Abstract
Mutations in the myotubularin (MTM)-related protein 2 (MTMR2) gene are responsible for the severe autosomal recessive neuropathy Charcot–Marie–Tooth disease type 4B1. MTMR2 belongs to the MTM family of dual-specific phosphatases that use phosphatidylinositol (PI) 3,5-bisphosphate [PI(3,5)P2] and PI 3-phosphate [PI(3)P] as their substrate. Because these substrates are localized in the membrane bilayer, membrane targeting of Mtmr2 is an important regulatory mechanism. In hypoosmotically stressed COS cells with increased levels of PI(3,5)P2, Mtmr2 is bound to the membrane of vacuoles formed under these conditions. Using several mutant forms of Mtmr2, we identified two domains that are necessary for membrane association: (i) A pleckstrin homology-GRAM domain; and (ii) a coiled-coil module. Protein-lipid overlay assays show that the pleckstrin homology-GRAM domain binds to PI(3,5)P2 and PI(5)P, a substrate and a product of the Mtmr2 enzyme, respectively. We also demonstrate that Mtmr2 forms a dimer and that the C-terminal coiled-coil is responsible for homodimerization, in addition to membrane association. Our data indicate that phosphoinositide–protein interactions, as well as protein–protein interactions, are necessary for the correct regulation of MTMR2.
Myotubularins (MTMs) represent one of the largest and most conserved families of dual-specific protein tyrosine phosphatase-like enzymes (1). To date, 14 MTM genes have been identified, three of which have been associated with human diseases. The MTM-related protein 2 (MTMR2) gene is mutated in Charcot–Marie–Tooth disease type 4B1 (CMT4B1), a severe autosomal recessive hereditary neuropathy (refs. 2–5; see ref. 6 for review), which is characterized by reduced nerve conduction velocity and focally folded myelin sheets in peripheral nerves (7). Mutations in sbf2/MTMR13 lead to a disease with identical pathology in the peripheral nervous system called CMT4B2, and are associated with early onset glaucomas in some families (8, 9). Mutations in MTM (MTM1), the founding member of the family, lead to X-linked myotubular myopathy (10). In affected individuals, myogenesis is arrested in a late phase of myofiber formation, leading to hypotonia and muscle weakness (11, 12).
The MTM family is defined by the presence of a phosphatase, a SET-interacting domain (SID), and a recently proposed, putative coiled-coil domain (Fig. 1A) (1). In addition to these motifs, most family members have domains implicated in binding of phosphoinositides, such as pleckstrin homology (PH) or FYVE zinc-finger domains.
Fig. 1.
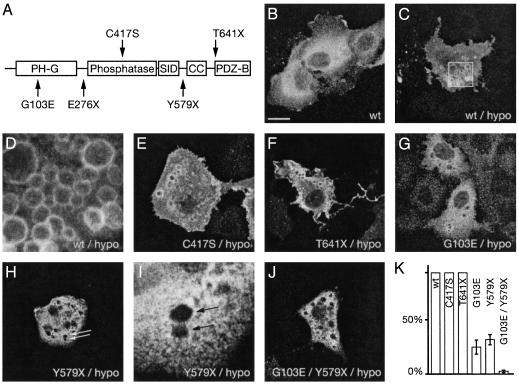
Membrane association of Mtmr2. (A) Domain structure of Mtmr2. Disease-associated mutations are indicated below the schematic structure and artificially designed mutations are indicated above. PH-G, PH-GRAM domain; SID, SET-interacting domain; CC, coiled-coil domain; PDZ-B, PDZ-binding motif. (B) A cytosolic localization of Mtmr2 with enrichment in the perinuclear region was observed in unstressed COS cells transfected with a construct expressing wild-type (wt) Mtmr2. (C) Under hypoosmotic (hypo) stress conditions, Mtmr2 binds to the membrane of large vacuoles. The boxed region is shown at higher magnification in D. (E and F) Mtmr2-C417S and Mtmr2-T641X also bind to membranes of the vacuoles. (G–J) Mtmr2-G103E, Mtmr2-Y579X, and Mtmr2-G103E-Y579X do not bind efficiently to these vacuoles. (I) A higher-power view of H.(K) The percentage of cells containing vacuoles with membrane staining. (Scale bars, 20 μm for B, C, E–H, and J; 4 μm for D and I.)
Phosphatidylinositol (PI) 3-phosphate [PI(3)P] was initially identified as the substrate of MTM1 (13, 14). Recently, MTM1, MTMR2, MTMR3, and MTMR6 were shown to dephosphorylate both PI 3,5-bisphosphate [PI(3,5)P2] and PI(3)P at position D3 pointing toward a broader substrate specificity, which is possibly common to all active family members (15–17). PI(3)P plays a key role in vesicular trafficking and membrane transport (18, 19). In addition, it enables the targeting of proteins with a FYVE domain to these membranes (20, 21). PI(3)P can be further phosphorylated on multiple sites of the inositol ring to generate distinct second messengers, which are involved in important cellular processes, such as cell survival, proliferation, differentiation, and cytoskeleton reorganization. PI(3,5)P2 is synthesized from PI(3)P by a phosphoinositide 5-kinase with a FYVE domain (PIKfyve) (22). It was first identified in COS cells in response to osmotic stress (23). PI(3,5)P2 is crucial for the recycling of membranes from vacuoles and lysosomes (24) and regulates the sorting of membrane protein cargo into the vacuole through endosomal multivesicular bodies (25).
