Abstract
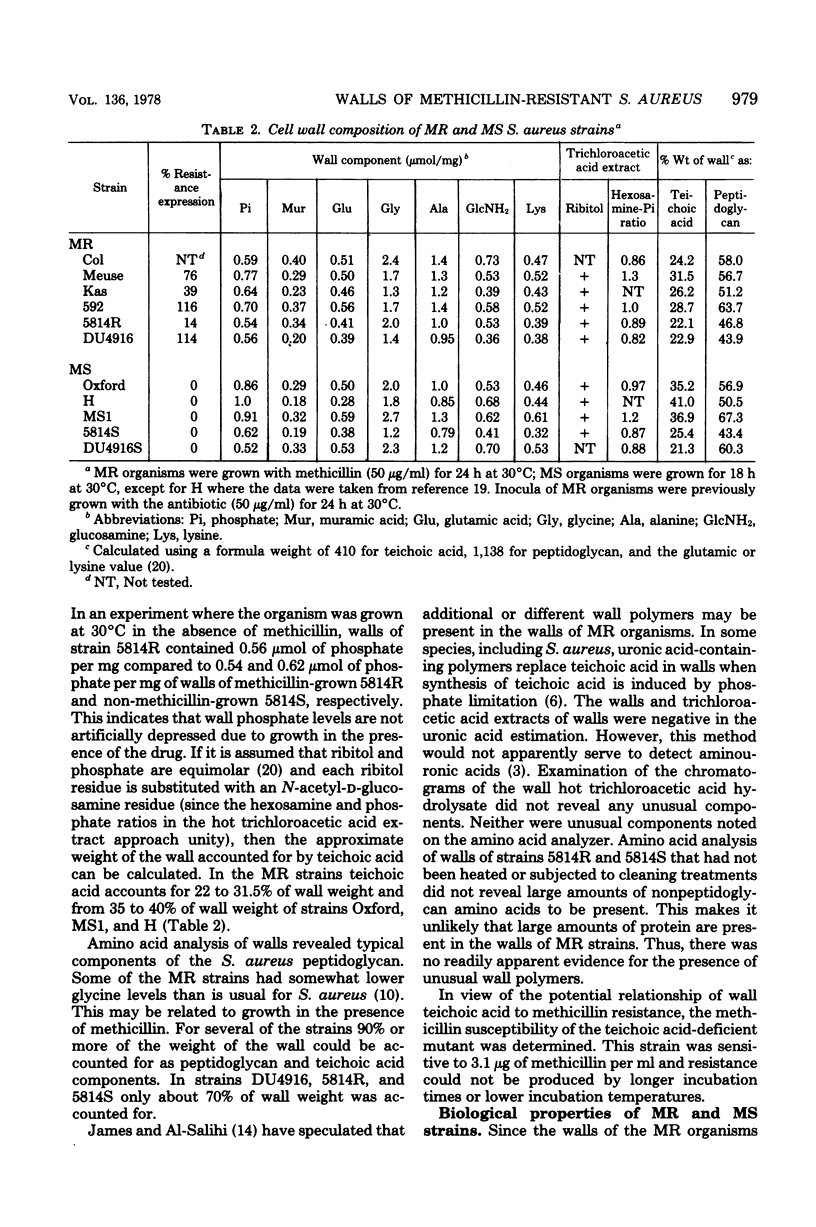
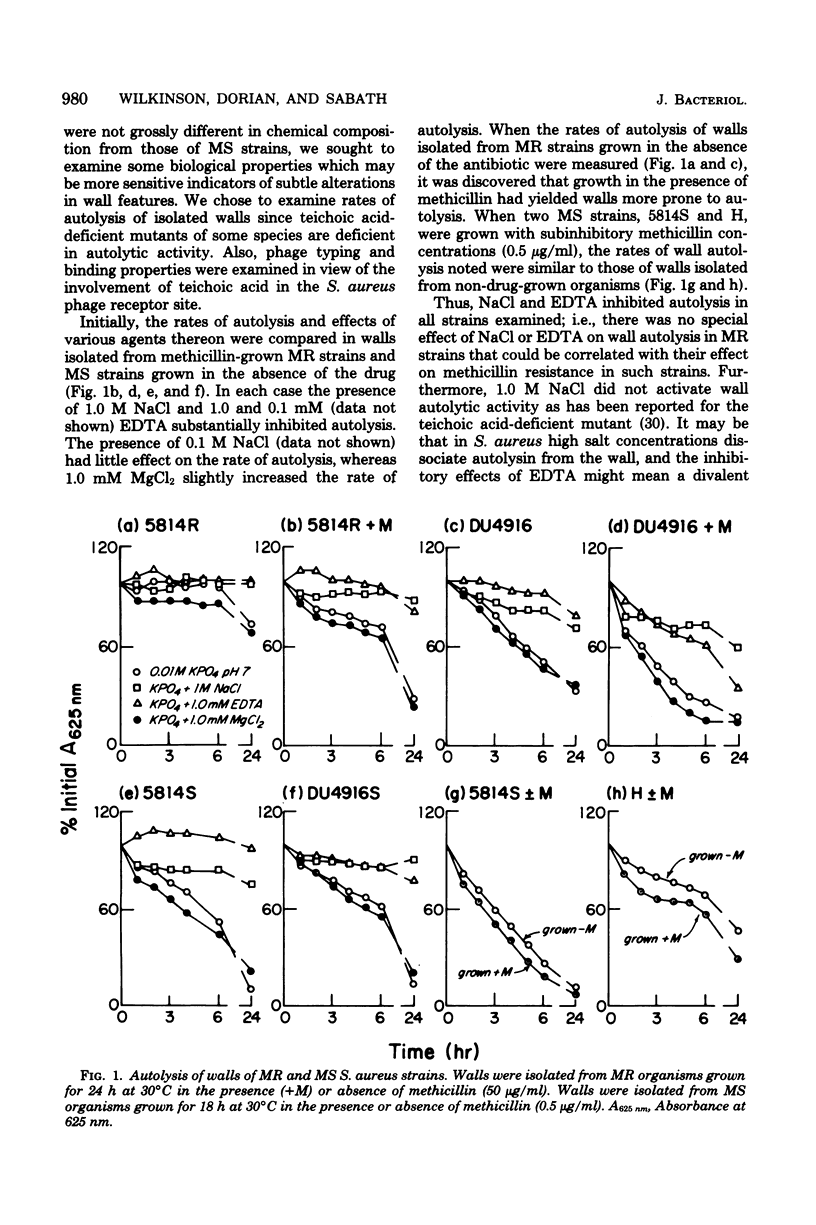
Methicillin-resistant (MR) Staphylococcus aureus strains have previously been reported to be deficient in surface negative charge; this has been correlated with methicillin resistance and ascribed to a deficiency of teichoic acid at the cell surface (A. W. Hill and A. M. James, Microbios 6:157-167, 1972). Teichoic acid was present in walls of MR organisms as revealed by appreciable phosphate levels and detection of ribitol residues. Phosphate levels in walls from five MR strains (0.54 to 0.77 μmol/mg of wall) were lower than in three unrelated methicillin-sensitive (MS) strains (0.86 to 1.0 μmol/mg of wall). However, two MS strains derived from two of the MR strains had wall phosphate levels very similar to those of the MR strains. No evidence for unusual wall polymers was found. Simple deficiency of wall teichoic acid does not result in methicillin resistance since an independently isolated teichoic acid-deficient strain (0.1 μmol of phosphate per mg of wall) was not methicillin resistant. In studies of biological properties possibly related to wall teichoic acid, it was discovered that walls isolated from MR organisms grown in the presence of methicillin autolyzed more rapidly than those isolated from organisms grown in the absence of the drug. Since methicillin resistance is enhanced by NaCl and suppressed by ethylenediaminetetraacetate, the effects of these compounds on autolysis of isolated walls were studied. NaCl (1.0 M) and ethylenediaminetetraacetate (1.0 mM) inhibited the autolysis of walls isolated from MR and MS strains. An MR strain bound phage 47, 52A, and 3A only slightly less well than their respective propagating strains.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chatterjee A. N. Use of bacteriophage-resistant mutants to study the nature of the bacteriophage receptor site of Staphylococcus aureus. J Bacteriol. 1969 May;98(2):519–527. doi: 10.1128/jb.98.2.519-527.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyke K. G., Jevons M. P., Parker M. T. Penicillinase production and intrinsic resistance to penicillins in Staphylococcus aures. Lancet. 1966 Apr 16;1(7442):835–838. doi: 10.1016/s0140-6736(66)90182-6. [DOI] [PubMed] [Google Scholar]
- Dyke K. G. Penicillinase production and intrinsic resistance to penicillins in methicillin-resistant cultures of Staphylococcus aureus. J Med Microbiol. 1969 Aug;2(3):261–278. doi: 10.1099/00222615-2-3-261. [DOI] [PubMed] [Google Scholar]
