate material under certain cell extraction conditions (Quail, 1983). Thus, it had remained obscure whether phytochrome indeed resides within organelles in vivo. More recently, we have shown that COOH-terminal fragments of phyB fused to GUS are localized to the nucleus (Sakamoto and Nagatani, 1996). Furthermore, a substantial amount of endogenous phyB has been detected in isolated nuclei. On the basis of these findings, we had tentatively proposed that phyB translocates to the nucleus to mediate the light responses (Sakamoto and Nagatani, 1996; Nagatani, 1997). However, this observation could have been due to a cryptic nuclear localization signal that is exposed only in the context of the fusion protein. It is also difficult to exclude the possibility that phyB detected in the isolated nuclei might be due to contamination during specimen preparation and staining.
This study provides more dramatic evidence for phyB nuclear localization. Localization of the phyB-GFP fusion protein in the nucleus has been observed in intact live cells without any pretreatment (Figs. 4 and 5). The optical sectioning by a confocal microscope clearly indicates that the fluorescence is distributed inside the nucleus (Fig. 6). Furthermore, the fusion protein appears to be fully functional as a photoreceptor (see above), suggesting that the structure of phyB is preserved in the phyB-GFP fusion context. However, it should be noted here that phyB-GFP is overexpressed under control of the 35S promoter. Hence, there remains the possibility that ectopic expression contributes somewhat to the observed intracellular distribution. To address this question, we are now screening for transgenic lines with lower accumulation levels.
In the light, fluorescence of the phyB-GFP protein was observed mainly in the nucleus in all the cell types examined. Hence, the nucleus is likely to be the major site of the phyB action, although it remains possible that a minor fraction of phyB-GFP is present in other compartments of the cell and might contribute to the overall response. All the phenotypes observed in this study, such as the shorter hypocotyls, overall dwarfing in mature plants, and late flowering, can ultimately be explained by alteration in gene expression patterns. Hence, it is an intriguing possibility that phyB-GFP translocates to the nucleus to affect the transcription of target genes. Although phytochrome appears to have neither DNA-binding nor transactivation domains, it could interact with other factors that directly regulate transcription. PIF3, a nuclear-localized basic helix-loop-helix protein that binds to the COOH-terminal domain of phytochrome (Ni et al., 1998), is a potential candidate for such a factor.
phyB-GFP Speckles
Interestingly, the phyB-GFP fusion protein forms speckles in the nucleus. The sizes of the speckles are mostly <1 μm. However, the size varies even within one nucleus under continuous light (Fig. 6). The number of speckles per nucleus also varies. Interestingly, the size of the speckle gradually increases during the dark to light transition (Fig. 8). Conversely, the number of speckles decreases.
Speckled structures similar to the one observed in this study have been reported in animal cells (Lamond and Earnshaw, 1998). Factors involved in the processing and transcription of RNA are found in those speckled structures in the nucleus. The promyelocytic leukemia (PML) nuclear body is another example of such a structure. However, the biological relevance of those structures remains unclear, although they may function as a repressor of transcription (Singer and Green, 1997; Lamond and Earnshaw, 1998). In plant cells, COP1, which is a negative regulator of plant photomorphogenesis or light responses (von Arnim and Deng, 1996), has been shown recently to form speckles in the nucleus (Ang et al., 1998). Hence, those speckles observed in plant cells might represent the site where the photoreceptor and other nuclear factors such as COP1 and PIF3 interact with each other to mediate light signals. Identification of proteins present in the phyB speckles is awaited.
It should be noted here that the phyB-GFP speckles could be due to an artifact caused by overaccumulation of the fusion protein at nonphysiological concentration. It is known that phytochrome in general tends to form aggregates in vitro (Quail, 1983). Hence, as with all studies using GFP fusion proteins, it is difficult at present to exclude this possibility. However, the light-dependent nature of the nuclear translocation supports the view that this distribution is indeed relevant to phytochrome function. Detailed analysis of transgenic lines that accumulate the fusion protein at lower levels is awaited. A search for mutations that abolish the speckles would greatly help to answer the question. For example, it has been shown in onion cells that GFP-HY5 fails to form speckles in the absence of COP1 expression (Ang et al., 1998). It will be of interest to investigate the effect of removing known light transduction components upon the subnuclear distribution of phyB.
phyB-GFP Is Distributed throughout the Cell in Darkness
In dark-grown seedlings, the phyB-GFP fusion protein appeared to be distributed evenly throughout the cytoplasm (Fig. 7). This is consistent with the previous observation that phyB is not detected in the nuclei isolated from the dark-adapted rosette leaves (Sakamoto and Nagatani, 1996). In addition, a similar result has been obtained in dark-grown pea seedlings (Nagatani, A., unpublished observations). However, the detailed distribution of phyB-GFP could not be determined because of the resolution limit within the small Arabidopsis cells. At present, it is not clear whether the fusion protein is excluded completely from the nucleus in darkness. Although fluorescence was observed in the nuclear region, this could be due to fluorescence from the cytoplasm surrounding the nucleus. It would be helpful to isolate intact cells or protoplasts to determine the localization pattern in greater resolution.
