Abstract
Green fluorescent protein has revolutionized cell labeling and molecular tagging, yet the driving force and mechanism for its spontaneous fluorophore synthesis are not established. Here we discover mutations that substantially slow the rate but not the yield of this posttranslational modification, determine structures of the trapped precyclization intermediate and oxidized postcyclization states, and identify unanticipated features critical to chromophore maturation. The protein architecture contains a dramatic ≈80° bend in the central helix, which focuses distortions at G67 to promote ring formation from amino acids S65, Y66, and G67. Significantly, these distortions eliminate potential helical hydrogen bonds that would otherwise have to be broken at an energetic cost during peptide cyclization and force the G67 nitrogen and S65 carbonyl oxygen atoms within van der Waals contact in preparation for covalent bond formation. Further, we determine that under aerobic, but not anaerobic, conditions the Gly-Gly-Gly chromophore sequence cyclizes and incorporates an oxygen atom. These results lead directly to a conjugation-trapping mechanism, in which a thermodynamically unfavorable cyclization reaction is coupled to an electronic conjugation trapping step, to drive chromophore maturation. Moreover, we propose primarily electrostatic roles for the R96 and E222 side chains in chromophore formation and suggest that the T62 carbonyl oxygen is the base that initiates the dehydration reaction. Our molecular mechanism provides the basis for understanding and eventually controlling chromophore creation.
The Aequorea victoria green fluorescent protein (GFP) undergoes a remarkable posttranslational modification to create a chromophore out of its amino acids (S65, Y66, and G67) (1–3). GFP is small (238 aa), tolerates both N- and C-terminal fusions, and can be targeted to specific cellular locations (4). Synthesis of the GFP fluorophore occurs spontaneously after protein folding without cofactors or accessory proteins (5), making GFP-protein fusions tractable in a variety of organisms. GFP mutants and homologs exhibit fluorescent emission maxima ranging from blue to red (3, 6–8), which allow concurrent surveillance of multiple targets. Together, these properties have fundamentally altered in vivo molecular tagging and cell labeling. In addition, GFP-based indicators monitor cellular redox potential (9), pH (10, 11), metal ion concentrations (12, 13), and halide levels (14, 15). Because of these applications and the novel fluorophore, there have been extensive structural, spectroscopic, and biochemical characterizations of the protein and its mutants, all in the mature chromophore state (4, 16). The crystallographic structure of GFP reveals that the overall fold is an 11-stranded antiparallel β-barrel protein with the chromophore located near the geometric center of the barrel on a distorted α-helix (1, 17). Few molecular details are known about chromophore maturation.
The proposed fluorophore formation mechanism entails three steps: peptide cyclization initiated by nucleophilic attack of the G67 amide nitrogen atom on the S65 carbonyl carbon to create a five-membered imidazolone ring, dehydration of the S65 carbonyl oxygen, and rate-limiting oxidation of the Y66 Cα—Cβ bond to conjugate the ring systems (2, 3, 18). The enzyme histidine ammonia lyase (HAL) undergoes a related posttranslational modification to generate an electrophile from the tripeptide loop sequence Ala-Ser-Gly (19). There are currently two proposals for the driving force of peptide cyclization in each system: the mechanical compression hypothesis, which suggests that cyclization relaxes an energetically unfavorable precyclized state (20, 21), and the alternative model where either tyrosine oxidation (GFP) (22) or serine dehydration (HAL) (23) precedes cyclization. In GFP, conserved residues R96 and E222 have been proposed to have key roles in chromophore synthesis but have not been experimentally evaluated. This lack of experimental data is likely caused by the difficulties of investigating posttranslational modification reactions that occur during or rapidly after protein folding.
Here we report mutations that substantially slow chromophore formation, determine GFP structures in trapped pre- and oxidized postcyclization states, identify unanticipated features critical for this posttranslational modification, and propose additional functional roles for residues R96 and E222 and the carbonyl oxygen of T62. These discoveries lead directly to a conjugation-trapping mechanism for GFP fluorophore synthesis.
Materials and Methods
Experimental Preparation. To create the R96A construct, we used the QuikChange method (Stratagene) to introduce the R96A/230stop and solubility optimizing (F99S, M153T, V163A) (24) mutations into the GFPmut1 (F64L, S65T) pKEN2 vector (25). Similarly, we created the Gly-Gly-Gly construct, by placing the S65G/Y66G mutations into GFPsol (GFPmut1 + solubility mutations), which had been subcloned into a pET11 vector (Novagen). The resulting plasmids were transformed into either BL21-CodonPlus(DE3)-RIL (for pET11a) or JM109 (for pKEN2) Escherichia coli cells (Stratagene), which were grown at 25°C in 9-liter batches. At an optical density of 0.5 at 600 nm, protein expression was induced with 0.2 mM isopropyl-β-d-thiogalactoside. The bacteria cells were pelleted 6–12 h later and frozen in liquid nitrogen until purification. Proteins were purified aerobically by modifying a published protocol (26) to incorporate HQ (26 mm × 30 cm) (PerSeptive Biosystems, Framingham, MA) and S-100 (26 mm × 60 cm) (Pharmacia) columns (27). To prepare the Gly-Gly-Gly anaerobic sample, the protein was purified and crystallized in an anaerobic glove box (Vacuum Atmospheres, Hawthorne, CA). Yields were 200–1,000 mg of >95% pure protein in 3–5 days.
