Abstract
How the innate immune system tailors specific responses to diverse microbial infections is not well understood. Cells use a limited number of host receptors and signaling pathways to both discriminate among extracellular and intracellular microbes, and also to generate responses commensurate to each threat. Here, we have addressed these questions by using DNA microarrays to monitor the macrophage transcriptional response to the intracellular bacterial pathogen Listeria monocytogenes. By utilizing combinations of host and bacterial mutants, we have defined the host transcriptional responses to vacuolar and cytosolic bacteria. These compartment-specific host responses induced significantly different sets of target genes, despite activating similar transcription factors. Vacuolar signaling was entirely MyD88-dependent, and induced the transcription of pro-inflammatory cytokines. The IRF3-dependent cytosolic response induced a distinct set of target genes, including IFNβ. Many of these cytosolic response genes were induced by secreted cytokines, so we further identified those host genes induced independent of secondary signaling. The host response to cytosolic bacteria was reconstituted by the cytosolic delivery of L. monocytogenes genomic DNA, but we observed an amplification of this response by NOD2 signaling in response to MDP. Correspondingly, the induction of IFNβ was reduced in nod2 −/− macrophages during infection with either L. monocytogenes or Mycobacterium tuberculosis. Combinatorial control of IFNβ induction by recognition of both DNA and MDP may highlight a mechanism by which the innate immune system integrates the responses to multiple ligands presented in the cytosol by intracellular pathogens.
Author Summary
Macrophages are critical cells of the innate immune system, contributing to immediate and robust defense against microbial infections. Macrophages detect pathogens using host receptors located on the cell surface, in phagosomal vacuoles, and in the cytosol. While fundamental to innate immunity, it is not clear if these different receptors merely provide redundant mechanisms for sensing microbial infection, or if instead they induce distinct gene expression programs that may allow for threat-specific host responses. We addressed this question by dissecting the macrophage transcriptional responses to the model intracellular bacterial pathogen Listeria monocytogenes. Using genetic and genomic approaches, we found that the macrophage response to L. monocytogenes trapped in phagosomal compartments was distinct and separable from the response to live bacteria replicating in the host cytosol. The macrophage response to cytosolic bacteria was recapitulated by bacterial nucleic acid and cell wall fragments, and induced surprisingly few primary response genes. These findings highlight a mechanism by which the innate immune system may specifically sense intracellular bacteria, as the macrophage response to Mycobacterium tuberculosis was similarly regulated.
Introduction
As sentinels of the immune system, macrophages must be able to determine the nature and scope of microbial threats to mount appropriate transcriptional responses [1,2]. Macrophages need to discriminate not only viral from bacterial infection, but also extracellular and possibly killed microbes from intracellular and replicating pathogens [3]. Cells receive information regarding infection using a limited number of pattern recognition receptors (PRRs) to sense conserved motifs presented by microbes [1,4–6]. Two major classes of PRRs include membrane-bound Toll-like receptors (TLRs) and soluble, cytosolic NOD-like receptors (NLRs). TLRs monitor the extracellular environment and phagolysosomal compartments, and recognize pathogen associated molecular patterns (PAMPs) that include lipopolysaccharide, flagella, CpG DNA, and bacterial lipoprotein [7]. NLRs complement this host defense by providing surveillance of the cytosol. The nucleotide-binding and oligomerization domain containing (NOD) proteins, for which this class of receptors is named, recognize cell wall fragments from both Gram-negative and Gram-positive bacteria [8–10]. NOD1 recognizes a specific peptidoglycan (PGN) fragment containing diaminopimelic acid, while NOD2 recognizes a muramyl dipeptide (MDP) fragment of PGN. The MDA5 and RIG-I NLRs detect cytosolic dsRNA [11], while DAI detects cytosolic dsDNA [12–14].
How cells initiate a threat-specific transcriptional response is poorly understood, as many PRRs, in response to different stimuli, utilize the MyD88 and TRIF signaling adaptor molecules to activate the same transcription factors [15]. For instance, activation of both TLRs and NLRs causes degradation of the repressor IκB, thereby freeing the transcription factor NFκB to enter the nucleus and bind to target promoters of genes important for host defense. Some target specificity is generated by the phosphorylation and activation of the IRF3 transcription factor by only a subset of PRRs, including those that recognize nucleic acids. The induction of certain host genes, including the Type I interferons α (IFNα) and β (IFNβ), requires both NFκB and IRF3 [16–18]. Secreted Type I interferons then induce many additional genes by secondary signaling through the Type I interferon receptor (IFNAR) [15].
L. monocytogenes is a ubiquitous Gram-positive intracellular bacterium that can cause serious illness in pregnant women and immunocompromised individuals [19], and is ideal for the study of host innate immune responses. Mutants defective in precise stages of the intracellular life cycle have been isolated, and in vitro infection of primary bone marrow–derived macrophages allows dissection of host signaling pathways. Approximately 30 minutes after initial phagocytic uptake into a membrane-bound vacuole, bacteria escape to the host cytosol by perforating the vacuolar membrane, using the pore-forming toxin listeriolysin O (LLO, encoded by the gene hly) [20]. Once in the host cytosol, L. monocytogenes replicates robustly, and uses a system of actin-based motility to spread from the initially infected macrophage to colonize neighboring cells [21]. Both heat-killed and hly− L. monocytogenes induce inflammatory cytokines, but in non-activated macrophages only wild-type (WT) bacteria that are able to access the host cytosol induce Type I interferons [22–24].
Although the L. monocytogenes Type I interferon-stimulating ligand has not been conclusively identified, evidence suggests that bacterial DNA possesses IFNβ-inducing activity and might be the relevant PAMP [12,25]. PGN fragments have also been shown to induce host transcriptional responses [3,8,26–28], and MDP is present both in digested L. monocytogenes PGN fragments [29,30] and in polymer-linked form in intact PGN [31]. The role of MDP during the host response to L. monocytogenes is not clear, however [3,24,32].
In this study, we have comprehensively determined the macrophage transcriptional responses to L. monocytogenes using DNA microarrays. Using macrophages deficient for defined host signaling pathways, and bacteria residing in different subcellular localizations, we have delineated distinct host responses to vacuolar and cytosolic bacteria, and addressed the mechanisms underlying the specificity of these responses. We have additionally determined the direct transcriptional targets of host NLR signaling in response to cytosolic bacteria. These genes are co-regulated with IFNβ, and are uniquely induced in infected cells, as their induction is independent of any secondary signaling. These primary targets have the potential to modify host signaling, and may therefore critically impact the host response to infection. Cytosolic delivery of purified bacterial genomic DNA reconstituted the host response to cytosolic L. monocytogenes. This response was synergistically amplified by NOD2 signaling in response to MDP. We find a similar role for NOD2 signaling in the host response to both L. monocytogenes and M. tuberculosis, and this may represent a mechanism by which cells integrate multiple PRR signals to accurately identify bacteria able to access the host cytosol.
Results
Identification of Distinct Macrophage Transcriptional Responses to Vacuolar and Cytosolic L. monocytogenes
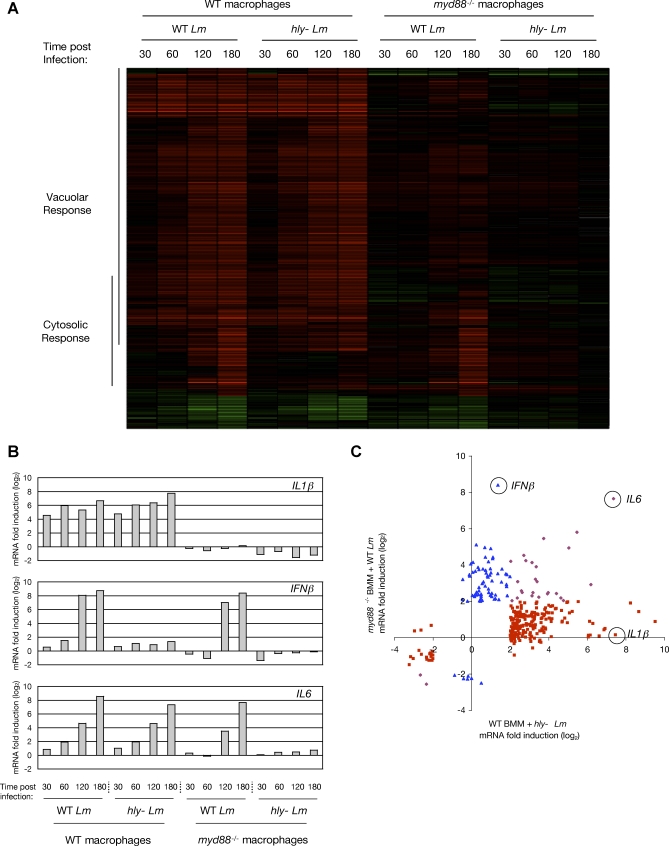
We first determined the global transcriptional response of WT and myd88 −/− macrophages infected with WT bacteria and hly− L. monocytogenes, using high-density oligonucleotide microarrays [33]. To identify genes induced by vacuolar bacteria, the transcriptional response of WT macrophages after 180 minutes of infection with hly− L. monocytogenes was subject to Significance Analysis of Microarrays (SAM) [34] (see Materials and Methods), and the resulting genes further selected to identify those with at least a 4-fold change in abundance. Using this approach, we identified 252 macrophage genes that met these criteria, which we defined as the “Vacuolar Response” of macrophages to L. monocytogenes (Figure 1A, “Vacuolar Response”; for full target gene list, see Dataset S1). This class of genes included many pro-inflammatory cytokines and chemokines, such as Interleukins 1α (IL1α) and 1β (IL1β), tumor necrosis factor (TNF), KC, and MIP2. Strikingly, none of the 252 genes of the Vacuolar Response were significantly induced in myd88 −/− macrophages (Figure S1). IL1β is hereafter used as a representative target gene of the Vacuolar Response (Figure 1B, top panel).