- Gavan T. L., Town M. A. A microdilution method for antibiotic susceptibility testing: an evaluation. Am J Clin Pathol. 1970 Jun;53(6):880–885. doi: 10.1093/ajcp/53.6.880. [DOI] [PubMed] [Google Scholar]
- Gilpin R. W., Chatterjee A. N., Young F. E. Autolysis of microbial cells: salt activation of autolytic enzymes in a mutant of Staphylococcus aureus. J Bacteriol. 1972 Jul;111(1):272–283. doi: 10.1128/jb.111.1.272-283.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gots J. S. THE DETECTION OF PENICILLINASE-PRODUCING PROPERTIES OF MICROORGANISMS. Science. 1945 Sep 21;102(2647):309–309. doi: 10.1126/science.102.2647.309. [DOI] [PubMed] [Google Scholar]
- Gramoli J. L., Wilkinson B. J. Characterization and identification of coagulase-negative, heat-stable deoxyribonuclease-positive staphylococci. J Gen Microbiol. 1978 Apr;105(2):275–285. doi: 10.1099/00221287-105-2-275. [DOI] [PubMed] [Google Scholar]
- HAIGHT T. H., FINLAND M. Modified Gots test for penicillinase production. Am J Clin Pathol. 1952 Aug;22(8):806–808. doi: 10.1093/ajcp/22.8_ts.806. [DOI] [PubMed] [Google Scholar]
- Hill A. W., James A. M. Effect of growth temperature on the surface properties of cells of Staphylococcus aureus with particular reference to methicillin-resistance. Microbios. 1972 Sep-Oct;6(22):169–178. [PubMed] [Google Scholar]
- Hill A. W., James A. M. Surface properties of cells of methicillin-sensitive and methicillin-resistant strains of Staphylococcus aureus grown at 37 degrees C. Microbios. 1972 Sep-Oct;6(22):157–167. [PubMed] [Google Scholar]
- LEVVY G. A., MCALLAN A. The N-acetylation and estimation of hexosamines. Biochem J. 1959 Sep;73:127–132. doi: 10.1042/bj0730127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey R. W. Antibiotic resistance plasmids of Staphylococcus aureus and their clinical importance. Bacteriol Rev. 1975 Mar;39(1):1–32. doi: 10.1128/br.39.1.1-32.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey R. W. Staphylococcus aureus strain DU4916--an atypical methicillin-resistant isolate? J Gen Microbiol. 1974 Sep;84(1):1–10. doi: 10.1099/00221287-84-1-1. [DOI] [PubMed] [Google Scholar]
- Marshall N. J., James A. M. Surface properties of methicillin-resistant cells of Staphylococcus aureus. Microbios. 1971 Dec;4(15):217–225. [PubMed] [Google Scholar]