Crystallization, Diffraction Data Collection, and Refinement. GFP variants were crystallized at 10–15 mg/ml in hanging drops, by modifying a published protocol (1, 27). Initial crystal clusters were crushed, serially diluted in a stabilizing solution (50 mM Hepes, pH 8.0/50 mM MgCl2/19% polyethylene glycol 4000) and used as microseeds to grow large single crystals. Diffraction data were collected from crystals that were cryocooled immediately after immersion in the stabilizing solution plus 20% ethylene glycol. The R96A mature data set was collected at the Advanced Photon Source (Argonne, IL) (APS14-BM-C) at a wavelength of 1.00 Å. The R96A precyclization structure A and S65G Y66G (aerobic) data sets were collected at the Stanford Synchrotron Radiation Laboratory (Stanford, CA) at beam lines 9–1(λ = 0.78 Å) and 11–1(λ = 0.965 Å), respectively. The R96A precyclization structure B and anaerobic Gly-Gly-Gly data sets were collected on a Siemens (Iselin, NJ) SRA direct drive rotating anode x-ray generator with a graphite monochromator and a MAR-Research (Hamburg, Germany) 34.5-cm image plate area detector. Data sets were indexed and reduced in the P212121 space group with the hkl package (28), and phases were determined by molecular replacement with amore (29). The search model was a refined 1.0-Å GFPsol structure, determined with molecular replacement from a previous GFP structure (1). The search model was modified for uncyclized variants by modeling the chromophore as its substituent amino acids; position 96 was truncated to alanine for the R96A variant structures. Difference electron density and omit maps were manually fit with the xtalview package (30) and refined in either cns (31) or shelx-97 (32) with all diffraction data, except for 5% used for Rfree calculations (33). Standard uncertainties were determined by inverting the full least-squares covariance matrix in shelx-97 (32).
All structures were superimposed with sequoia (34). Images for the cyclized product and reduced intermediate in the cartoon were made by placing a tyrosine side chain onto the nondehydrated Gly-Gly-Gly cyclized ring and reducing the Y66 Cα—Cβ bond of the mature chromophore, respectively.
Results
Structures of the R96A GFPsol Variant Before and After Peptide Cyclization. Because R96 has been proposed to either activate the S65 carbonyl for nucleophilic attack (20) or directly deprotonate the G67 backbone amide (22), we constructed an R96A variant and discovered that this point mutation slows the cyclization reaction from minutes to months. After purification of the initially colorless R96A protein, chromophore maturation was achieved by incubation for 3 months at 37°C. Our 1.50-Å resolution crystallographic structure of this matured R96A protein (Table 1) is highly similar to that of its solubility-optimized GFPsol parent, with an overall Cα rms deviation of 0.20 Å. In GFP, R96 forms a hydrogen bond with the imidazolone oxygen of the mature chromophore. In the R96A mutant, three water molecules fill the volume normally occupied by the R96 side chain but fail to form hydrogen bonds with the imidazolone oxygen. This may explain the shifts in fluorescence maxima for the R96A variant (468-nm excitation, 503-nm emission) compared with GFPsol (489-nm excitation, 508-nm emission), suggesting that R96 lowers the excited state energy of the chromophore, consistent with unpublished results on the R96C variant (4). Importantly, the chromophore of the R96A variant is fully matured (Fig. 1a), demonstrating that this mutant retains all components essential for chromophore formation.
Table 1. Data collection and refinement statistics.
| R96A Mature | R96A Pre. A | R96A Pre. B | S65G Y66G Aerobic | S65G Y66G Anaerobic | |
|---|---|---|---|---|---|
| Resolution, Å | 40.0-1.50 | 20.0-2.00 | 20.0-2.00 | 20.0-1.80 | 20.0-1.80 |
| Last shell, Å | 1.55-1.50 | 2.07-2.00 | 2.07-2.00 | 1.86-1.80 | 1.86-1.80 |
| Observations | 164,842 | 54,856 | 71,722 | 59,616 | 70,279 |
| Unique observations | 34,756 | 15,550 | 16,081 | 20,401 | 21,170 |
| Rsym, %*† | 5.2 (25.3) | 6.6 (37.2) | 9.9 (29.1) | 6.3 (28.9) | 7.2 (36.6) |
| Completeness, % | 92.4 (60.7) | 99.0 (99.8) | 99.7 (98.2) | 97.7 (99.2) | 96.5 (85.8) |
| I/σI | 27.4 (3.0) | 18.2 (3.3) | 15.0 (4.0) | 18.5 (3.4) | 20.2 (3.4) |
| Refinement parameters | 18,771 | 7,732 | 8,016 | 7,780 | 8,624 |
| Rwork/Rfree, %‡ | 14.5/21.4 | 21.8/25.8 | 20.3/24.1 | 21.0/25.1 | 20.0/22.7 |
Values in parentheses are the statistics for the highest resolution shell of data.