Figure 1. Macrophages Have Distinct Transcriptional Responses to Vacuolar and Cytosolic L. monocytogenes Infection.
(A) Cluster analysis of the microarray determination of all mouse macrophage genes with at least a 4-fold change in abundance during infection of WT and myd88−/− macrophages with either WT or hly− L. monocytogenes, at the indicated times post-infection (in minutes). Red indicates an increase in RNA abundance relative to uninfected macrophages, and green indicates a decrease. Genes identified by SAM and at least 4-fold induced in WT macrophages by hly− L. monocytogenes are indicated as targets of the “Vacuolar Response”. Genes identified by SAM and at least 4-fold induced in myd88 −/− macrophages by WT L. monocytogenes are indicated as targets of the “Cytosolic Response”. For target gene lists, see Datasets S1 and S2. The determination of Vacuolar Response and Cytosolic Response target genes was from multiple arrays representing four independent experiments (e.g. four independent dishes of uninfected myd88 −/− macrophages and four independent dishes of myd88 −/− macrophages infected with WT L. monocytogenes for 180 minutes were used for Cytosolic Response determination).
(B) Summary of the transcriptional responses of IL1β, IFNβ, and IL6 in WT and myd88 −/− macrophages during infection with either WT or hly− L. monocytogenes, at the indicated times post-infection (in minutes), determined by microarray analyses (Dataset S5).
(C) Scatter plot representation of the transcriptional response of all genes defined as targets of the Vacuolar Response or the Cytosolic Response. Microarray data from myd88 −/− macrophages infected with WT L. monocytogenes (Cytosolic Response-stimulating conditions) is plotted on the Y-axis, and microarray data from WT macrophages infected with hly− L. monocytogenes (Vacuolar Response-stimulating conditions) is plotted on the X-axis. Targets of only the Vacuolar Response are depicted as red squares (e.g. IL1β), targets of only the Cytosolic Response as blue triangles (e.g. IFNβ), and targets of both the Vacuolar Response and Cytosolic Response as purple diamonds (e.g. IL6).
To identify genes induced by cytosolic bacteria, the transcriptional response of myd88 −/− macrophages after 180 minutes of infection with WT L. monocytogenes was subject to SAM, and the resulting genes further selected to identify those with at least a 4-fold change in abundance. Using this approach, we identified 106 macrophage genes, which we defined as the “Cytosolic Response” of macrophages to L. monocytogenes (Figure 1A, “Cytosolic Response”; for full target gene list, see Dataset S2). These genes were strongly induced starting at 2 hours post-infection, by which point WT bacteria were replicating robustly in the cytosol of infected cells. Among the genes most highly induced by the Cytosolic Response were Type I interferons, including IFNβ and multiple IFNα genes, and many known interferon-regulated genes. IFNβ is hereafter used as a representative target gene of the Cytosolic Response (Figure 1B, middle panel).
For 27 genes we observed induction by both the Vacuolar Response and the Cytosolic Response, as these genes were induced both in WT macrophages infected with hly− L. monocytogenes and in myd88 −/− macrophages infected with WT L. monocytogenes. IL6 is hereafter used as a representative target gene of this class (Figure 1B, bottom panel).
Identification of the Primary Cytosolic Response
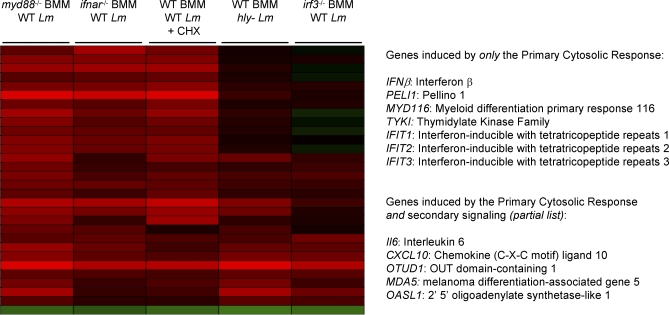
Cytosolic Response targets included Type I interferons and Interleukin 6 (IL6), both of which induce the transcription of many additional genes in neighboring cells [15,35,36]. Five additional analyses were performed to identify genes directly induced by the host response to cytosolic bacteria, and not by secondary signaling. First, we required significant induction in infected ifnar −/− macrophages, which cannot respond to secreted Type I interferons. Second, induction in infected ifnar −/− macrophages must not have been significantly less than that in infected WT macrophages. These two filters removed genes significantly induced as a result of secondary Type I interferon signaling. Third, we required significant induction in infected WT macrophages treated with the protein synthesis inhibitor cycloheximide. Fourth, induction in cycloheximide-treated infected WT macrophages must not have been significantly less than induction in untreated infected WT macrophages. These two filters removed genes significantly induced as a result of secondary signaling by any other translated and secreted cytokine. Finally, we required that genes not have also been targets of the Vacuolar Response, as these genes would be induced in uninfected cells responding to extracellular TLR ligands. Ultimately, we identified only seven genes that met these five strict criteria, which we defined as targets of the Primary Cytosolic Response. These genes included IFNβ, PELI1, MYD116, TYKI, and three members of the IFIT (interferon-inducible with tetratricopeptide repeats) family (Figure 2). None of these genes were significantly induced in irf3 −/− macrophages (data not shown). To identify genes induced by the Primary Cytosolic Response, but also induced by either secondary signaling or the Vacuolar Response, only criteria 1 and 3 (above) were used. By these relaxed criteria 20 additional genes were identified, including IL6, CXCL10, OTUD1, MDA5, IGTP, and OASL1 (for full target gene list, see Dataset S3).
Figure 2. Determination of the Primary Cytosolic Response.
Cluster analysis of the microarray data used to determine the targets of the Primary Cytosolic Response, which are directly induced by NLR signaling in response to L. monocytogenes. The analyses used to determine these targets are described in Results. For the full target list, see Dataset S3.
IRF3 Determines the Specificity of the Cytosolic Response
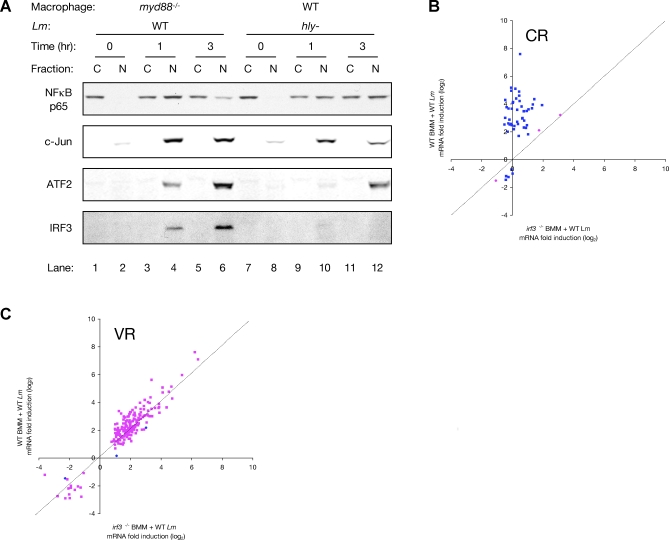
The minimal overlap between the targets of the Vacuolar Response and the Cytosolic Response indicated a high degree of target gene specificity in these responses (Figure 1C). Stimulation of the Cytosolic Response during infection of myd88 −/− macrophages with WT L. monocytogenes resulted in the activation and nuclear translocation of both NFκB and IRF3 (Figure 3A, lanes 1–6). The p65 subunit of NFκB was initially restricted to the cytosol of uninfected cells (lanes 1 and 2), but it accumulated in the nucleus as early as 1 hour post-infection (lanes 3 and 4), where it was still localized 3 hours post-infection (lanes 5 and 6). Similarly, IRF3 was observed in the nucleus of infected cells after 1 hour of infection (lanes 3 and 4), and by 3 hours accumulation of IRF3 in the nucleus had sharply increased (lanes 5 and 6). The transcription factors c-Jun and ATF2, both of which bind to the IFNβ promoter [16], also localized to the nucleus of infected cells during the Cytosolic Response (lanes 4 and 6). Stimulation of the Vacuolar Response during infection of WT macrophages with hly− L. monocytogenes (lanes 7–12) did not lead to nuclear localization of IRF3, which is consistent with failure of the Vacuolar Response to induce IFNβ. NFκB, c-Jun, and ATF2, however, were localized to the nucleus during the Vacuolar Response (lanes 10 and 12).