To observe GFP fluorescence, the seedlings received relatively intense actinic blue light. In theory, this could alter the localization pattern of phyB-GFP. The apparent nuclear fluorescence observed in the dark-grown seedlings might be due to this effect. However, the distribution of phyB-GFP did not change significantly during the observation period (10–20 min). This is consistent with the relatively slow kinetics of phyB-GFP accumulation in the nucleus induced by continuous red light (Fig. 8). In addition, no change was observed in light-grown seedlings during the observation. Hence, the actinic blue light does not appear to disturb the distribution pattern at least during the observation period of 10–20 min.
Light-induced Nuclear Accumulation of phyB-GFP
Continuous red light induced accumulation of phyB-GFP in the nucleus (Fig. 8). Relatively slow kinetics of this process is consistent with the observation made in pea seedlings (Nagatani, A., unpublished observations). phyB was not detected in the nuclei isolated from dark-grown pea seedlings. However, treatment of the seedlings with continuous red light induced nuclear localization of phyB. The level of nuclear phyB reached a plateau ∼4 h after the onset of light treatment.
Phytochrome is known to regulate expression of various genes, of which CAB is the best characterized (Terzaghi and Cashmore, 1995). In Arabidopsis, the time course of the CAB gene induction by a red light pulse has been examined at high time resolution by using luciferase as a reporter (Millar et al., 1992; Millar and Kay, 1996; Anderson et al., 1997). The results indicate that the induction is multiphasic. An acute response occurs in Arabidopsis with the peak at 2 h after the pulse treatment. Subsequently, the expression oscillates under control of the biological clock. Namely, the level falls to a trough at 6.5–8 h and peaks again at 15.5 h. Analysis of the phyB mutant has demonstrated that phyB contributes to both the acute and clock-dependent responses (Anderson et al., 1997).
As shown in Fig. 8, it takes ∼4 h for phyB-GFP to complete the translocation from the cytoplasm to the nucleus, which is significantly slower than the acute response of the CAB gene expression. This may indicate that the level of nuclear phyB attained 2 h after the onset of light might be sufficient to induce the acute response. It is also possible that phyB-GFP migrates more slowly than the authentic phyB. Alternatively, the signal transduction from phyB to the downstream components may take place in the cytoplasm in the early phase of the response. Interestingly, it seems here that the speckle formation is not required for the acute response of the CAB gene expression. Almost no speckles could be observed 2 h after the onset of red light treatment (Fig. 8).
It is more probable that the clock-dependent induction of the CAB gene expression is under control of nuclear-localized phyB. The extent of clock-dependent expression is maximal under continuous light. In such a condition, phyB-GFP is localized to the nucleus almost exclusively (Figs. 4–6 and 8). In addition, we have confirmed in pea seedlings that a pulse of red light can induce long-lasting accumulation of phyB in the nucleus (Nagatani, A., unpublished observations). It has been proposed that the circadian clock confines the ability of light to induce CAB expression (Kay and Millar, 1992). Hence, it is an intriguing possibility that phyB and components of the biological clock directly interact with each other within the nucleus.
Possible Functions of phyB in the Nucleus
Together with the previous report (Sakamoto and Nagatani, 1996), the present results provide compelling evidence that the nucleus is at least one of the sites of phyB action. In animal and yeast cells, many signal transduction factors are known to translocate to the nucleus upon receipt of the signal (Nagatani, 1998). For example, steroid hormone receptors are targeted to the nucleus upon binding of the hormonal ligands (Mangelsdorf et al., 1995). Therefore, it is not surprising that phyB translocates to the nucleus upon light stimulation.
The det, cop, and fus mutants of Arabidopsis exhibit the constitutive photomorphogenic phenotypes in darkness (von Arnim and Deng, 1996; Fankhauser and Chory, 1997). The DET1, COP1, COP9, and FUS6 proteins can be localized to the nucleus. It is especially interesting that the nuclear localization of COP1 is light dependent (von Arnim and Deng, 1996). Conversely, the hy5 mutant is impaired in the light signal transduction. The HY5 gene has been cloned recently (Oyama et al., 1997). The gene encodes a putative transcription factor which is constitutively localized to the nucleus (Chattopadhyay et al., 1998). The HY5 protein binds to the light-responsive promoters of the CAB and CHS genes (Ang et al., 1998; Chattopadhyay et al., 1998). More recently, a putative transcription factor, PIF3, has been identified as a phytochrome-interacting protein (Ni et al., 1998). Hence, it is an attractive possibility that phyB interacts with those proteins in the nucleus to modify transcription of the target genes.