Rsym = ΣIhkl — 〈I〉 |/|Σ 〈I〉, where 〈I〉 is the average individual measurement of Ihkl.
Rwork = (Σ |Fobs — Fcalc|)/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively.
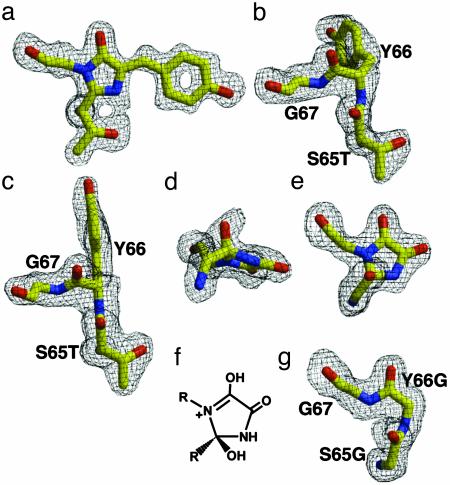
Fig. 1.
Posttranslational modifications revealed by structures of GFP variants before and after backbone cyclization. Omit |Fo – Fc| electron density maps for the chromophore residues contoured at 3 σ (black). (a) The 1.50-Å cyclized R96A structure. (b) The 2.00-Å precyclization R96A intermediate A structure. (c) The 2.00-Å precyclization R96A intermediate B structure. (d and e) Orthogonal views of the 1.80-Å Gly-Gly-Gly aerobic oxidized postcyclization structure. (f) Proposed molecular structure of the Gly-Gly-Gly cyclized ring. (g) The 1.80-Å Gly-Gly-Gly anaerobic precyclization structure. All are illustrated inraster 3d (44).
We used the slow maturation rate of the R96A variant to isolate GFP intermediates before cyclization and thereby determine two independent crystallographic structures (Table 1) of GFP before posttranslational modifications (Fig. 1 b and c). In the mechanical compression hypothesis, steric interactions generated by the GFP architecture are proposed to raise the energy of the precyclization state above that of the cyclized intermediate and relaxing this strained conformation drives chromophore formation. Thus, we examined the 2.0-Å resolution R96A precyclization structures for evidence of steric interactions that could be relaxed upon chromophore formation. Instead, the electron density reveals favorable conformations for the chromophore residues with no significant van der Waals collisions. Outside of the chromophore residues, the two structures are highly similar (Fig. 2a). The Cα atoms superimpose with a rms deviation of 0.28 Å, and side-chain conformational rearrangements are minor. Interestingly, these independently determined structures exhibit distinct Y66 side-chain conformations, lying on either side of the fourth β-strand (Fig. 1 b and c). Each stacks with Q94 and occupies part of the cavity created by truncating R96. The crystals used to determine these different precyclization intermediate structures grew under the same conditions. Thus, we propose that the protein possesses isoenergetic states for Y66 with low interconversion energy barriers and that subtle crystal packing differences may propagate to the protein core to select the observed conformations.
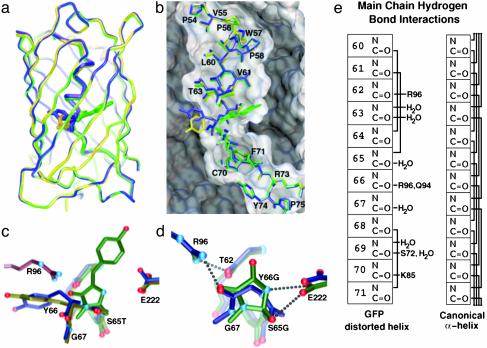
Fig. 2.
Architectural distortions and structural comparisons between precyclization and postcyclization states. (a) Superposition of R96A structures, emphasizing large conformational change for Y66 but otherwise small Cα differences between precyclization (A in yellow, B in blue) and postcyclization (green) states. (b) Central helix for three R96A structures displayed with the surface of the R96A mature structure, emphasizing helical bend. (c) Structural overlay of R96A precyclization intermediates A (yellow) and B (blue) with the mature R96A (green) structure, showing large main-chain movements in forming the chromophore. Modeled R96 (purple) indicates steric interactions with the Y66 side-chain position of the precyclization intermediate structure. (d) Superposition of the Gly-Gly-Gly structures before (blue, anaerobic) and after (green, aerobic) peptide cyclization shows functional group interactions between the R96, E222, and T62 carbonyl oxygen atoms and the chromophore residues. (e) Schematic of distortions in main-chain hydrogen-bonding interactions for the WT, Gly-Gly-Gly precyclization, and postcyclization structures (Left) displayed in comparison to a canonical α-helix (Right). Solid lines between main-chain atoms indicate presence of a hydrogen bond. a–d are illustrated with avs (45).