Hypoosmotically stressed COS cells are a well suited biological system to study membrane binding and cellular function of the MTM family because the stressed cells have an increased level of PI(3,5)P2. In this article, we show that, after hypoosmotic shock, Mtmr2 is bound to the membrane of vacuoles, which are immediately formed under these conditions. By domain mapping using several disease-associated and designed mutations, we demonstrate that two domains are responsible for membrane attachment; namely, the pleckstrin homology-GRAM (PH-G) domain located toward the N terminus of Mtmr2, and the putative coiled-coil domain in the C-terminal part of the protein. We also show that in a protein-lipid overlay assay for lipid-binding specificity, the PH-G domain binds to a substrate [PI(3,5)P2] and a product [PI(5)P] of the Mtmr2 enzyme reaction. We provide experimental evidence that the C-terminal region of Mtmr2 indeed adopts a coiled-coil structure and is responsible for dimerization of Mtmr2, in addition to mediating membrane attachment. These findings identify the PH-G and the coiled-coil domains as crucially important modules for the localization and the regulation of Mtmr2.
Materials and Methods
Constructs for Eukaryotic and Prokaryotic Expression of Mtmr2. The mouse Mtmr2 cDNA was cloned into SacI/SalI cut pQE30 vector (Qiagen, Hilden, Germany) by using an internal SacI site and a SalI site, which was generated by PCR 3′ of the stop codon. The resulting protein consists of an N-terminal His-tag, followed by amino acids 4–643 of Mtmr2. From this vector, the EcoRI/SalI fragment containing the complete coding region was subcloned into EcoRI/XhoI cleaved pcDNA3.1 zeo(+) vector (Invitrogen). For some experiments, untagged constructs were used. The mutations were introduced by PCR by using Taq Plus Precision (Stratagene), and were confirmed by DNA sequencing.
Cell Culture. COS cells were maintained at 37°C with 6% CO2 in DMEM containing 10% FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were transfected in 3.5-cm dishes by using Superfect, according to the recommendations of the manufacturer (Qiagen). Forty hours after transfection, cells were hypoosmotically shocked for 15 min with 0.25× medium (diluted with water). Cells were then fixed with 2% paraformaldehyde in PBS for 20 min, followed by a 1-h incubation in blocking buffer (10% goat serum/0.05% saponin/PBS). As primary antibodies, a 1:200 diluted anti-His antibody (Qiagen) or 1:1,500 diluted anti-Mtmr2 antibody were used. As secondary antibodies, Cy3-conjugated anti-mouse or anti-rabbit antibodies were used (Jackson ImmunoResearch). Fluorescence analysis was performed by a Leica TCS SP confocal microscope. For quantitative analysis, 3 × 100 cells in nonoverlapping fields of two independent transfections were counted.
Purification of Mtmr2. His-tagged Mtmr2 was expressed in Escherichia coli XL1Blue cells. After 16 h induction with 1 mM IPTG, the cells were harvested, resuspended in lysis buffer (25 mM Tris·HCl, pH 8.6/250 mM NaCl/10 mM imidazole), and passed through a French press. The cleared lysate was applied to a nickel-nitrilotriacetic acid column (Qiagen) equilibrated in lysis buffer. The protein was eluted with 125 mM imidazole, dialyzed against buffer 1 (15 mM Tris·HCl, pH 9.4/50 mM NaCl/2 mM DTT/0.01% Tween 20) and loaded on a MonoQ column (Amersham Biosciences). Mtmr2 was in the flow-through fraction, whereas E. coli proteins remained bound to the column. The flow-through fraction was concentrated and applied to a Superdex S200 HR column (Amersham Biosciences) equilibrated with buffer 2 (20 mM Tris·HCl, pH 8.8/150 mM NaCl/2 mM DTT/0.01% Tween 20) and calibrated by using a high molecular weight gel filtration kit (Amersham Biosciences).
Characterization of Mtmr2 by Sedimentation Velocity. Analytical ultracentrifugation was carried out in a Beckmann Optima XL-1 centrifuge by using an An50 Ti eight-hole rotor and the interference optical detection system. Double-sector charcoal-filled epon centerpieces were filled with 350 μl of 10 μM Mtmr2. The sedimentation velocity experiment was performed at 4°C, at 42,000 rpm, for 6 h. Fringe profiles were recorded at 210-s intervals. Data were analyzed with sedfit software by direct boundary modeling by solutions of the Lamm equation (26).