Figure 3. Specificity of the Cytosolic and Vacuolar Responses.
(A) Western blot analysis of the distribution of transcription factors NFκB p65, phospho-c-Jun, phospho-ATF2, and IRF3 in fractionated lysates of myd88 −/− or WT macrophages infected with WT or hly− L. monocytogenes, respectively, for the indicated times (in hours). “C” indicates cytosolic fractions and “N” indicates nuclear fractions. Data is from the pooled lysates of two independent dishes.
(B) Scatter plot representation of the transcriptional response of WT and irf3 −/− macrophages infected for 3 hours with WT L. monocytogenes, as determined by microarrays. Shown are the responses of Cytosolic Response-specific target genes. Y-axis values are log2 fold change in RNA abundance in WT macrophages infected with WT L. monocytogenes. X-axis values are log2 fold change in RNA abundance in irf3 −/− macrophages infected with WT L. monocytogenes. The superimposed dashed line has a slope = 1. Spots in blue are those identified by SAM as being significantly differently induced in the two conditions, while spots in pink are not different.
(C) Scatter plot representation of the transcriptional response of WT and irf3 −/− macrophages infected for 3 hours with WT L. monocytogenes, as determined by microarrays. Shown are the responses of Vacuolar Response-specific target genes. Y-axis values are log2 fold change in RNA abundance in WT macrophages infected with WT L. monocytogenes. X-axis values are log2 fold change in RNA abundance in irf3 −/− macrophages infected with WT L. monocytogenes. The superimposed dashed line has a slope = 1. Spots in blue are those identified by SAM as being significantly differently induced in the two conditions, while spots in pink are not different.
The induction of IFNβ during infection of macrophages with WT L. monocytogenes requires IRF3 [24,37]. We further found that the majority of the Cytosolic Response is IRF3-dependent, as 94% of Cytosolic Response-specific target genes (i.e. genes that are not also targets of the Vacuolar Response, such as IL6) were significantly less induced in infected irf3 −/− macrophages compared to infection of WT macrophages (Figure 3B).
WT bacteria in the cytosol of myd88 −/− macrophages failed to induce the vast majority of Vacuolar Response genes, despite the robust nuclear localization of NFκB in these infected cells. The Cytosolic Response did not lead to feedback inhibition of Vacuolar Response target gene induction, unlike what has been reported during similar innate immune signaling in Drosophila [38]. Only 1.7% of Vacuolar Response-specific target genes showed greater induction during infection of irf3 −/− macrophages, in which the Cytosolic Response was almost entirely absent, compared to infected WT macrophages (Figure 3C).
Induction of the Cytosolic Response by Cytosolic Delivery of L. monocytogenes Genomic DNA Is Amplified by Co-Delivery of MDP
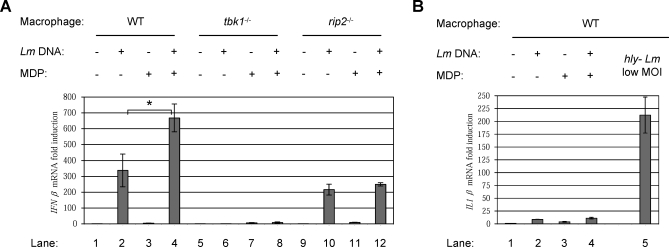
To examine and dissect the Cytosolic Response, we attempted to recapitulate the host response to cytosolic bacteria by instead treating macrophages with purified bacterial ligands, using L. monocytogenes genomic DNA and synthetic MDP. When delivered directly into the cytosol of WT macrophages by transfection, L. monocytogenes DNA strongly induced the expression of IFNβ (Figure 4A, lane 2). However, co-transfection of macrophages with both L. monocytogenes genomic DNA and synthetic MDP yielded double the induction of IFNβ than that observed with DNA alone (lane 4), even though MDP alone yielded minimal IFNβ induction (lane 3). IFNβ induction was absolutely dependent on TBK1 (lanes 6 and 8), a kinase required by all identified nucleic acid recognition receptors to activate the IRF3 transcription factor [37,39]. This defect was specific to the Cytosolic Response, as induction of Vacuolar Response target genes in response to hly− L. monocytogenes was unaffected in tbk1−/− cells (data not shown). Macrophages deficient for RIP2, an adaptor molecule required for NOD2-dependent NFκB activation [40,41], still induced IFNβ in response to DNA (lane 10), but co-transfection with MDP no longer produced any additional synergistic induction (lane 12). This response was independent of both MyD88 and TRIF (Figure S2A), and was not due to contamination of the L. monocytogenes genomic DNA with other bacterial ligands, as we observed identical results with synthetic poly(dAT-dTA) DNA (Figure S2B). While virtually all Cytosolic Response-specific target genes were induced by transfection of macrophages with L. monocytogenes DNA, 94% of these genes were induced to an even greater magnitude by co-transfection of macrophages with both DNA and MDP (Figure S2C). Cytosolic delivery of DNA and MDP specifically induced Cytosolic Response genes without inducing Vacuolar Response genes, as these ligands only weakly induced IL1β (Figure 4B, compare lanes 2–4 to lane 5; Dataset S4).
Figure 4. Cytosolic Delivery of L. monocytogenes DNA and Synthetic MDP Synergistically Induce the Cytosolic Response.
(A) Analysis by qPCR of IFNβ transcriptional induction in WT, tbk1 −/−, and rip2 −/− macrophages 6 hours after transfection with the indicated combinations of L. monocytogenes genomic DNA (250 ng/ml) and synthetic MDP (10 μg/ml). The “*” indicates that these values are significantly different statistically with a p-value of 0.07.
(B) Analysis by qPCR of IL1β transcriptional induction in WT macrophages 6 hours after transfection with the indicated combinations of L. monocytogenes genomic DNA and synthetic MDP, or 4 hours after infection with hly− L. monocytogenes at a low MOI.
Nuclear NFκB Abundance Controls Synergistic Induction of the Cytosolic Response
Western blots were used to assess the activation of Cytosolic Response transcription factors by delivery of DNA and MDP. Transfection of WT macrophages with DNA alone (Figure 5A, lane 4) or MDP alone (lane 6) strongly activated NFκB to similar levels. When macrophages were transfected with both DNA and MDP, NFκB accumulated in the nucleus to a magnitude equal to the sum of that observed for the two ligands individually (lane 8). Similar results were observed for nuclear translocation of c-Jun (lanes 4, 6, and 8). No additive nuclear translocation of ATF2 was observed by co-delivery of both ligands (data not shown). MDP has never been found to activate IRF3 [8,24,28], consistent with our finding that MDP delivery alone does not induce IFNβ (Figure 4A, lanes 3 and 7).
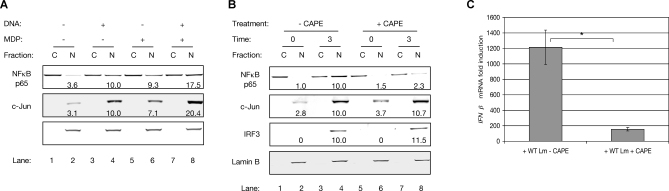
Figure 5. Induction of the Cytosolic Response Requires Nuclear NFκB.
(A) Quantitative Western blot analysis of NFκB p65, c-Jun, and ATF2 distribution in fractionated lysates of WT macrophages transfected with the indicated combinations of L. monocytogenes genomic DNA and synthetic MDP. “C” indicates cytosolic fractions and “N” indicates nuclear fractions. Nuclear abundance of proteins (indicated below the nuclear fraction within each blot) was normalized to the nuclear protein Lamin B, which also serves as fractionation control. For each transcription factor, abundance in the nucleus of macrophages transfected with L. monocytogenes DNA was arbitrarily set to 10.0, and all other abundances were calculated relative to this. Data was collected from two blots, with each blot using pooled lysates of two independent dishes.