Comparisons of the precyclization and mature chromophore states for the R96A structures identify both global and local features that drive peptide cyclization. Despite dramatic motions of chromophore-forming residues, in which the Y66 phenolic oxygen atom moves 14 Å and backbone atoms shift 2.6–3.1 Å, residues outside of the chromophore superimpose well for the three R96A structures (Fig. 2a). The chromophore is anchored both by adjacent hydrophobic residues and hydrophobic interactions at the ends of the central distorted helix (Fig. 2b). In a sequence alignment of 48 GFP homologs (Table 2, which is published as supporting information on the PNAS web site, www.pnas.org), residue 64, which immediately precedes the chromophore, is essentially a hydrophobic (41 sequences; F, L, V) or cysteine (6 sequences) residue. Interestingly, all of the sequences that contain C64 also contain C29. Mapping these cysteine residues onto the GFP structure (data not shown) places them in a reasonable orientation to form a disulfide bond and provide an alternate anchoring method (to hydrophobic interactions). Moreover, despite right-handed helical conformations in a Ramachandran plot, the residues of this distorted “helix” in mature GFP make only three main-chain hydrogen bonds, between residue pairs L60-L64 (α-helix), V61-S65T (α-helix), and V68-F71 (310-helix). The R96A precyclization structures add the T62-Y66 (α-helix) and S65T-V68 (310-helix) hydrogen bonds. Thus, most of the distortions in the central helix are not a consequence of chromophore formation, but rather are imposed by the protein scaffold. In all structures of GFP (1, 17) and its red fluorescent protein homologs (35, 36) a dramatic ≈80° bend in the helix (Fig. 2b), generated by the protein architecture, is focused at the chromophore (Fig. 2c). The severe bend exposes the T62 and Y66 carbonyl oxygen atoms for interactions with R96 (see below) and forces the G67 nitrogen nucleophile and S65T carbonyl oxygen into closer contact (3.0 and 3.2 Å in the two precyclization structures) than the sum (3.25 Å) of their van der Waals radii (37), in preparation for covalent bond formation during peptide cyclization (Fig. 2c). Significantly, these distortions eliminate potential helical hydrogen bonds that would otherwise have to be broken at an energetic cost during cyclization. Together, the R96A structures suggest that the GFP architecture enforces destabilizing distortions, precluding a stable α-helical conformation and creating a state closer to the transition state for peptide cyclization. This is analogous to the entatic state proposal for metalloproteins (38), in which the protein scaffold constrains a metal center in a destabilized geometric conformation to lower reorganization energy barriers and increase reaction rates.
Structures of the S65G Y66G Variant Under Aerobic and Anaerobic Conditions. To examine whether side-chain interactions of the chromophore residues are critical to backbone cyclization, we constructed and characterized the colorless S65G Y66G variant (sequence Gly-Gly-Gly for chromophore residues). Mutational results have established that almost any substitution for S65, aromatic substitutions for Y66, and the WT G67 form mature chromophores (4). However, if the mechanical compression model for peptide cyclization (20) or the proposal that side-chain oxidation is required before cyclization (22) were correct, truncation to the Gly-Gly-Gly variant should hinder or preclude backbone cyclization. Remarkably, the electron density for a 1.80-Å resolution structure of this variant (Table 1) reveals that the backbone is cyclized. Simulated annealing omit maps for this cyclized Gly-Gly-Gly variant (Fig. 1 d and e) show that the imidazolone ring is shifted ≈0.7 Å from its position in the GFPsol structure and modified by two nonhydrogen atoms. The first atom, the S65G carbonyl oxygen, has not been lost as water, as is the case for WT GFP. Instead, this oxygen remains attached to the imidazolone ring and forms a hydrogen bond with the side chain of E222 (Fig. 2d). The second nonhydrogen atom is covalently bound to the Y66G Cα of the cyclized ring and is most likely an oxygen atom incorporated through an oxidation reaction (see Fig. 4, which is published as supporting information on the PNAS web site, for proposed mechanism). We confirmed that the extra peaks in our omit maps were real and not derived from the protein variant by resequencing the plasmid, purifying a second batch of protein, solving a second 2.00-Å structure, and observing the same peaks in new omit maps. The electron density (Fig. 1d) is consistent with a five-π-electron, nonaromatic ring system that contains a tetrahedral S65G carbonyl carbon atom (puckering the cyclized ring), an enol tautomer for the Y66G carbonyl, and a keto tautomer for the newly incorporated oxygen atom (Fig. 1f). Cyclization of Gly-Gly-Gly, which contains no side-chain atoms, argues against the proposed model in which side-chain oxidation precedes cyclization (22).