Thioredoxin Fusion of the Coiled-Coil Domain. A C-terminal region of Mtmr2 containing the putative coiled-coil domain was amplified from pQE30-Mtmr2 by using oligonucleotides Coilfor (5′-CGGGCCCGGGGGATCCATCCACAGCAGATACAAAG-3′) and Coilrev (5′-GGGGTACCCGGGCTATCACTCTGAAGAGGAGGTTG-3′). The product was cloned by means of BamHI and KpnI into a pRET3a-HisTRXN vector, which encodes a thioredoxin fused to an N-terminal His6-tag and a C-terminal glycine/serine-rich linker, followed by a tobacco etch virus (TEV) protease cleavage site (T. J. Richmond, unpublished work). The coiled-coil domain is thus produced as a fusion protein downstream of the protease cleavage site. His6TRXNCoil was expressed in E. coli BL21(DE3) cells and purified by cobalt affinity chromatography by using Talon resin (Clontech), following the manufacturer's recommendations. Eluted protein was subsequently passed in Buffer C (20 mM Tris·HCl, pH 8.0/150 mM NaCl/2 mM DTT) through a Superdex S75 HR column (Amersham Biosciences), calibrated by using the low molecular weight gel filtration kit (Amersham Biosciences). His6TRXNCoil (3.5 mg) were cleaved by TEV protease, as recommended by the manufacturer (Invitrogen). A part of the cleavage reaction was passed over an analytical Superdex S75 HR column equilibrated in Buffer C, whereas the remainder was dialyzed in Buffer D (10 mM Tris·HCl, pH 8.0/100 mM NaCl) and subjected to Talon affinity chromatography. The liberated coiled-coil domain eluted in the flow-through was applied to a MonoQ column. Coiled-coil was eluted in the flow-through and was concentrated to 1.2 mg/ml (270 mM). Protein concentrations were determined by UV absorption at 280 nm, assuming an extinction coefficient of ε280 = 16,500 M–1·cm–1 (His6 TRXNCoil), and ε280 = 1,280 M–1·cm–1(Coil), respectively (27).
CD Spectroscopy. CD spectra were recorded at 20°C by using a J710 spectropolarimeter (Jasco, Easton, MD), which was equipped with a thermoelectric temperature controller and interfaced to a personal computer. The CD spectrum of the coiled-coil domain was measured at a band width of 2 nm, with a step size of 0.5 nm, and with 4-s averaging time per point in a 0.1-cm cuvette. Spectra were signal averaged by adding three scans, which were baseline-corrected, and smoothed by using the Jasco software.
Fluorescence Resonance Energy Transfer (FRET) Experiment. The coiled-coil domain was amplified from pQE30-Mtmr2 by using oligonucleotides Coilfor and Coilrev. The PCR product was digested with XmaI and cloned into pBFP-GFP (28), digested with BspEI, yielding a plasmid coding for BFP-Coil. For the N-terminal fusion, PCR primers Xcoilfor (5′-CGCTCTAGAATCCACAGCAGATACAAAG-3′) and Xcoilrev (5′-GCGTCTAGACTCTGAAGAGGAGGTTG-3′) were used. PCR product was digested with XbaI and cloned into pBFP plus GFP vector (28) through the NheI sites, yielding plasmids coding for Coil-BFP and Coil-GFP. To minimize geometrical constraints on fluorophore orientation, we provided a SSKG-linker for N-terminal fusions and a SGGS-linker for the C-terminal fusion between the fluorescent proteins and the coiled-coil domain. Further details of generating the dual vectors expressing Coil-BFP and Coil-GFP, on one hand, as well as BFP-Coil and Coil-GFP, on the other, will be published elsewhere. Plasmids were transformed into E. coli JM83 cells and plated on dYT-agar containing 100 μg/ml ampicillin and 1 mM IPTG, followed by incubation at 30°C. Cells were irradiated with UV light at 366 nm. Fluorescence was recorded with a Nikon Coolpix 995 digital camera.
Protein Lipid Overlay Assay. The overlay assay was performed as described, by using prespotted phosphoinositide arrays (Echelon Biosciences, Salt Lake City) (29). Membranes were incubated overnight with 500 ng/ml purified protein containing a GST-tag at the N terminus, and a His-tag at the C terminus (16). As primary antibody for the detection of Mtmr2 or GST, the membranes were incubated with a 1:1,000 diluted rabbit anti-Mtmr2 or rabbit anti-GST antibody. As secondary antibody, a 1:25,000 diluted horse radish peroxidase-coupled goat anti-rabbit antibody (Santa Cruz Biotechnology) was used, followed by chemiluminescence detection with a Renaissance Western blot Reagent Plus kit (Perkin–Elmer Life Sciences, Boston).