The biological relevance of the phyB-GFP speckles in the nucleus is unknown as discussed above. The time course analysis in this study suggests that speckles are not required for the acute response of the CAB gene expression (Fig. 8). However, the speckled structure may contribute to long-term effects of phyB. It is intriguing here that GFP-COP1 fusion protein forms speckles in the nucleus (Ang et al., 1998). Furthermore, GFP-HY5 is recruited to the speckles if it is coexpressed with COP1. Hence, the speckles of COP1 may represent the site where nuclear factors interact with each other to mediate light signals. In this connection, it would be particularly interesting to know whether phyB colocalizes with other factors in the nucleus.
Concluding Remarks
As discussed above, the present results suggest that phyB translocates to the nucleus upon light stimulation. In the nucleus, phyB may interact with other nuclear factors to transduce the light signal to alter transcription of target genes. However, the kinetics of the light-induced accumulation of phyB-GFP in the nucleus is clearly too slow to explain rapid phytochrome responses such as light-induced changes in intracellular Ca2+ levels (Roux, 1994). Hence, phyB might be functioning not only in the nucleus but also in the cytoplasm. Alternatively, different molecular species of phytochromes may function at different sites within the cell. In this connection, it will be important to know whether other molecular species are localized to the nucleus.
Acknowledgments
We thank Drs. Jen Sheen and Kenzo Nakamura for providing the GFP gene construct and the transformation vector pBI101-Hm, respectively. We also thank Dr. Naoto Yabe for advice on cDNA isolation.
This work was supported in part by a grant from the Program for Promotion of Basic Research Activities for Innovative Biosciences (to A. Nagatani and N. Mochizuki), Special Coordination Funds for Promoting Science and Technology from the Science and Technology Agency of Japan (to A. Nagatani), a Grant-in-Aid for Scientific Research (B) (No. 08454253) from the Ministry of Education, Science, Sports and Culture of Japan (to A. Nagatani), a Grant-in-Aid for Scientific Research on Priority Areas (A) (no. 10182206) from the Ministry of Education, Science, Sports and Culture of Japan (to A. Nagatani), and National Institutes of Health grant GM56006 (to S.A. Kay).
Abbreviations used in this paper
- DIC
differential interference contrast
- GFP
green fluorescent protein
- Pfr
far-red light absorbing form of phytochrome
- phy
phytochrome
References
- Anderson SL, Somers DE, Millar AJ, Hanson K, Chory J, Kay SA. Attenuation of phytochrome A and B signaling pathways by the Arabidopsiscircadian clock. Plant Cell. 1997;9:1727–1743. doi: 10.1105/tpc.9.10.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L-H, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng X-W. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsisdevelopment. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA. A deletion in the PHYD gene of the ArabidopsisWassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell. 1997;9:1317–1326. doi: 10.1105/tpc.9.8.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thalianaplants. CR Acad Sci Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- Chattopadhyay S, Ang L-H, Puente P, Deng X-W, Wei N. ArabidopsisbZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998;10:673–684. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W-I, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC. Phytochrome E influences internode elongation and flowering time in Arabidopsis. . Plant Cell. 1998;10:1479–1488. doi: 10.1105/tpc.10.9.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. Light control of plant development. Annu Rev Cell Dev Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- Furuya M. Phytochromes: their molecular species, gene families, and functions. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:617–645. [Google Scholar]
- Furuya M, Schaefer E. Photoperception and signalling of induction reactions by different phytochromes. Trends Plant Sci. 1996;1:301–307. [Google Scholar]
- Halliday KJ, Thomas B, Whitelam GC. Expression of heterologous phytochromes A, B, or C in transgenic tobacco plants alters vegetative development and flowering time. Plant J. 1997;12:1079–1090. doi: 10.1046/j.1365-313x.1997.12051079.x. [DOI] [PubMed] [Google Scholar]
- Hughes J, Lamparter T, Mittmann F, Hartmann E, Gartner W, Wilde A, Borner T. A prokaryotic phytochrome. Nature. 1997;386:663. doi: 10.1038/386663a0. [DOI] [PubMed] [Google Scholar]
- Kay, S.A., and A.J. Millar. 1992. Circadian regulated Cab gene transcription in higher plants. In The Molecular Biology of Circadian Rhythms. M. Youn, editor. Marcel Dekker, New York. 73–89.
- Kehoe DM, Grossman AR. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science. 1996;273:1409–1412. doi: 10.1126/science.273.5280.1409. [DOI] [PubMed] [Google Scholar]
- Kendrick, R.E., and G.H.M. Kronenberg. 1994. Photomorphogenesis in Plants. 2nd ed. Kluwer Academic Publishers, Dordrecht. 828 pp.