We prepared and crystallized the S65G Y66G variant under anaerobic conditions to explore the unexpected incorporation of oxygen at Y66G Cα. Surprisingly, the anaerobic structure at 1.80-Å resolution (Table 1) revealed uncyclized chromophore residues (Fig. 1g). Outside of the chromophore, the precyclization and postcyclization Gly-Gly-Gly structures are remarkable similar (Fig. 2d) and share with WT GFP the same limited hydrogen bonding (6 of 24 possible main-chain interactions) for the central helix (Fig. 2e). Thus, even the added backbone flexibility conferred by the substitution of Gly for the chromophore amino acids does not result in the formation of additional main-chain hydrogen bonds. The lack of main-chain hydrogen bonds for the chromophore residues contributes to the apparent low interconversion energy barriers and large local rearrangements for cyclization observed in the R96A structures (above). Distortions in the precyclization state are maintained in the postcyclization state, rather than relieved by chromophore formation. This argues against the mechanical compression hypothesis, but underscores the importance of the GFP architecture in creating a specific conformation that favors peptide cyclization.
The Gly-Gly-Gly structural results reveal conformational and energetic features critical to peptide cyclization. Standard main-chain conformations for G67 in both precyclization (Φ = –90°, Ψ = –16°) and postcyclization (Φ = –90°, Ψ = –35°) states suggests that the apparent requirement for G67 in chromophore formation (4) results from steric rather than conformational restrictions. In fact, a modeled Ala side chain for residue 67 has significant van der Waals collisions with the T63 carbonyl oxygen. More importantly, the failure of the Gly-Gly-Gly sequence to cyclize anaerobically indicates that cyclization in this mutant is coupled to oxidation. The lack of any partial occupancy of a cyclized product under anaerobic conditions suggests that the precyclization structure is thermodynamically more stable than a cyclized but not yet oxidized state. Thus, oxidation serves to increase the electronic conjugation of the Gly-Gly-Gly variant and trap a thermodynamically unfavorable cyclization product according to Le Chatelier's principle as a resonance-stabilized, five-π-electron, nonaromatic species.
Discussion
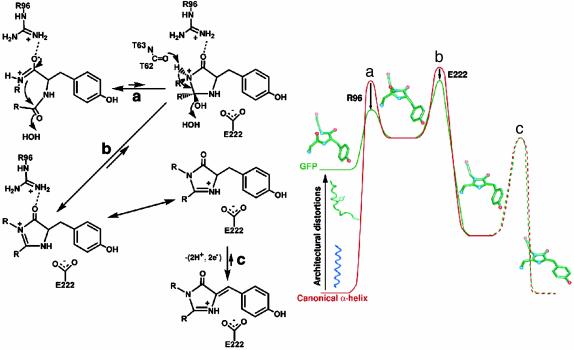
Conjugation-Trapping Mechanism. We propose that the initial cyclized intermediate in WT GFP, as in Gly-Gly-Gly, is higher in energy than the precyclization state and is trapped through conjugation (Fig. 3). Modeling the missing Y66 side chain into the precyclization Gly-Gly-Gly structure produces a favorable conformer without significant van der Waals collisions, arguing that the precyclization state of WT GFP is not significantly destabilized relative to Gly-Gly-Gly. Further, the cyclized product before dehydration and oxidation is not likely to be stabilized for WT GFP relative to Gly-Gly-Gly. Thus, for GFP, as for the Gly-Gly-Gly variant, the cyclization reaction appears thermodynamically unfavorable, consistent with the relative thermodynamic stabilities calculated with density functional theory (endothermic cyclization, ≈10 kcal/mol) (22). Previously, this was interpreted to suggest that side-chain oxidation may precede cyclization in GFP (22). The driving force for this unprecedented Cα—Cβ bond oxidation before peptide cyclization is unclear. Instead, we suggest a fundamentally different molecular mechanism in which the unfavorable cyclization reaction is driven by a subsequent trapping reaction, likely dehydration of the residue 65 carbonyl oxygen. Dehydration (Fig. 3b) would generate an α-β unsaturated ketone (a resonance-stabilized five-π-electron, nonaromatic species like Gly-Gly-Gly; see below) and serve to drive the cyclization equilibrium (Fig. 3a) toward the higher energy cyclization product. This conjugation-trapping mechanism for chromophore formation would further provide the driving force for subsequent Cα—Cβ bond oxidation (Fig. 3c) by generating an aromatic imidazolone and conjugating the newly formed backbone ring system and the aromatic side chain of position 66.
Fig. 3.