Results and Discussion
Membrane Association of Mtmr2. We used transfected COS cells to study the subcellular localization of MTMR2. In resting cells, Mtmr2 is localized mainly in the cytosol, with a stronger staining in the perinuclear region (Fig. 1B) (30, 31). Studies with markers for the endoplasmic reticulum and the Golgi revealed partial colocalization, suggesting that some Mtmr2 may be associated with membranes of those organelles (data not shown). In COS cells and other cell types, the level of PI(3,5)P2, which is a main substrate of MTMR2, is usually low (23, 32), thus hampering functional studies. On the other hand, in hypoosmotically stressed COS cells, as well as in T lymphocytes after IL-2 or UV treatment (33), the level of PI(3,5)P2 is transiently up-regulated. Therefore, we used hypoosmotically stressed COS cells as a model system for our studies. Under hypoosmotic conditions, COS cells immediately start to form large vacuoles, and Mtmr2 is bound to the membranes of these vacuoles (Fig. 1 C and D). To map the domains relevant for membrane binding, we used several mutant forms of Mtmr2, which were identified either in CMT4B1 patients, or affect specific functional domains (Fig. 1 A). Mutation G103E is located in the PH-G domain and causes strongly reduced phosphatase activity (16). The catalytic activity in the MTMR2 mutant C417S is entirely abolished (16). A stop codon at position 641 leads to a shortened protein (T641X) lacking the putative PDZ-binding motif. A stop codon at position 579 (Y579X) deletes both the coiled-coil domain and the PDZ-binding motif, which is present in the C-terminal part of the protein. In hypoosmotically stressed COS cells, two of these mutants, Mtmr2-C417S and Mtmr2-T641X, bind to the vacuoles like wild type, indicating that membrane association does not depend on phosphatase activity or the PDZ-binding motif (Fig. 1 E and F). In contrast, binding of Mtmr2-G103E and Mtmr2-Y579X is severely impaired (Fig. 1 G–I). Only 20–40% of the cells with vacuoles showed vacuolar membrane staining (Fig. 1K). Membrane binding was almost completely abolished when the double mutant Mtmr2-G103E-Y579X was used (Fig. 1J). This reduced binding capability cannot be explained by the slightly reduced expression level of the mutant forms, because membrane association was also observed in COS cells, which express wild-type Mtmr2 at low levels (data not shown). These results indicate that both the PH-G and the putative coiled-coil domains, and thus both N- and C-terminal regions of Mtmr2, are involved in vacuole membrane association.
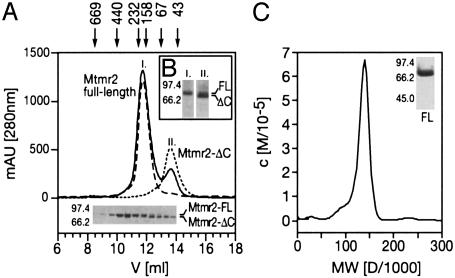
The Coiled-Coil Domain Mediates Mtmr2 Homodimerization, in Addition to Membrane Binding. We expressed and purified Mtmr2 and subjected the protein to analytical gel filtration. Mtmr2 consistently eluted in two distinct peaks, corresponding to an apparent molecular mass of 170 (Fig. 2A; Mtmr2 full-length) and 70 kDa (Fig. 2 A; Mtmr2-ΔC), respectively. SDS/PAGE analysis revealed a single protein band migrating at 73 kDa (Fig. 2 A Lower; Mtmr2-FL), which is present in the first peak, and a somewhat shorter species (70 kDa) (Fig. 2 A Lower; Mtmr2-ΔC) in the second peak. Western blot analysis with an anti-His-tag antibody showed that both the 73-kDa (Fig. 2B; FL) and 70-kDa (Fig. 2B; ΔC) proteins contain the His-tag that was added to the N terminus of the recombinant protein. This result indicates that the 70-kDa protein is a C-terminally truncated version of Mtmr2, probably arising through cleavage by endogenous protease activity in the E. coli lysate. Furthermore, the gel filtration results implicate the C-terminal region in the oligomerization of Mtmr2, because the full-length protein migrates at an apparent molecular mass of more than twice its expected size. Mass spectroscopy of the 70-kDa species revealed that the cleavage occurs after lysine 603 (data not shown). This cleavage site is located within a putative coiled-coil domain as predicted by multicoil (34). Coiled-coil domains are known to mediate protein–protein interactions. We determined the precise mass of native Mtmr2 by ultracentrifugation, yielding a value of 140 kDa (Fig. 2C). Thus, we conclude that Mtmr2 exists as a homodimer in solution, and that the dimerization is mediated by the C-terminal region of Mtmr2 containing the putative coiled-coil domain.
Fig. 2.
Homodimerization of Mtmr2. (A) A gel filtration elution diagram of His-tagged Mtmr2 protein and SDS/PAGE analysis is shown (Lower). The two peaks elute with molecular masses of ≈170 and 70 kDa, respectively. The first peak contains full-length protein (73 kDa) (Mtmr2 full-length), whereas the protein in the second peak is truncated (Mtmr2-ΔC). (B) Western blot analysis with anti-His antibody of peak fractions (lanes I and II) reveals that the N-terminal His-tag is present in both protein species (FL and ΔC). (C) Analytical ultracentrifugation of full-length Mtmr2 indicates a molecular mass of 140 kDa, which is consistent with Mtmr2 forming a homodimer. Sample purity is shown by SDS/PAGE and staining with Coomassie blue (Inset).