- Peterson P. K., Wilkinson B. J., Kim Y., Schmeling D., Douglas S. D., Quie P. G., Verhoef J. The key role of peptidoglycan in the opsonization of Staphylococcus aureus. J Clin Invest. 1978 Mar;61(3):597–609. doi: 10.1172/JCI108971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozgonyi F. Genotypic stability of methicillin resistance in Staphylococcus aureus at supraoptimal temperature. Antimicrob Agents Chemother. 1976 Aug;10(2):377–379. doi: 10.1128/aac.10.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath L. D. Chemical and physical factors influencing methicillin resistance of Staphylococcus aureus and Staphylococcus epidermidis. J Antimicrob Chemother. 1977 Nov;3 (Suppl 100):47–51. doi: 10.1093/jac/3.suppl_c.47. [DOI] [PubMed] [Google Scholar]
- Sabath L. D., Leaf C. D., Gerstein D. A., Finland M. Altered cell walls of Staphylococcus aureus resistant to methicillin. Nature. 1970 Mar 14;225(5237):1074–1074. doi: 10.1038/2251074a0. [DOI] [PubMed] [Google Scholar]
- Sabath L. D., Wallace S. J., Gerstein D. A. Suppression of intrinsic resistance to methicillin and other penicillins in Staphylococcus aureus. Antimicrob Agents Chemother. 1972 Nov;2(5):350–355. doi: 10.1128/aac.2.5.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman S. J. Penicillinase-negative variants of methicillin-resistant Staphylococcus aureus. Nature. 1966 Mar 5;209(5027):994–996. doi: 10.1038/209994a0. [DOI] [PubMed] [Google Scholar]
- Shafer W. M., Iandolo J. J. Chromosomal locus for staphylococcal enterotoxin B. Infect Immun. 1978 Apr;20(1):273–278. doi: 10.1128/iai.20.1.273-278.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon G. N., Russell A. D. Surface properties of cells of some methicillin-resistant strains of Staphylococcus aureus. J Antibiot (Tokyo) 1977 Nov;30(11):974–979. doi: 10.7164/antibiotics.30.974. [DOI] [PubMed] [Google Scholar]
- Wilkinson B. J., White P. J. The effect of antibiotics on synthesis of mucopeptide and teichoic acid by Pediococcus cerevisiae and by a substrain that requires methicillin for growth. J Gen Microbiol. 1973 Dec;79(2):195–204. doi: 10.1099/00221287-79-2-195. [DOI] [PubMed] [Google Scholar]
- Winblad S., Ericson C. Sensitized sheep red cells as a reactant for Staphylococcus aureus protein A. Methodology and epidemiology with special reference to weakly reacting methicillin-resistant strains. Acta Pathol Microbiol Scand B Microbiol Immunol. 1973 Feb;81(1):150–156. [PubMed] [Google Scholar]
- Wong W., Chatterjee A. N., Young F. E. Regulation of bacterial cell walls: correlation between autolytic activity and cell wall turnover in Staphylococcus aureus. J Bacteriol. 1978 May;134(2):555–561. doi: 10.1128/jb.134.2.555-561.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]