The proposed conjugation-trapping mechanism for GFP chromophore formation. The chemical mechanism for GFP chromophore formation (Left) is displayed along with a cartoon representation of the corresponding reaction coordinate (Right). The reaction coordinates (x axis) for GFP (green) and a canonical α-helix (red) are displayed against increasing energy for the chromophore residues (y axis), to highlight the three features favoring ring synthesis in the GFP scaffold: architectural distortions, R96 enhancement of the G67 nucleophile, and E222 stabilization of the dehydration transition state. (a) Peptide cyclization to generate a destabilized intermediate. (b) Dehydration, initiated by the T62 carbonyl, to trap the cyclized product through conjugation. (c) Oxidation to generate an aromatic imidazolone and conjugate the two ring systems. The chromophore images superimposed onto the cartoon are (from left to right) the R96A precyclization structure, model of cyclized intermediate, model of reduced intermediate and the R96A mature chromophore structure. Our data do not address the oxidation transition state (displayed as dashed lines).
Role of R96 in GFP Peptide Cyclization. We propose that R96 contributes to the architectural distortions important for peptide cyclization and increases the nucleophilicity of the attacking G67 nitrogen. The primary difference between the Gly-Gly-Gly and R96A precyclization structures can be attributed to the R96 side chain, which prevents the formation of the T62-Y66 and S65T-V68 main-chain hydrogen bonds that must otherwise be broken at an energetic cost during chromophore formation. Thus, R96 may contribute modestly to generating the distortions required for the entatic state. However, the positively charged guanidinium group of R96 also interacts with the Y66G carbonyl in both precyclization and postcyclization Gly-Gly-Gly structures. Thus, the conserved R96 (see Table 2) may also form an ion pair with the Y66 carbonyl in WT GFP to favor the peptide bond resonance form with a carbon-nitrogen double bond (Fig. 3a). Initially, increasing the nucleophilicity of the Gly-67 nitrogen by favoring the enolate resonance form, which formally has a positively charged nitrogen atom, seemed paradoxical. However, ab initio calculations suggest that in the peptide bond mimic formamide the partial charge is more negative on the nitrogen atom in the enolate resonance form (39). In this partial charge effect or “back transfer,” the π-electron shift from the lone pair on the nitrogen toward the carbonyl carbon is more than compensated for by shifts from the carbon to nitrogen atoms through σ-orbitals (40–42). Previously, R96 was proposed to assist in chromophore formation by activating the S65 carbonyl for nucleophilic attack (20) or by directly deprotonating the G67 backbone amide (22). Our precyclization structure of the Gly-Gly-Gly variant reveals that R96 is distant (4.8 and 7.0 Å from the nitrogen and carbonyl oxygen atoms) and, more importantly, in the wrong orientation to have either function.
Roles of R96, the T62 Carbonyl Oxygen, and E222 in the GFP Dehydration Reaction. We propose that R96 acidifies the G67 nitrogen of the cyclized intermediate, the T62 carbonyl oxygen is the base that abstracts this proton to initiate main-chain dehydration, and E222 accelerates this trapping reaction and stabilizes the resulting intermediate (Fig. 3b). E222 forms hydrogen bonds to the Y66G nitrogen and T65G oxygen atoms in the Gly-Gly-Gly variant and appears properly positioned to initiate the dehydration reaction (Fig. 2d) by abstracting a proton from the Y66G nitrogen and donating a proton to the emerging water molecule, as proposed (22). However, the Gly-Gly-Gly structure clearly shows that this variant is dehydration-compromised. To explain the inability of this mutant to dehydrate, we propose an alternative dehydration mechanism that is initiated by the abstraction of a proton from the G67 nitrogen (Fig. 3b). The only functional group in the proper orientation and distance (3.0 Å) for this task appears to be the T62 carbonyl oxygen. Although this is not a strong base, the nearby positively charged R96 (≈4.8 Å between the guanidinium and G67 nitrogen atoms) would enhance the acidity of the G67 nitrogen atom. Moreover, in the oxidized Gly-Gly-Gly structure the shifted imidazolone ring increases the distance between the G67 nitrogen and the proposed base (3.3 Å), explaining the inability of this mutant to dehydrate. Instead, the T62 carbonyl oxygen is closer to the Y66G Cα atom (3.1 Å) and in the proper orientation to abstract a proton and initiate the proposed oxygen incorporation reactions (Fig. 2d). Red fluorescent protein shares with GFP this geometric arrangement of the G67 nitrogen atom with the R96 side chain and the residue 62 carbonyl oxygen atom (35, 36). Thus, this proton abstraction mechanism may be common to this class of fluorescent proteins. Further, we suggest that E222 assists in the trapping reaction (Fig. 3b) by electrostatically complementing the positive charge that develops on the S65T carbonyl oxygen atom in the transition state and then stabilizing the resonance form of the imidazolone ring with the positive charge on the Y66 nitrogen.