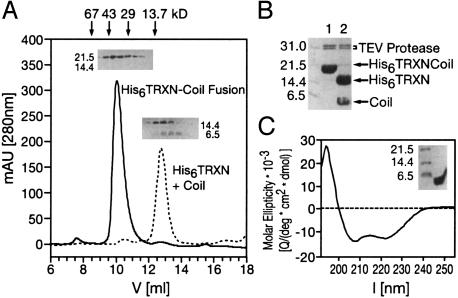
The Coiled-Coil Domain Is Sufficient for Mtmr2 Dimerization. We wanted to confirm that the predicted region in the C terminus of Mtmr2 is indeed a coiled-coil domain, and mediates dimerization. Therefore, amino acids 593–627 of Mtmr2, corresponding to the putative coil-coil domain, were fused to thioredoxin. Gel filtration of the purified fusion protein revealed that it elutes as a dimer (Fig. 3A; His6TRXN-Coil Fusion). Specific cleavage with TEV protease between thioredoxin and the putative coiled-coil domain yielded monomeric thioredoxin and a dimeric coiled-coil domain (Fig. 3A; His6TRXN plus Coil), thus confirming that the coiled-coil domain of Mtmr2 is responsible for the dimerization of the fusion protein (Fig. 3 A and B). CD spectroscopy of the purified coiled-coil domain showed a pronounced α-helical spectrum with minima present at 207 and 222 nm (Fig. 3C). Taken together, our results provide strong experimental evidence for a coiled-coil domain in the C-terminal region of Mtmr2, and show that this domain is responsible for homodimerization.
Fig. 3.
The Mtmr2 coiled-coil domain is a dimerization module. (A) Size-exclusion chromatography of His6TRXNCoil, His6TRXN, and the coiled-coil domain of Mtmr2. The A280 nm trace of the His6TRXNCoil (solid line) is superimposed on the trace of the individual proteins after TEV cleavage (dashed line) marked His6TRXN + Coil. Homogeneity of His6TRXNCoil was confirmed by SDS/PAGE (Inset). The fusion protein elutes at an apparent molecular mass of 37.6 kDa, indicating dimer formation. The TEV cleavage products, His6TRXN and the coiled coil, elute in one peak with an apparent molecular mass of 11.9 kDa. The presence of both proteins was confirmed by SDS/PAGE, using a 20% acrylamide gel (Inset). His6TRXN (13.9 kDa) elutes as a monomer, and the coiled-coil domain (monomer size: 4.3 kDa) elutes as a dimer. (B) TEV protease cleavage of His6TRXNCoil fusion protein. Reaction was complete after 1 h, as shown by SDS/PAGE (lane 1, directly after addition of TEV protease; lane 2, after 1 h incubation). (C) The CD signal (solid line) of the coiled-coil domain shows the spectrum of an α-helix. The coiled-coil domain was purified to homogeneity as confirmed by SDS/PAGE (Inset).
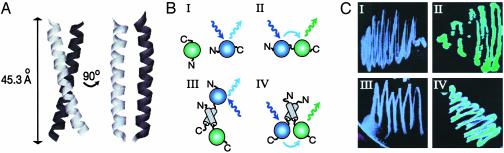
The Orientation of the Coiled Coil in the Dimer. We investigated the orientation of the Mtmr2 coiled-coil module in the dimer by FRET. Our system is based on the simultaneous expression of the Mtmr2 coiled-coil fused to the N terminus of GFP (Coil-GFP), and blue fluorescent protein (BFP) with either a C-terminal (BFP-Coil) or N-terminal coiled-coil domain (Coil-BFP). When BFP is excited at 366 nm, light is emitted by BFP, which coincides with the absorption spectrum of GFP (centered ≈450 nm). Then, if BFP and GFP are in sufficiently close proximity, efficient FRET from BFP to GFP is observed in vivo in E. coli cells, leading to green fluorescence. The theoretically predicted Ro value (i.e., the distance between donor and acceptor where FRET efficiency is half-maximal) for the BFP-GFP donor-acceptor couple is 41 Å (28). The distance between the buried chromophores and the surface of the fluorescent proteins amounts to ≈8 Å (28). The Mtmr2 coiled-coil module extends to 45.3 Å (Fig. 4A). FRET efficiency has R–6 dependence on the distance, R, between the chromophores. Therefore, one would predict that FRET efficiency drops below the presumed detection limit of 10% in our experiments if the fluorescent proteins are located at opposite ends of the coiled-coil dimer (Fig. 4B III). Conversely, if the fluorescent proteins are located at neighboring ends of the coiled-coil dimer, efficient FRET occurs, giving rise to green color (Fig. 4B IV). We observed strong green fluorescence when coexpressing Coil-BFP with Coil-GFP (Fig. 4C IV). In contrast, coexpression of BFP-Coil and Coil-GFP did not result in green fluorescence levels above background (Fig. 4 B III and C III). Our experiments, therefore, suggest that the coiled-coil modules of two Mtmr2 proteins adopt a parallel configuration in the Mtmr2 homodimer.
Fig. 4.