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Mancinelli, A.L. 1994. The physiology of phytochrome action. In Photomorphogenesis in Plants. 2nd ed. R.E. Kendrick and G.H.M. Kronenberg, editors. Kluwer Academic Publishers, Dordrecht. 211–269.
- Mangelsdorf D, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R. The nuclear receptor superfamily: the second decade. Annu Rev Med. 1995;46:443–453. [Google Scholar]
- Mathews S, Sharrock RA. Phytochrome gene diversity. Plant Cell Environ. 1997;20:666–671. [Google Scholar]
- McCormac AC, Wagner D, Boylan MT, Quail PH, Smith H, Whitelam GC. Photoresponses of transgenic Arabidopsisseedlings expressing introduced phytochrome-B-encoding cDNAs: evidence that phytochrome-A and phytochrome-B have distinct photoregulatory functions. Plant J. 1993;4:19–27. [Google Scholar]
- Millar AJ, Kay SA. Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. . Proc Natl Acad Sci USA. 1996;93:15491–15496. doi: 10.1073/pnas.93.26.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Chua N-H, Kay SA. A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell. 1992;4:1075–1087. doi: 10.1105/tpc.4.9.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A. Spatial distribution of phytochromes. J Plant Res. 1997;110:123–130. doi: 10.1007/BF02506851. [DOI] [PubMed] [Google Scholar]
- Nagatani A. Regulated nuclear targeting. Curr Opin Plant Biol. 1998;1:470–474. doi: 10.1016/s1369-5266(98)80037-x. [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt, L.H. 1994. Distribution and localization of phytochrome within the plant. In Photomorphogenesis in Plants. 2nd ed. R.E. Kendrick and G.H.M. Kronenberg, editors. Kluwer Academic Publishers, Dordrecht. 163–185.
- Qin M, Kuhn R, Moran S, Quail PH. Overexpressed phytochrome C has similar photosensory specificity to phytochrome B but a distinctive capacity to enhance primary leaf expansion. Plant J. 1997;12:1163–1172. doi: 10.1046/j.1365-313x.1997.12051163.x. [DOI] [PubMed] [Google Scholar]
- Quail, P.H. 1983. Rapid action of phytochrome in photomorphogenesis. In Photomorphogenesis, Encyclopedia of Plant Physiology. Vol. New Ser. 16A. W. Shropshire, Jr., and H. Mohr, editors. Springer-Verlag, Berlin. 178–212.
- Quail PH. An emerging molecular map of the phytochromes. Plant Cell Environ. 1997;20:657–665. [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D. Phytochromes: photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red far-red light receptor phytochrome-B alter cell elongation and physiological responses throughout Arabidopsisdevelopment. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, S.J. 1994. Signal transduction in phytochrome responses. In Photomorphogenesis in Plants. 2nd ed. R.E. Kendrick and G.H.M. Kronenberg, editors. Kluwer Academic Publishers, Dordrecht. 187–209.
- Sage, L.C. 1992. Pigment of the Imagination: A History of Phytochrome Research. Academic Press, San Diego. 562 pp.
- Sakamoto K, Nagatani A. Nuclear localization activity of phytochrome B. Plant J. 1996;10:859–868. doi: 10.1046/j.1365-313x.1996.10050859.x. [DOI] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. . Proc Natl Acad Sci USA. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer RH, Green MR. Compartmentalization of eukaryotic gene expression: causes and effects. Cell. 1997;91:291–294. doi: 10.1016/s0092-8674(00)80411-0. [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol. 1995;46:445–474. [Google Scholar]
- von Arnim AG, Deng X-W. Light control of seedling development. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:215–243. doi: 10.1146/annurev.arplant.47.1.215. [DOI] [PubMed] [Google Scholar]
- Wada M, Grolig F, Haupt W. Light-oriented chloroplast positioning: contribution to progress in photobiology. J Photochem Photobiol B Biol. 1993;17:3–25. [Google Scholar]
- Wagner D, Tepperman JM, Quail PH. Overexpression of phytochrome-B induces a short hypocotyl phenotype in transgenic Arabidopsis. . Plant Cell. 1991;3:1275–1288. doi: 10.1105/tpc.3.12.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester L, Somers DE, Clack T, Sharrock RA. Transgenic complementation of the hy3 phytochrome B mutation and response to phyB gene copy number in Arabidopsis. . Plant J. 1994;5:261–272. doi: 10.1046/j.1365-313x.1994.05020261.x. [DOI] [PubMed] [Google Scholar]
- Yeh K-C, Lagarias JC. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K-C, Wu S-H, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]