(B) Quantitative Western blot analysis of NFκB p65, c-Jun, and IRF3 protein distribution in fractionated lysates of WT macrophages infected with WT L. monocytogenes for the indicated times (in hours). Where indicated, cells were additionally treated with 10 μg/ml CAPE.
(C) Analysis by qPCR of IFNβ transcriptional induction in WT macrophages infected with WT L. monocytogenes for 3 hours. Where indicated, cells were additionally treated with 10 μg/ml CAPE. The “*” indicates that these values are significantly different statistically with a p-value of 0.03.
To investigate the role of NFκB abundance in the synergistic induction of IFNβ, WT macrophages infected with WT L. monocytogenes were treated with 10 μg/ml caffeic acid phenyl ester (CAPE), a pharmacological inhibitor of NFκB nuclear trafficking [42]. CAPE treatment of infected macrophages caused a greater than 4-fold reduction in nuclear NFκB p65 accumulation, but had no effect on either c-Jun or IRF3 nuclear localization (Figure 5B). Compared to untreated macrophages, induction of IFNβ by L. monocytogenes in CAPE-treated macrophages was reduced greater than 7-fold (Figure 5C). Using DNA microarrays, we observed that this inhibition of NFκB nuclear translocation affected the entire Cytosolic Response, as 99% of Cytosolic Response-specific target genes were significantly less induced in infected CAPE-treated macrophages, compared to infected macrophages not CAPE-treated (Figure S3).
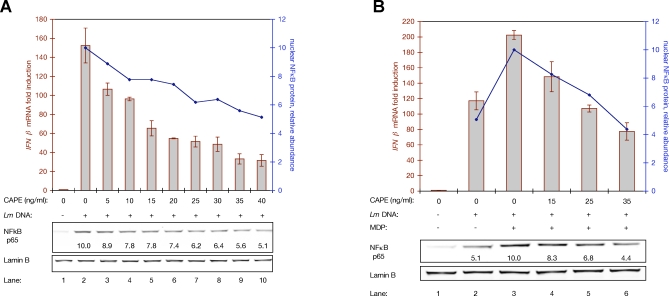
Additionally, macrophages transfected with L. monocytogenes genomic DNA were treated with concentrations of CAPE ranging from 5 ng/ml to 40 ng/ml, which reduced nuclear NFκB abundance from 10%–50% compared to untreated cells (Figure 6A, lanes 3–10). These lower concentrations of CAPE allowed precise titration of nuclear NFκB, instead of the near complete inhibition observed previously with higher doses (Figure 5). Coincident with this reduction in nuclear NFκB abundance, induction of IFNβ decreased 30%–80%. At each increase in CAPE dosage, translocation of NFκB to the nucleus was reduced and induction of IFNβ declined. Similarly, macrophages transfected with either DNA alone or with both DNA and MDP were treated with a titration of CAPE. Without CAPE treatment, co-transfection of macrophages with both DNA and MDP yielded both twice the induction of IFNβ and twice the relocalization of NFκB to the nucleus, compared to transfection with DNA alone (Figure 6B, lanes 2 and 3). At increasing concentrations of CAPE, macrophages co-transfected with DNA and MDP induced less IFNβ (lanes 4–6). When macrophages co-transfected with DNA and MDP were treated with 25–35 ng/ml CAPE, the magnitude of nuclear NFκB matched that observed in untreated macrophages transfected only with DNA (compare lanes 5 and 6 to lane 2). Under these two conditions with equivalent NFκB activation—one with DNA alone and one with DNA, MDP, and CAPE—IFNβ induction was nearly identical.
Figure 6. Synergistic Induction of IFNβ Is Sensitive to NFκB Nuclear Abundance.
(A) Dual analysis of IFNβ transcriptional induction and nuclear NFκB abundance in WT macrophages transfected with L. monocytogenes DNA, and additionally treated with CAPE at the indicated concentrations. IFNβ transcriptional analysis (red columns and left Y-axis) was by qPCR. Nuclear NFκB abundance (blue line and right Y-axis) was by quantitative Western blot analysis of the nuclear fractions of fractionated cells; blot is shown at the bottom, along with calculated relative NFκB abundances.
(B) Dual analysis of IFNβ transcriptional induction and nuclear NFκB abundance in WT macrophages transfected with the indicated combinations of L. monocytogenes genomic DNA and MDP, and additionally treated with CAPE at the indicated concentrations. IFNβ transcriptional analysis (columns and left Y-axis, in red) was by qPCR. Nuclear NFκB abundance (line and right Y-axis, in blue) was by quantitative Western blot analysis; blot is shown at the bottom, along with calculated relative NFκB abundances.
NOD2 Is Required for Full Induction of IFNβ during Infection of Macrophages with Either L. monocytogenes or M. tuberculosis
To determine if L. monocytogenes release MDP during intracellular infection of macrophages, and if this is sensed by NOD2, IFNβ induction in nod2 −/− macrophages infected with WT L. monocytogenes was assessed. Residual vacuolar signaling in these macrophages continued to activate NFκB, however, obscuring the contribution of NOD2 signaling to nuclear NFκB abundance (Figure S4A). This residual TLR signaling was likely in response to the high concentrations of bacterial fragments delivered during the initial infection inoculum, and would be absent during a natural infection by a single bacterium. Therefore, to reduce NFκB activation by TLR overstimulation, macrophages were first tolerized by prior exposure to the synthetic TLR2 agonist Pam3CSK4 [43] (Figure S4B and S4C).
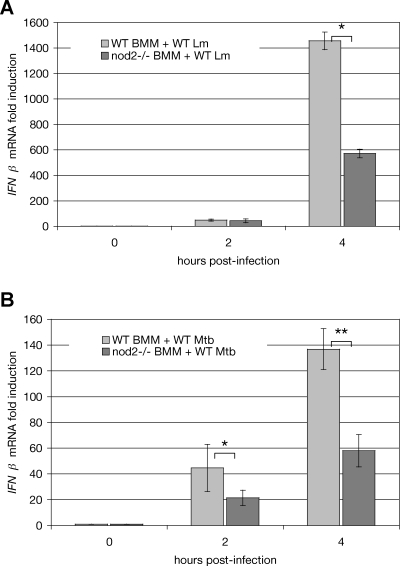
In tolerized macrophages infected with WT L. monocytogenes, at 4 hours post-infection nod2 −/− macrophages exhibited a greater than 2-fold reduction in IFNβ induction, as compared to WT macrophages (Figure 7A). Thus, NOD2 signaling doubled the induction of IFNβ under these conditions. This exactly mirrored the effects of cytosolic co-delivery of DNA and MDP—a 2-fold amplification of IFNβ induction (Figure 4A) and a 2-fold amplification of NFκB nuclear abundance (Figure 5A), compared to the response elicited by bacterial DNA alone. To determine if NOD2 signaling was necessary for the full induction of IFNβ in response to infection with other intracellular bacteria, we infected tolerized WT and nod2 −/− macrophages with the pathogenic Gram-positive intracellular bacterium M. tuberculosis [44,45]. We observed that induction of IFNβ in nod2 −/− macrophages infected with M. tuberculosis for 4 hours was less than half that observed in infected WT macrophages (Figure 7B).
Figure 7. NOD2 Is Required for Full IFNβ Induction during Infection with Either L. monocytogenes or M. tuberculosis .
(A) Analysis by qPCR of IFNβ transcriptional induction in tolerized WT and nod2 −/− macrophages infected with WT L. monocytogenes. The “*” indicates that these values are significantly different statistically with a p-value of 0.003.
(B) Analysis by qPCR of IFNβ transcriptional induction in tolerized WT and nod2 −/− macrophages infected with WT M. tuberculosis. The “*” indicates that these values are significantly different statistically with a p-value of 0.05. The “**” indicates that these values are significantly different statistically with a p-value of 0.001.
Discussion
Identification of the Vacuolar and Cytosolic Responses to L. monocytogenes
In this study we have rigorously and comprehensively identified the Vacuolar and Cytosolic Responses of macrophages to infection with an intracellular bacterial pathogen. Our model system—primary bone marrow–derived macrophages of multiple genotypes infected with both WT L. monocytogenes and hly− bacteria—allowed precise separation of host responses to cytosolic and vacuolar bacteria. Previous genomic analyses of the macrophage transcriptional response to L. monocytogenes did not use myd88 −/− macrophages to fully separate TLR and NLR signaling [23].