The Mtmr2 coiled-coil domain forms a parallel dimer. (A) A model of the Mtmr2 coiled-coil module, comprising 35 amino acids in two orthogonal views is shown. A dimer of two Mtmr2 coiled-coils extends to ≈45 Å.(B) FRET experiments are shown in a schematic representation. Coexpression of isolated BFP and GFP molecules (I) yields a blue color on excitation at 366 nm (blue arrows). Covalent linking of BFP and GFP (II) leads to FRET, resulting in emission of green light by GFP on excitation at 366 nm (green arrow). (III) Coil-GFP and BFP-Coil adopting a parallel arrangement are shown. Green light is not emitted, due to the location of the fluorophores at opposite ends of the coiled-coil dimer. (IV) Coil-GFP and Coil-BFP forming a parallel coiled-coil dimer are shown. The fluorophores are now located at close proximity, and FRET occurs, resulting in a green color. Results illustrated in I–IV are depicted in C, showing bacteria expressing the respective protein, which is plated on IPTG-containing dYT-agar.
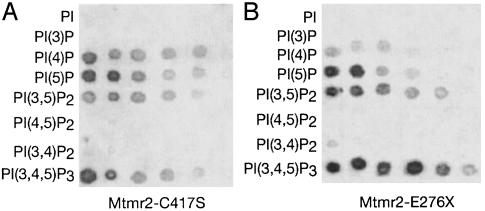
Phosphoinositide Binding of the PH-G Domain. Protein-lipid overlay assays are a suitable tool for the identification of ligands of lipid-binding proteins (29). To prevent dephosphorylation of the spotted lipids in these experiments, we used a GST-Mtmr2 fusion protein with a mutation in the phosphatase domain (Mtmr2-C417S) that abolishes catalytic activity. This protein bound to PI(4)P, PI(5)P, PI(3,5)P2, and PI 3,4,5-trisphosphate [PI(3,4,5)P3], but not to PI, PI(3)P, PI(3,4)P2, and PI 4,5-bisphosphate [PI(4,5)P2] (Fig. 5A). The same specificity was observed when the N-terminal part containing the PH-G domain (Mtmr2-E276X; Fig. 1 A) was used to test its contribution to the phosphoinositide binding (Fig. 5B). The complementary protein lacking the first 266 amino acids (Mtmr2-I267-V643/C417S) and an Mtmr2 variant protein with a mutated PH-G domain (Mtmr2-G103E; Fig. 1 A), both containing the phosphatase, the SID, and the coiled-coil domains, showed drastically reduced binding to phosphoinositides. Furthermore, a protein containing the C-terminal part of Mtmr2 (Mtmr2-S477-V643), including the SID and the coiled-coil domain, did not reveal detectable phosphoinositide binding (see Fig. 7). Using a different approach, Schaletzky et al. (17) recently showed that binding of PI(5)P to MTM1 plays an important role in its regulation. After binding to PI(5)P, MTM1 changes from a monomeric to a heptameric state with higher phosphatase activity. Furthermore, a disease-causing mutation in the PH-G domain of MTM1 reduces the ability of MTM1 to bind to PI(5)P.
Fig. 5.
Protein-lipid overlay assay. (A) Serial dilutions of spotted phospholipids (100–3 pmol in two-fold dilution steps) were incubated with Mtmr2-C417S, a protein without phosphatase activity, to prevent dephosphorylation of the lipids. Mtmr2 binds to PI(4)P, PI(5)P, PI(3,5)P2, and PI(3,4,5)P3, but not to PI, PI(3)P, PI(3,4)P2, or PI(4,5)P2.(B) Phospholipids were incubated with Mtmr2-E276X, which contains only the N-terminal part with the PH-G domain. The same specificity as with the full-length protein is observed. See Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org, for additional blots.
Implications for Human Disease. Membrane association of Mtmr2 is crucial, because the substrates of Mtmr2, PI(3)P, and PI(3,5)P2 are localized in the membrane bilayer. We show that two domains mediate this functionality. Both are necessary for full levels of vacuolar membrane attachment, a PH-G domain, and a coiled-coil domain. Phosphoinositide-binding domains like the PH-G domain are well known adaptors of proteins to membranes (35, 36). The PH-G domain of Mtmr2 binds to PI(3,5)P2, a substrate of Mtmr2, and to PI(5)P, the corresponding product of the dephosphorylation reaction. Substrate binding is a well established mechanism for enzyme targeting. PIKfyve for example, the kinase that produces PI(3,5)P2, binds to PI(3)P with its FYVE domain (37). Binding of the product, as also observed in our assays, might provide a mechanism to concentrate an enzyme at a specific place for a subsequent function. Alternatively, PI(5)P binding to Mtmr2 may lead to structural changes, or PI(5)P may act as a specific allosteric activator as described for MTM1 (17). The significance of the observed binding to PI(3,4,5)P3 and PI(4)P remains to be elucidated.