Conclusions. Our structures of precyclized intermediate and oxidized postcyclized protein states not only explain the known features of GFP chromophore formation, but suggest an additional conjugation-trapping mechanism. The protein architecture creates a dramatic bend at the chromophore of the central helix that removes specific main-chain hydrogen bonds, which must otherwise be broken during maturation. In our conjugation-trapping mechanism, these distortions, which are present in both precyclization and postcyclization structures, serve to lower the energetic cost of peptide cyclization. Our data are inconsistent with the mechanical compression hypothesis, in which steric distortions imposed by the protein scaffold relax upon chromophore formation. Additionally, the Gly-Gly-Gly structures indicate the initial cyclization reaction is thermodynamically unfavorable and that a subsequent trapping reaction is required to drive chromophore maturation. This result is also at odds with the mechanical compression hypothesis, in which the initial cyclization reaction is thermodynamically favorable. Further, our architecturally driven conjugation-trapping mechanism accounts for the robustness of chromophore maturation despite many mutations at or surrounding the chromophore (4), including R96A and E222G (43), and provides a scaffold for specific functional group chemistry to accelerate chromophore maturation. Toward that end, we suggest that conserved residues R96 and E222 have primarily electrostatic roles in chromophore formation and the T62 carbonyl oxygen is the base that initiates the dehydration reaction (Fig. 3). Similarities between the peptide cyclization products in GFP and histidine ammonia lyase raise the possibility that conjugation-trapping serves as a common driving force for both of these posttranslational modifications. These structural results and hypotheses establish a testable molecular mechanism relevant to understanding and ultimately controlling chromophore synthesis from amino acids with widespread applications in biology and medicine.
Supplementary Material
Acknowledgments
We are grateful to F. Henderson for assistance in purifying the R96A variant and V. A. Roberts, D. S. Daniels, M.G. Finn, T. I. Wood, C. D. Stout, J. L. Tubbs, J. L. Huffman, and C. A. Mullen for scientific discussions. This work was supported by La Jolla Interfaces in Sciences and National Institutes of Health (GM19290) Postdoctoral Fellowships (to D.P.B.), the Damon Runyon Cancer Research Foundation (Robert Black Fellow DRG1699-02 to C.D.P.), National Institutes of Health Grant GM37684, the Stanford Synchrotron Radiation Laboratory, and the Advanced Photon Source.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1QXT, 1QY3, 1QYF, 1QYO, and 1QYQ).
References
- 1.Ormö, M., Cubitt, A. B., Kallio, K., Gross, L. A., Tsien, R. Y. & Remington, S. J. (1996) Science 273, 1392–1395. [DOI] [PubMed] [Google Scholar]
- 2.Cubitt, A. B., Heim, R., Adams, S. R., Boyd, A. E., Gross, L. A. & Tsien, R. Y. (1995) Trends Biochem. Sci. 20, 448–455. [DOI] [PubMed] [Google Scholar]
- 3.Heim, R., Prasher, D. C. & Tsien, R. Y. (1994) Proc. Natl. Acad. Sci. USA 91, 12501–12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsien, R. Y. (1998) Annu. Rev. Biochem. 67, 509–544. [DOI] [PubMed] [Google Scholar]
- 5.Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W. & Prasher, D. C. (1994) Science 263, 802–805. [DOI] [PubMed] [Google Scholar]
- 6.Heim, R. & Tsien, R. Y. (1996) Curr. Biol. 6, 178–182. [DOI] [PubMed] [Google Scholar]
- 7.Matz, M. V., Fradkov, A. F., Labas, Y. A., Savitsky, A. P., Zaraisky, A. G., Markelov, M. L. & Lukyanov, S. A. (1999) Nat. Biotechnol. 17, 969–973. [DOI] [PubMed] [Google Scholar]
- 8.Labas, Y. A., Gurskaya, N. G., Yanushevich, Y. G., Fradkov, A. F., Lukyanov, K. A., Lukyanov, S. A. & Matz, M. V. (2002) Proc. Natl. Acad. Sci. USA 99, 4256–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostergaard, H., Henriksen, A., Hansen, F. G. & Winther, J. R. (2001) EMBO J. 20, 5853–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robey, R. B., Ruiz, O., Santos, A. V. P., Ma, J., Kear, J., Wang, L., Li, C., Bernardo, A. A. & Arruda, J. A. L. (1998) Biochemistry 37, 9894–9901. [DOI] [PubMed] [Google Scholar]