We have found that the Vacuolar Response was entirely MyD88-dependent, suggesting that the role of the TLR adaptor TRIF may be specific to innate immune responses to Gram-negative bacteria. The Vacuolar Response controlled the transcriptional induction of many pro-inflammatory cytokines, such as IL1α, IL1β, and TNF. In contrast, the IRF3-dependent Cytosolic Response induced a distinct and significantly non-overlapping set of 106 host response genes, including Type I interferons. We have further identified the 27 targets of the Cytosolic Response that were directly induced in response to cytosolic L. monocytogenes, and did not require secondary cytokine signaling for induction. Seven of these Primary Cytosolic Response target genes were induced entirely independent of any secondary signaling, and therefore were only induced directly in infected cells. Of these 7 genes, only IFNβ encodes a secreted protein, while many of the others encode potential regulators of signaling. For instance, MYD116 is a homolog of GADD34, which can form a complex with protein phosphatase 1α to dephosphorylate eIF2α, thereby inhibiting protein synthesis [46]. PELI1 is an E3 ubiquitin ligase that has been shown to modulate both TLR and IL1 signaling [47]. Consequently, the significant representation and robust induction of members of the IFIT family (IFIT1–3; induced 29.9-, 21.1-, and 13.9-fold, respectively) in this select group of genes warrants further study. Very little is known about IFIT proteins other than the potential of their tetratricopeptide repeats to mediate protein–protein interactions [48]. Despite their name, the IFIT genes responded directly to NLR signaling in response to L. monocytogenes.
We propose that these 7 targets of the Primary Cytosolic Response may provide a mechanism by which infected and uninfected cells could respond differently to the Type I interferons and other cytokines secreted during infection. Cytokines would trigger secondary signaling in all cells near the site of infection, but induction of MYD116, PELI1, TYKI, and IFIT1–3 only in infected cells might modify this secondary signaling to trigger a different response. In this manner, infected cells could be specifically re-programmed to help contain infection.
We have found that two separable pathways coordinately control the Cytosolic Response. The first pathway consists of recognition of bacterial nucleic acid by a cytosolic pattern recognition receptor, perhaps DAI, that activates the transcription factors IRF3 and NFκB. We have demonstrated that L. monocytogenes genomic DNA induced Cytosolic Response target genes, and others have also shown that L. monocytogenes DNA induces IFNβ [12]. The response to both genomic DNA and live bacteria was TBK1-dependent (this study and [37]), and DAI has been shown to associate with TBK1 [14].
The second pathway consists of peptidoglycan fragment recognition by NOD2, which activates NFκB but not IRF3. We have demonstrated that the nuclear abundance of NFκB was limiting for the induction of IFNβ, and our data suggest that the two pathways converge by coordinate control of NFκB nuclear abundance. First, co-delivery of bacterial DNA and synthetic MDP doubled the nuclear abundance of NFκB, and induced twice as much IFNβ, compared to transfection of macrophages with DNA alone. Second, CAPE inhibited induction of IFNβ, and blocked only the nuclear translocation of NFκB. Third, under two conditions yielding equivalent NFκB activation—one with DNA alone and one with DNA, MDP, and CAPE—IFNβ induction was nearly identical, suggesting that control of NFκB abundance was the principal mechanism by which NOD2 amplified IFNβ induction. The further activation of NFκB by NOD2 signaling explains how this pathway contributed to IFNβ induction without activating IRF3.
We further found that nod2 −/− macrophages, when TLR signaling was eliminated, induced significantly less IFNβ during infection with either L. monocytogenes or M. tuberculosis. We speculate that NOD2 may play a similar role in response to other IFNβ-inducing intracellular bacteria whose peptidoglycan contains MDP, such as Francisella tularensis [36]. The convergence of NLR signaling pathways at the level of transcription factor abundance might allow complex signal integration in cells using a limited number of PRRs.
Specificity in Target Gene Induction by the Vacuolar and Cytosolic Responses
The Vacuolar Response and Cytosolic Response controlled the transcription of largely distinct sets of target genes. The Vacuolar Response activated NFκB, and thereby induced pro-inflammatory cytokines such as IL1β, but did not activate IRF3, and hence did not activate IRF3-dependent targets including IFNβ. The Vacuolar Response was completely MyD88-dependent, and it has been demonstrated that a TLR-dependent but NFκB-independent remodeling of nucleosomes at the promoters of certain pro-inflammatory cytokines is required for induction [49–51]. Chromatin modifications are increasingly viewed as critical modulators ensuring appropriate control of inflammation [43], and may be a mechanism for determining target specificity of the Vacuolar and Cytosolic Responses. This may explain why activation of both NFκB and IRF3 during the Cytosolic Response did not induce IL1β, even though IFNβ was induced. Infection of myd88 −/− macrophages with WT bacteria and the cytosolic delivery of purified ligands both bypassed TLR signaling, and therefore may not have triggered this nucleosome remodeling.
The OspF virulence factor injected by the bacterial pathogen Shigella flexneri modifies host chromatin during infection to block the activation of certain NFκB target genes [52], but we have no evidence that L. monocytogenes possesses analogous effector molecules. Instead, by escaping the vacuole, L. monocytogenes may avoid induction of inflammation by taking advantage of the inherent Cytosolic Response target specificity. Previous studies in which MDP was observed to induce pro-inflammatory cytokines used non-primary, non-immune system cells, and often assessed host response as late as 24 hours after MDP delivery, all of which could have resulted in significantly altered chromatin states [8,9,53–55].
The Role of NOD2 in Bacterial Pathogenesis
The role of NOD2 in innate immunity is controversial [56,57]. A previous study found that NOD2 was involved in the mouse innate immune response to L. monocytogenes, but only during intragastric infection [32]. This in vivo infection model may be particularly important for understanding the pathogenesis of food-borne bacteria such as L. monocytogenes. In certain cells of the gut epithelium, through which L. monocytogenes must pass in an oral model of infection, expression of many TLRs is naturally downregulated, possibly to reduce inappropriate responses to commensal flora [58]. This is consistent with our finding that NOD2-mediated synergistic induction of IFNβ in response to intracellular bacterial infection was only manifest when TLR signaling was pharmacologically suppressed, and may also explain the results of a previous study in which rip2 −/− macrophages with intact TLR signaling had no defect in their response to L. monocytogenes [37]. Curiously, while irf3 −/− and ifnar −/− mice infected intravenously with L. monocytogenes were more resistant to infection [59–61], nod2 −/− mice infected intragastrically were more susceptible [32]. Given our results showing that infected nod2 −/− macrophages induced less IFNβ than infected WT macrophages, the role of Type I interferons in intracellular bacterial pathogenesis may differ depending on the in vivo model used.
Materials and Methods
For more detailed versions of many of the following methods, please see: http://microbiology.berkeley.edu/ppmicroarrayhybridprotoc.htm.
Cell culture and bacteria.
Macrophages were derived from the bone marrow of mice over 7 days in media composed of DMEM, 2 mM glutamine, 1 mM pyruvate, 10% CSF from 3T3 cells, 20% heat inactivated FBS, and penicillin-streptomycin. For all experiments, macrophages were grown in identical media without penicillin-streptomycin. For infections, WT 10403S (DP-L184) and hly− (DP-L2161) L. monocytogenes were grown to mid-log in BHI media at 30 °C with shaking. Bacteria were then PBS-washed, resuspended in PBS at a normalized OD600 = 1.2, and added to macrophages at a 1:50 volume:volume ratio, resulting in > 75% of macrophages infected with at least one bacterium. For experiments with “low MOI” infection, and experiments involving Pam3CSK4-pre-treated macrophages, resuspended bacteria were added at a 1:1000 ratio. At 30 minutes post-infection macrophages were washed 3 times with fresh pre-warmed media, and at 60 minutes post-infection gentamicin was added to a final concentration of 50 μg/ml. For experiments containing a 30 minute post-infection time point, macrophages were instead washed at 20 minutes and gentamicin added at 30 minutes. CAPE (Calbiochem) (or EtOH for mock-treated controls) was added 60 minutes post-infection. For tolerization of cells, Pam3CSK4 (Invivogen) was added at a final concentration of 100 ng/ml, 24 hours prior to the subsequent experiment. Where indicated, cycloheximide (or water for mock-treated controls) was added at a final concentration of 10 μg/ml 30 minutes before infection with bacteria, and added back after the washes at t = 30 minutes. This treatment reduces macrophage translation by 94.5% (determined by 35S-Met incorporation, data not shown).
For infections with M. tuberculosis, macrophages were infected with the Erdman strain as previously described [45]. Briefly, M. tuberculosis cultures were washed with PBS and resuspended in DMEM supplemented with 10% horse serum. Pam3CSK4-pre-treated macrophages were incubated with bacteria in DMEM + 10% horse serum for 2 hours at an MOI of 10. Cells were then washed three times with PBS and returned to macrophage media.