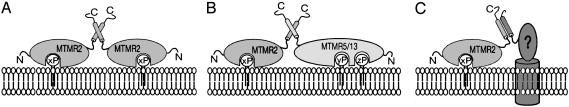
Our findings show that the PH-G domain binds phosphoinositides, but is not sufficient for vacuole binding in hypoosmotically shocked COS cells. The latter appears to require in addition the coiled-coil domain of Mtmr2. Homodimerization mediated by the coiled-coil structure offers a possibility for enhancement of membrane binding by combining two PH-G domains (Fig. 6A). One PH-G domain, then, may be sufficient to bind phosphoinositides, but several domains are required for efficient membrane targeting, and this result is achieved by dimerization. In a recent study, Kim et al. (31) showed that MTMR2 can associate with sbf1/MTMR5, leading to enhanced phosphatase activity. The inactive phosphatase sbf1/MTMR5 provides additional lipid-binding motifs (Fig. 6B) to the complex, thus allowing targeting to specific membrane subdomains. The likely loss-of-function mutations of MTMR2 in CMT4B1 indicate that this protein is functionally nonredundant in peripheral nerves. In this setting, sbf1/MTMR5 may not be the most relevant interaction partner of MTMR2. The identification of the inactive phosphatase sbf2/MTMR13 as the disease-causing gene of CMT4B2, however, indicates that sbf2/MTMR13 may form a crucial complex with MTMR2 in this tissue (Fig. 6B). CMT4B2 and CMT4B1 are clinically indistinguishable, and we have previously shown (16) that loss of phosphatase activity of Mtmr2 is correlated with CMT4B1. Mutations in sbf2/MTMR13 may cause impaired localization of MTMR2 phosphatase activity within a cell, or alter this activity. Inactive adaptors may guide the phosphatases to the correct target in addition, or as an alternative, to competing for the substrate (38). Finally, the coiled-coil domain may also allow for distinction between MTM family members in multimer formation and function. For example, MTMR2 cannot form a heterodimer with MTM1, due to differences in the coiled-coil domain (31). Alternative binding partners could potentially explain the lack of functional redundancy within the MTM family, particularly in muscle in X-linked myotubular myopathy, and in peripheral nerves in CMT4B1, despite MTM1 and MTMR2 sharing substrate specificity of the phosphatase domains, as well as possibly the binding specificity of the PH-G domains.
Fig. 6.
Proposed models for membrane association mediated by the PH-G and coiled-coil domains. (A) Homodimerization provides two PH-G domains, leading to enhanced affinity. xP, specifically bound phosphoinositides (see text). (B) Heterodimerization with sbf1/MTMR5 or hypothetically sbf2/MTMR13 allows interaction with other as-yet-unidentified phosphoinositides (yP and zP). (C) A hypothetical interaction with a potential membrane-bound interaction partner.
We show that the coiled-coil domain of MTMR2 is involved in membrane attachment, in addition to mediating dimerization. This observation may lead to the speculation that there may be other membrane-anchored proteins available as potential interaction partners (Fig. 6C). Phosphoinositides, as well as coiled-coil-containing SNARE proteins and sorting nexins, are important mediators of vesicular trafficking. A recent study (39) revealed that MTMR2 also interacts with neurofilament light-chain protein, providing a link to the cytoskeleton. Future experiments will determine whether, and how, MTMR2 can mediate vesicular trafficking along the cytoskeleton.
Supplementary Material
Acknowledgments
We thank Ned Mantei for reading the manuscript, Matthias Wymann for fruitful discussions, Rene Brunisholz and Peter Kleinert for mass spectroscopy and Edman degradation, Thomas Schalch for help with analytical centrifugation, and Timothy J. Richmond and Rudi Glockshuber for plasmids. This work was supported by the Swiss National Science Foundation and the National Competence Center in Research “Neural Plasticity and Repair.” C.S. was supported by an Ernst Schering Research Foundation postdoctoral fellowship. I.B. was the recipient of a Liebig fellowship from the Fonds der Chemischen Industrie (Germany).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MTM, myotubularin; MTMR, MTM-related protein; CMT, Charcot–Marie–Tooth disease; PH, pleckstrin homology; PH-G, PH-GRAM; PI, phosphatidylinositol; PI 3,5-bisphosphate, PI(3,5)P2; PI 3-phosphate, PI(3)P; TEV, tobacco etch virus; BFP, blue fluorescent protein.