- 11.Kneen, M., Farinas, J., Li, Y. & Verkman, A. (1998) Biophys. J. 74, 1591–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pozzan, T. (1997) Nature 388, 834–835. [DOI] [PubMed] [Google Scholar]
- 13.Baird, G. S., Zacharias, D. A. & Tsien, R. Y. (1999) Proc. Natl. Acad. Sci. USA 96, 11241–11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wachter, R. M. & Remington, S. J. (2000) Curr. Biol. 9, R628–R639. [DOI] [PubMed] [Google Scholar]
- 15.Jayaramn, S., Haggie, P., Wachter, R. M., Remington, S. J. & Verkman, A. S. (2000) J. Biol. Chem. 275, 6047–6050. [DOI] [PubMed] [Google Scholar]
- 16.Zimmer, M. (2002) Chem. Rev. 102, 759–782. [DOI] [PubMed] [Google Scholar]
- 17.Yang, F., Moss, L. G. & Phillips, G. N., Jr. (1996) Nat. Biotechnol. 14, 1246–1251. [DOI] [PubMed] [Google Scholar]
- 18.Reid, B. G. & Flynn, G. C. (1997) Biochemistry 36, 6786–6791. [DOI] [PubMed] [Google Scholar]
- 19.Schwede, T. F., Retey, J. & Schulz, G. E. (1999) Biochemistry 38, 5355–5361. [DOI] [PubMed] [Google Scholar]
- 20.Branchini, B. R., Nemser, A. R. & Zimmer, M. (1998) J. Am. Chem. Soc. 120, 1–6. [Google Scholar]
- 21.Baedeker, M. & Schulz, G. E. (2002) Structure (London) 10, 61–67. [DOI] [PubMed] [Google Scholar]
- 22.Siegbahn, P. E. M., Wirstam, M. & Zimmer, M. (2001) Int. J. Quantum Chem. 81, 169–186. [Google Scholar]
- 23.Donnelly, M., Fedeles, F., Wirstam, M., Siegbahn, P. E. & Zimmer, M. (2001) J. Am. Chem. Soc. 123, 4679–4686. [DOI] [PubMed] [Google Scholar]
- 24.Crameri, A., Whitehorn, E. A., Tate, E. & Stemmer, W. P. C. (1996) Nat. Biotechnol. 14, 315–319. [DOI] [PubMed] [Google Scholar]
- 25.Cormack, B. P., Valdiva, R. H. & Falkow, S. (1996) Gene 173, 33–38. [DOI] [PubMed] [Google Scholar]
- 26.Deschamps, J. R., Miller, C. E. & Ward, K. B. (1995) Protein Expression Purif. 6, 555–558. [DOI] [PubMed] [Google Scholar]
- 27.Barondeau, D. P., Kassmann, C. J., Tainer, J. A. & Getzoff, E. D. (2002) J. Am. Chem. Soc. 124, 3522–3524. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski, Z. & Minor, W. (1997) Macromol. Crystallogr. A 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 29.Navaza, L. (1994) Acta Crystallogr. A 50, 157–163. [Google Scholar]
- 30.McRee, D. E. (1999) J. Struct. Biol. 125, 156–165. [DOI] [PubMed] [Google Scholar]
- 31.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, N., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 32.Sheldrick, G. M. & Schneider, T. R. (1997) Methods Enzymol. 277, 319–343. [PubMed] [Google Scholar]
- 33.Brunger, A. T. (1992) Nature 355, 472–474. [DOI] [PubMed] [Google Scholar]
- 34.Bruns, C. M., Hubatsch, I., Ridderstrom, M., Mannervik, B. & Tainer, J. A. (1999) J. Mol. Biol. 288, 427–439. [DOI] [PubMed] [Google Scholar]
- 35.Wall, M. A., Socolich, M. & Ranganathan, R. (2000) Nat. Struct. Biol. 7, 1133–1138. [DOI] [PubMed] [Google Scholar]
- 36.Yarbrough, D., Wachter, R. M., Kallio, K., Matz, M. V. & Remington, S. J. (2001) Proc. Natl. Acad. Sci. USA 98, 462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bondi, A. (1964) J. Phys. Chem. 441–451.
- 38.Vallee, B. L. & Williams, R. J. (1968) Proc. Natl. Acad. Sci. USA 59, 498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiberg, K. B. & Breneman, C. M. (1992) J. Am. Chem. Soc. 123, 831–840. [Google Scholar]
- 40.Milner-White, E. J. (1997) Protein Sci. 6, 2477–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webster, B. (1990) Chemical Bonding Theory (Blackwell Scientific, Oxford).
- 42.Polard, D. & Scheraga, H. A. (1967) Biochemistry 6, 3791–3800. [Google Scholar]
- 43.Ehrig, T., O'Kane, D. J. & Prendergast, F. G. (1995) FEBS Lett. 367, 163–166. [DOI] [PubMed] [Google Scholar]
- 44.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]
- 45.Upson, C., Faulhaber, T., Jr., Kamins, D., Laidlaw, D., Schlegel, D., Vroom, J., Gurwitz, R. & van Dam, A. (1989) IEEE Comp. Graph. Appl. 9, 30–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.