Mice.
All macrophages were from mice in the C57BL/6 genetic background, including femurs from knockout mice (see Acknowledgments).
Macrophage RNA preparation.
Macrophage RNA was isolated with the Ambion RNAqueous kit (Applied Biosystems) according to the manufacturer's protocol, after first treating the cells with Ambion RNAlater (Applied Biosystems). For microarray experiments, RNA was amplified to generate amplified RNA (aRNA) using the Ambion Amino-Allyl Message Amp II aRNA Amplification Kit (Applied Biosystems) according to the manufacturer's protocol. For qPCR experiments, RNA was DNase treated with the Ambion TURBO DNA-free kit (Applied Biosystems).
Microarrays.
Microarrays were printed at the UCSF Center for Advanced Technology, using the MEEBO 70-mer oligonucleotide set (Illumina; for more details see http://alizadehlab.stanford.edu/). Microarray probes were generated by coupling aRNA to Cy dye mono-functional NHS esters (Amersham) using 100 mM sodium bicarbonate (pH 9.0) and 50% DMSO. 5 μg of Cy5-coupled sample and 5 μg Cy3-coupled reference (generated by pooling an equal volume of every sample used in a given set of arrays) were hybridized to MEEBO microarrays at 63 °C for 2 days. After washing, arrays were scanned on a GenePix 4000B scanner (Molecular Devices). Arrays were gridded with SpotReader software (Niles Scientific). Acquisition of data was performed using the GenePix Pro 6 software package (Molecular Devices). Data was normalized by first using stringent criteria to identify a subset of features of highest quality, and then calculating a normalization factor such that the ratio of medians of the Cy5 and Cy3 values for these features = 1. This ratio was then applied to all the features. Features not meeting minimum criteria to assure quality were removed from the datasets. These criteria are available upon request. Hierarchical Pearson clustering and other analyses were performed with the Acuity 4 software package (Molecular Devices). The determination of Vacuolar Response and Cytosolic Response target genes was each from multiple arrays representing four independent experiments (e.g. 4 independent dishes of uninfected myd88 −/− macrophages and 4 independent dishes of myd88 −/− macrophages infected with WT L. monocytogenes for 180 minutes were used for Cytosolic Response determination), and other determinations were from 2–4 independent experiments. Fold change in RNA abundance is relative to uninfected samples. SAM analysis (Stanford University) [34] was performed with two-class unpaired designs to identify genes that were differently expressed in infected versus uninfected macrophages. For initial target gene discovery (defining the Vacuolar Response and Cytosolic Response), the false discovery rate (FDR) was set to 1%. For subsequent SAM analyses the FDR was set to 10%. Information linked to each unique Oligo ID can be found at http://meebo.ucsf.edu:8080/meebo/meeboInfo.jsp?oligoid=(insert Oligo ID here). The GEO accession number for the primary array data is GSE8104. Accession numbers for genes of the Primary Cytosolic Response are as follows: NM_010510 (IFNβ), NM_008331 (IFIT1), NM_008332 (IFIT2), NM_010501 (IFIT3), NM_020557 (TYKI), NM_030015 (PELI1), NM_008654 (MYD116), NM_031168 (IL6), BC030067 (CXCL10), XM_130015 (OTUD1), NM_027835 (MDA5), NM_018738 (IGTP), and NM_145209 (OASL1). See Dataset S3 for more information on the Primary Cytosolic Response target genes.
qPCR.
DNase-treated macrophage RNA was reverse transcribed and subject to quantitative PCR using DyNAmo SYBR Green 2-step qRT-PCR reagents (NEB/Finnzyme) and was performed on an Mx3000P machine and analyzed using MxPro software (Stratagene). The sequences of gene-specific primers are as follows: tggcattgttaccaactgggacg (5' β-actin), gcttctctttgatgtcacgcacg (3' β-actin), gcactgggtggaatgagactattg (5' IFNβ), ttctgaggcatcaactgacaggtc (3' IFNβ), gacctgttctttgaagttgacgg (5' IL1β), and tgtcgttgcttggttctccttg (3' IL1β). All RNA abundances are normalized to β-actin, and fold induction is relative to mock-infected samples. Data shown is the mean of at least three, and in most cases four, independent experiments (except for the data shown in Figure 5C, which represents the average of two independent experiments), and error bars represent the standard error of the mean. Statistical analyses (t-Tests with equal variances assumed) were performed using the Analysis Package Add-In feature of Microsoft Excel, and p-values are noted in figure legends.
Cell fractionation and Western blots.
Macrophages were fractionated using NE-PER reagents (Pierce) supplemented with HALT protease inhibitors (Pierce) following the manufacturer's protocol. Protein concentration was determined using BCA reagents (Pierce) and equal masses of protein were run on 10% NuPAGE bis-tris gels (Invitrogen). Blots were probed with the following primary antibodies, all from Santa Cruz Biotech: anti-NFκB p65 (sc-372X), anti-IRF3 (sc-9082X), anti phospho-c-Jun (sc-16312X), anti-phospho-ATF2 (sc-8398X), and anti-Lamin B (sc-6217). Protein blots were probed with a secondary antibody covalently attached to an infrared emitting fluorphor (IRdye-680 and IRdye-800, Li-Cor Biosciences). Blots were scanned using the Odyssey Infrared Imaging System (Li-Cor Biosciences), and quantitated using the accompanying software package. Relative protein abundances were normalized to Lamin B. Data was collected from two blots, with each blot using pooled lysates of two independent dishes (four dishes total, except for the data shown in Figure 2A, which is from two independent dishes). The non-quantitative Western blot shown in Figure 2A was instead probed with an HRP-coupled secondary antibody, and developed with the ECL Plus Western Blot Detection System (GE Healthcare).
Transfections.
Macrophages were transfected using FuGene 6 Transfection Reagent (Roche) according to the manufacturer's protocol. L. monocytogenes genomic DNA was prepared by manual disruption of mid-log WT 10403S bacteria using glass beads and phenol:chloroform. DNA was extracted 3 times with phenol:chloroform, treated with RNases A and H, and extracted again with phenol:chloroform. DNA was then digested to completion using EcoRI and BamHI, yielding fragments averaging 1–5 kb, extracted with phenol:chloroform, and precipitated. All phenol:chloroform extractions were done using PhaseLock gel (Eppendorf). DNA was dissolved in pyrogen-free water and used at a final concentration of 250 ng/ml. MDP (Calbiochem) was used at final concentration of 100 μg/ml. Poly(dAT-dTA) double-stranded nucleic acid (Amersham) was dissolved in 100 mM NaCl and used at a final concentration of 250 ng/ml. All transfection experiments were done for 6 hours. CAPE (Calbiochem) (or EtOH for mock-treated controls) was added at the indicated concentration 180 minutes post-infection. Cycloheximide (or water for mock-treated controls) was added at a final concentration of 10 μg/ml 180 minutes after initial ligand transfection.
Supporting Information
(34 KB XLS)
(18 KB XLS)
(20 KB XLS)
(48 KB XLS)
(124 KB XLS)
Scatter plot representation of microarray data from myd88 −/− macrophages infected with hly− L. monocytogenes (see Dataset S5). Shown are the responses of Vacuolar Response-specific target genes. Y-axis values are log2 fold change in RNA abundance in WT macrophages infected with hly− L. monocytogenes. X-axis values are log2 fold change in RNA abundance in myd88 −/− macrophages infected with hly− L. monocytogenes. Spots in blue are those identified by SAM as being significantly differently induced in the two conditions (all spots).
(40 KB PDF)
(A) Analysis by qPCR of IFNβ transcriptional induction in myd88 −/− trif −/− macrophages transfected with the indicated combinations of L. monocytogenes genomic DNA and synthetic MDP.
(B) Analysis by qPCR of IFNβ transcriptional induction in myd88 −/− trif −/− macrophages transfected with the indicated combinations of synthetic poly(dAT-dTA) double-stranded DNA and synthetic MDP.
(C) Scatter plot representation of the transcriptional response of WT macrophages transfected with either L. monocytogenes genomic DNA alone, or with both L. monocytogenes DNA and MDP, as determined by microarrays. Shown are the responses of Cytosolic Response-specific target genes. Y-axis values are log2 fold change in RNA abundance in WT macrophages transfected with L. monocytogenes DNA, and X-axis values are log2 fold change in RNA abundance in WT macrophages transfected with both L. monocytogenes DNA and synthetic MDP. Spots in blue are those identified by SAM as being significantly differently induced in the two conditions, while spots in pink are not different. The superimposed dashed line has a slope = 1. The superimposed solid black line represents the best fit of the observed transcriptional responses, and has a slope = 0.71 (see Dataset S4 for complete array data).
(56 KB PDF)
Scatter plot representation of the transcriptional response of WT macrophages infected for 3 hours with WT L. monocytogenes, and where indicated additionally treated with 10 μg/ml CAPE. Shown are the responses of Cytosolic Response-specific target genes. Transcription responses were determined by microarrays. Y-axis values are log2 fold change in RNA abundance in WT macrophages infected with WT L. monocytogenes. X-axis values are log2 fold change in RNA abundance in WT macrophages infected with WT L. monocytogenes and additionally treated with CAPE. The superimposed dashed line has a slope = 1. Spots in blue are those identified by SAM as being significantly differently induced in the two conditions, while spots in pink are not different.
(12 KB PDF)
We previously observed that WT macrophages infected with WT L. monocytogenes exhibited persistent TLR-mediated signaling, even at later time points. From our microarray analyses, we found that L. monocytogenes induced IL1β exclusively by MyD88-dependent signaling pathways (summarized in Figure 1B). However, IL1β was still strongly induced at later time points in WT macrophages infected with WT L. monocytogenes, when the bacteria were in the cytosol. This suggested that these cells continued TLR signaling from the vacuole even after the bacteria had escaped from this compartment. As TLR signaling strongly activated NFκB (Figure 2A), we considered it likely that this residual TLR signaling masked any contribution by NOD2 signaling to nuclear NFκB. Indeed, we found that both WT and nod2 −/− macrophages infected with WT L. monocytogenes induced IFNβ to nearly identical levels (Figure S4A). To remove residual TLR signaling in these cells we treated macrophages with Pam3CSK4, a synthetic TLR2 agonist. Pretreatment for 24 hours with Pam3CSK4 had previously been shown to “tolerize” macrophages, causing their TLR signaling to dampen after transitory stimulation [43]. Compared to non-tolerized macrophages, Pam3CSK4-treated WT macrophages almost fully ceased the induction of IL1β 2 hours after infection with WT L. monocytogenes (Figure S4B). These tolerized macrophages were still capable of appropriate TLR-mediated signaling, as we observed robust and sustained induction of IL1β during infection of Pam3CSK4-treated WT macrophages with hly− L. monocytogenes, which remain in the vacuole (Figure S4C).
(A) Analysis by qPCR of IFNβ transcriptional induction in non-tolerized WT and nod2 −/− macrophages infected with WT L. monocytogenes.
(B) Analysis by qPCR of IL1β transcriptional induction in WT macrophages infected with WT L. monocytogenes, and where indicated additionally tolerized 24 hours pre-infection with 100 ng/ml Pam3CSK4.
(C) Analysis by qPCR of IL1β transcriptional induction in WT macrophages tolerized 24 hours pre-infection with 100 ng/ml Pam3CSK4, and infected with either WT L. monocytogenes or hly− L. monocytogenes.
(43 KB PDF)
Acknowledgments
We thank the following for generous contributions of mouse femurs: A. Shahangian, B. Guo, and G. Cheng (UCLA) for irf3 −/− and tbk1 −/− tnfr −/− femurs; S. Mariathasan and V. Dixit, (Genentech), for nod2 −/− femurs; D. Stetson and R. Medzhitov (Yale University) for rip2 −/− femurs; and G. Barton (UC Berkeley) for myd88 −/− trif −/− femurs. We thank A. Carroll, P. Nittler, and K. Hermens (UCSF Center for Advanced Technology) for help with printing MEEBO arrays. We thank J. D. Sauer (UC Berkeley) for helpful discussions and critical review of the manuscript, and R. Nir-Paz (UC Berkeley) for help with statistical analyses.
Footnotes
Author contributions. JHL, GTC, SR, NPMM, JSC, and DAP conceived and designed the experiments. JHL, GTC, SR, and NPMM performed the experiments. JHL, GTC, SR, NPMM, JSC, and DAP analyzed the data. JHL and DAP wrote the paper.
Funding. This research was supported by NIH grants AI063302 (DAP and JSC) and AI51667 (JSC). JHL is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG 1801–04). JSC gratefully acknowledges the support of the Sandler Family Supporting Foundation and the W.M. Keck Foundation. DAP is a Senior Scholar of the Ellison Medical Foundation.
Competing interests. DAP has a consulting relationship with and a financial interest in Cerus Corporation, which may benefit from commercialization of the results of this research.
References
- Aderem A. Phagocytosis and the inflammatory response. J Infect Dis. 2003;187(Suppl 2):S340–S345. doi: 10.1086/374747. [DOI] [PubMed] [Google Scholar]
- Stuart LM, Ezekowitz RA. Phagocytosis: elegant complexity. Immunity. 2005;22:539–550. doi: 10.1016/j.immuni.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Herskovits AA, Auerbuch V, Portnoy DA. Bacterial ligands generated in a phagosome are targets of the cytosolic innate immune system. PLoS Pathog. 2007;3:e51. doi: 10.1371/journal.ppat.0030051. doi: 10.1371/journal.ppat.0030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Janeway C., Jr. Innate immune recognition: mechanisms and pathways. Immunol Rev. 2000;173:89–97. doi: 10.1034/j.1600-065x.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, et al. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- Chamaillard M, Girardin SE, Viala J, Philpott DJ. Nods, Nalps and Naip: intracellular regulators of bacterial-induced inflammation. Cell Microbiol. 2003;5:581–592. doi: 10.1046/j.1462-5822.2003.00304.x. [DOI] [PubMed] [Google Scholar]
- Inohara Chamaillard, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Hugot JP, Sansonetti PJ. Lessons from Nod2 studies: towards a link between Crohn's disease and bacterial sensing. Trends Immunol. 2003;24:652–658. doi: 10.1016/j.it.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2005. [DOI] [PubMed]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Falvo JV, Kim TH, Kim TK, Lin CH, et al. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, et al. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- Silverman N, Maniatis T. NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001;15:2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- Lorber B. Listeriosis. Clin Infect Dis. 1997;24:1–9. 10–11. doi: 10.1093/clinids/24.1.1. quiz. [DOI] [PubMed] [Google Scholar]
- Portnoy DA, Jacks PS, Hinrichs DJ. Role of hemolysin for the intracellular growth of Listeria monocytogenes . J Exp Med. 1988;167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes . J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Riordan M, Yi CH, Gonzales R, Lee KD, Portnoy DA. Innate recognition of bacteria by a macrophage cytosolic surveillance pathway. Proc Natl Acad Sci U S A. 2002;99:13861–13866. doi: 10.1073/pnas.202476699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey RL, Fawcett P, O'Riordan M, Lee KD, Havell EA, et al. A specific gene expression program triggered by Gram-positive bacteria in the cytosol. Proc Natl Acad Sci U S A. 2004;101:11386–11391. doi: 10.1073/pnas.0403215101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger S, Reutterer B, Schaljo B, Schellack C, Brunner S, et al. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J Immunol. 2004;173:7416–7425. doi: 10.4049/jimmunol.173.12.7416. [DOI] [PubMed] [Google Scholar]
- Soulat D, Bauch A, Stockinger S, Superti-Furga G, Decker T. Cytoplasmic Listeria monocytogenes stimulates IFN-beta synthesis without requiring the adapter protein MAVS. FEBS Lett. 2006;580:2341–2346. doi: 10.1016/j.febslet.2006.03.057. [DOI] [PubMed] [Google Scholar]
- Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- Flak TA, Heiss LN, Engle JT, Goldman WE. Synergistic epithelial responses to endotoxin and a naturally occurring muramyl peptide. Infect Immun. 2000;68:1235–1242. doi: 10.1128/iai.68.3.1235-1242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci U S A. 2007;104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Yang K, Hashimoto M, Park JH, Kim YG, et al. Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. J Biol Chem. 2006;281:29054–29063. doi: 10.1074/jbc.M602638200. [DOI] [PubMed] [Google Scholar]
- Madigan MT, Martinko JM. Brock biology of microorganisms. Pearson Prentice Hall; 2006. [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- Verdugo RA, Medrano JF. Comparison of gene coverage of mouse oligonucleotide microarray platforms. BMC Genomics. 2006;7:58. doi: 10.1186/1471-2164-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner JE, Starr R, Cornish AL, Zhang JG, Metcalf D, et al. Suppressor of cytokine signaling 1 regulates the immune response to infection by a unique inhibition of type I interferon activity. Nat Immunol. 2006;7:33–39. doi: 10.1038/ni1287. [DOI] [PubMed] [Google Scholar]
- Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Vaidya SA, Perry AK, Saha SK, Dempsey PW, et al. Immune activation of type I IFNs by Listeria monocytogenes occurs independently of TLR4, TLR2, and receptor interacting protein 2 but involves TNFR-associated NF kappa B kinase-binding kinase 1. J Immunol. 2005;174:1602–1607. doi: 10.4049/jimmunol.174.3.1602. [DOI] [PubMed] [Google Scholar]
- Kim LK, Choi UY, Cho HS, Lee JS, Lee WB, et al. Down-regulation of NF-kappaB target genes by the AP-1 and STAT complex during the innate immune response in Drosophila. PLoS Biol. 2007;5:e238. doi: 10.1371/journal.pbio.0050238. doi: 10.1371/journal.pbio.0050238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry AK, Chow EK, Goodnough JB, Yeh WC, Cheng G. Differential requirement for TANK-binding kinase-1 in type I interferon responses to toll-like receptor activation and viral infection. J Exp Med. 2004;199:1651–1658. doi: 10.1084/jem.20040528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott DW, Wilkins A, Asara JM, Cantley LC. The Crohn's disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr Biol. 2004;14:2217–2227. doi: 10.1016/j.cub.2004.12.032. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, et al. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- Losick VP, Isberg RR. NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila . J Exp Med. 2006;203:2177–2189. doi: 10.1084/jem.20060766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- Stanley SA, Raghavan S, Hwang WW, Cox JS. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci U S A. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Johndrow JE, Manzanillo P, Cox JS. The type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauvliege R, Janssens S, Beyaert R. Pellino proteins are more than scaffold proteins in TLR/IL-1R signalling: a role as novel RING E3-ubiquitin-ligases. FEBS Lett. 2006;580:4697–4702. doi: 10.1016/j.febslet.2006.07.046. [DOI] [PubMed] [Google Scholar]
- Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Sanjabi S, Williams KJ, Saccani S, Zhou L, Hoffmann A, et al. A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes Dev. 2005;19:2138–2151. doi: 10.1101/gad.1329805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann AS, Mitchell DM, Sanjabi S, Bradley MN, Hoffmann A, et al. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat Immunol. 2001;2:51–57. doi: 10.1038/83168. [DOI] [PubMed] [Google Scholar]
- Sanjabi S, Hoffmann A, Liou HC, Baltimore D, Smale ST. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc Natl Acad Sci U S A. 2000;97:12705–12710. doi: 10.1073/pnas.230436397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbibe L, Kim DW, Batsche E, Pedron T, Mateescu B, et al. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat Immunol. 2006;8:47–56. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- Netea MG, Ferwerda G, de Jong DJ, Girardin SE, Kullberg BJ, et al. NOD2 3020insC mutation and the pathogenesis of Crohn's disease: impaired IL-1beta production points to a loss-of-function phenotype. Neth J Med. 2005;63:305–308. [PubMed] [Google Scholar]
- Netea MG, Ferwerda G, de Jong DJ, Werts C, Boneca IG, et al. The frameshift mutation in Nod2 results in unresponsiveness not only to Nod2- but also Nod1-activating peptidoglycan agonists. J Biol Chem. 2005;280:35859–35867. doi: 10.1074/jbc.M504924200. [DOI] [PubMed] [Google Scholar]
- Maeda S, Hsu LC, Liu H, Bankston LA, Iimura M, et al. Nod2 mutation in Crohn's disease potentiates NF-kappaB activity and IL-1beta processing. Science. 2005;307:734–738. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- Strober W, Murray PJ, Kitani A, Watanabe T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat Rev Immunol. 2006;6:9–20. doi: 10.1038/nri1747. [DOI] [PubMed] [Google Scholar]
- Furrie E, Macfarlane S, Thomson G, Macfarlane GT. Toll-like receptors-2, −3 and −4 expression patterns on human colon and their regulation by mucosal-associated bacteria. Immunology. 2005;115:565–574. doi: 10.1111/j.1365-2567.2005.02200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbuch V, Brockstedt DG, Meyer-Morse N, O'Riordan M, Portnoy DA. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes . J Exp Med. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero JA, Calderon B, Unanue ER. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J Exp Med. 2004;200:535–540. doi: 10.1084/jem.20040769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(34 KB XLS)
(18 KB XLS)
(20 KB XLS)
(48 KB XLS)
(124 KB XLS)
Scatter plot representation of microarray data from myd88 −/− macrophages infected with hly− L. monocytogenes (see Dataset S5). Shown are the responses of Vacuolar Response-specific target genes. Y-axis values are log2 fold change in RNA abundance in WT macrophages infected with hly− L. monocytogenes. X-axis values are log2 fold change in RNA abundance in myd88 −/− macrophages infected with hly− L. monocytogenes. Spots in blue are those identified by SAM as being significantly differently induced in the two conditions (all spots).
(40 KB PDF)
(A) Analysis by qPCR of IFNβ transcriptional induction in myd88 −/− trif −/− macrophages transfected with the indicated combinations of L. monocytogenes genomic DNA and synthetic MDP.
(B) Analysis by qPCR of IFNβ transcriptional induction in myd88 −/− trif −/− macrophages transfected with the indicated combinations of synthetic poly(dAT-dTA) double-stranded DNA and synthetic MDP.
(C) Scatter plot representation of the transcriptional response of WT macrophages transfected with either L. monocytogenes genomic DNA alone, or with both L. monocytogenes DNA and MDP, as determined by microarrays. Shown are the responses of Cytosolic Response-specific target genes. Y-axis values are log2 fold change in RNA abundance in WT macrophages transfected with L. monocytogenes DNA, and X-axis values are log2 fold change in RNA abundance in WT macrophages transfected with both L. monocytogenes DNA and synthetic MDP. Spots in blue are those identified by SAM as being significantly differently induced in the two conditions, while spots in pink are not different. The superimposed dashed line has a slope = 1. The superimposed solid black line represents the best fit of the observed transcriptional responses, and has a slope = 0.71 (see Dataset S4 for complete array data).
(56 KB PDF)
Scatter plot representation of the transcriptional response of WT macrophages infected for 3 hours with WT L. monocytogenes, and where indicated additionally treated with 10 μg/ml CAPE. Shown are the responses of Cytosolic Response-specific target genes. Transcription responses were determined by microarrays. Y-axis values are log2 fold change in RNA abundance in WT macrophages infected with WT L. monocytogenes. X-axis values are log2 fold change in RNA abundance in WT macrophages infected with WT L. monocytogenes and additionally treated with CAPE. The superimposed dashed line has a slope = 1. Spots in blue are those identified by SAM as being significantly differently induced in the two conditions, while spots in pink are not different.
(12 KB PDF)
We previously observed that WT macrophages infected with WT L. monocytogenes exhibited persistent TLR-mediated signaling, even at later time points. From our microarray analyses, we found that L. monocytogenes induced IL1β exclusively by MyD88-dependent signaling pathways (summarized in Figure 1B). However, IL1β was still strongly induced at later time points in WT macrophages infected with WT L. monocytogenes, when the bacteria were in the cytosol. This suggested that these cells continued TLR signaling from the vacuole even after the bacteria had escaped from this compartment. As TLR signaling strongly activated NFκB (Figure 2A), we considered it likely that this residual TLR signaling masked any contribution by NOD2 signaling to nuclear NFκB. Indeed, we found that both WT and nod2 −/− macrophages infected with WT L. monocytogenes induced IFNβ to nearly identical levels (Figure S4A). To remove residual TLR signaling in these cells we treated macrophages with Pam3CSK4, a synthetic TLR2 agonist. Pretreatment for 24 hours with Pam3CSK4 had previously been shown to “tolerize” macrophages, causing their TLR signaling to dampen after transitory stimulation [43]. Compared to non-tolerized macrophages, Pam3CSK4-treated WT macrophages almost fully ceased the induction of IL1β 2 hours after infection with WT L. monocytogenes (Figure S4B). These tolerized macrophages were still capable of appropriate TLR-mediated signaling, as we observed robust and sustained induction of IL1β during infection of Pam3CSK4-treated WT macrophages with hly− L. monocytogenes, which remain in the vacuole (Figure S4C).
(A) Analysis by qPCR of IFNβ transcriptional induction in non-tolerized WT and nod2 −/− macrophages infected with WT L. monocytogenes.
(B) Analysis by qPCR of IL1β transcriptional induction in WT macrophages infected with WT L. monocytogenes, and where indicated additionally tolerized 24 hours pre-infection with 100 ng/ml Pam3CSK4.
(C) Analysis by qPCR of IL1β transcriptional induction in WT macrophages tolerized 24 hours pre-infection with 100 ng/ml Pam3CSK4, and infected with either WT L. monocytogenes or hly− L. monocytogenes.
(43 KB PDF)