References
- 1.Wishart, M. J. & Dixon, J. E. (2002) Trends Cell Biol. 12, 579–585. [DOI] [PubMed] [Google Scholar]
- 2.Bolino, A., Muglia, M., Conforti, F. L., LeGuern, E., Salih, M. A., Georgiou, D. M., Christodoulou, K., Hausmanowa-Petrusewicz, I., Mandich, P., Schenone, A., et al. (2000) Nat. Genet. 25, 17–19. [DOI] [PubMed] [Google Scholar]
- 3.Houlden, H., King, R. H., Wood, N. W., Thomas, P. K. & Reilly, M. M. (2001) Brain 124, 907–915. [DOI] [PubMed] [Google Scholar]
- 4.Bolino, A., Lonie, L. J., Zimmer, M., Boerkoel, C. F., Takashima, H., Monaco, A. P. & Lupski, J. R. (2001) Neurogenetics 3, 107–109. [DOI] [PubMed] [Google Scholar]
- 5.Nelis, E., Erdem, S., Tan, E., Lofgren, A., Ceuterick, C., De Jonghe, P., Van Broeckhoven, C., Timmerman, V. & Topaloglu, H. (2002) Neuromuscular Disord. 12, 869–873. [DOI] [PubMed] [Google Scholar]
- 6.Berger, P., Young, P. & Suter, U. (2002) Neurogenetics 4, 1–15. [DOI] [PubMed] [Google Scholar]
- 7.Quattrone, A., Gambardella, A., Bono, F., Aguglia, U., Bolino, A., Bruni, A. C., Montesi, M. P., Oliveri, R. L., Sabatelli, M., Tamburrini, O., et al. (1996) Neurology 46, 1318–1324. [DOI] [PubMed] [Google Scholar]
- 8.Senderek, J., Bergmann, C., Weber, S., Ketelsen, U. P., Schorle, H., Rudnik-Schoneborn, S., Buttner, R., Buchheim, E. & Zerres, K. (2003) Hum. Mol. Genet. 12, 349–356. [DOI] [PubMed] [Google Scholar]
- 9.Azzedine, H., Bolino, A., Taieb, T., Birouk, N., Di Duca, M., Bouhouche, A., Benamou, S., Mrabet, A., Hammadouche, T., Chkili, T., et al. (2003) Am. J. Hum. Genet. 72, 1141–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laporte, J., Hu, L. J., Kretz, C., Mandel, J. L., Kioschis, P., Coy, J. F., Klauck, S. M., Poustka, A. & Dahl, N. (1996) Nat. Genet. 13, 175–182. [DOI] [PubMed] [Google Scholar]
- 11.Sarnat, H. B. (1990) Can. J. Neurol. Sci. 17, 109–123. [DOI] [PubMed] [Google Scholar]
- 12.Laporte, J., Biancalana, V., Tanner, S. M., Kress, W., Schneider, V., Wallgren-Pettersson, C., Herger, F., Buj-Bello, A., Blondeau, F., Liechti-Gallati, S. & Mandel, J. L. (2000) Hum. Mutat. 15, 393–409. [DOI] [PubMed] [Google Scholar]
- 13.Taylor, G. S., Maehama, T. & Dixon, J. E. (2000) Proc. Natl. Acad. Sci. USA 97, 8910–8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blondeau, F., Laporte, J., Bodin, S., Superti-Furga, G., Payrastre, B. & Mandel, J. L. (2000) Hum. Mol. Genet. 9, 2223–2229. [DOI] [PubMed] [Google Scholar]
- 15.Walker, D. M., Urbe, S., Dove, S. K., Tenza, D., Raposo, G. & Clague, M. J. (2001) Curr. Biol. 11, 1600–1605. [DOI] [PubMed] [Google Scholar]
- 16.Berger, P., Bonneick, S., Willi, S., Wymann, M. & Suter, U. (2002) Hum. Mol. Genet. 11, 1569–1579. [DOI] [PubMed] [Google Scholar]
- 17.Schaletzky, J., Dove, S. K., Short, B., Lorenzo, O., Clague, M. J. & Barr, F. A. (2003) Curr. Biol. 13, 504–509. [DOI] [PubMed] [Google Scholar]
- 18.Corvera, S., D'Arrigo, A. & Stenmark, H. (1999) Curr. Opin. Cell Biol. 11, 460–465. [DOI] [PubMed] [Google Scholar]
- 19.Odorizzi, G., Babst, M. & Emr, S. D. (2000) Trends Biochem. Sci. 25, 229–235. [DOI] [PubMed] [Google Scholar]
- 20.Gaullier, J. M., Simonsen, A., D'Arrigo, A., Bremnes, B., Stenmark, H. & Aasland, R. (1998) Nature 394, 432–433. [DOI] [PubMed] [Google Scholar]
- 21.Gillooly, D. J., Morrow, I. C., Lindsay, M., Gould, R., Bryant, N. J., Gaullier, J. M., Parton, R. G. & Stenmark, H. (2000) EMBO J. 19, 4577–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sbrissa, D., Ikonomov, O. C. & Shisheva, A. (1999) J. Biol. Chem. 274, 21589–21597. [DOI] [PubMed] [Google Scholar]
- 23.Dove, S. K., Cooke, F. T., Douglas, M. R., Sayers, L. G., Parker, P. J. & Michell, R. H. (1997) Nature 390, 187–192. [DOI] [PubMed] [Google Scholar]
- 24.Gary, J. D., Wurmser, A. E., Bonangelino, C. J., Weisman, L. S. & Emr, S. D. (1998) J. Cell Biol. 143, 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odorizzi, G., Babst, M. & Emr, S. D. (1998) Cell 95, 847–858. [DOI] [PubMed] [Google Scholar]
- 26.Lebowitz, J., Lewis, M. S. & Schuck, P. (2002) Protein Sci. 11, 2067–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gill, S. C. & von Hippel, P. H. (1989) Anal. Biochem. 182, 319–326. [DOI] [PubMed] [Google Scholar]
- 28.Philipps, B., Hennecke, J. & Glockshuber, R. (2003) J. Mol. Biol. 327, 239–249. [DOI] [PubMed] [Google Scholar]
- 29.Dowler, S., Kular, G. & Alessi, D. R. (2002) Sci. STKE 129, PL6. [DOI] [PubMed] [Google Scholar]
- 30.Laporte, J., Blondeau, F., Gansmuller, A., Lutz, Y., Vonesch, J. L. & Mandel, J. L. (2002) J. Cell Sci. 115, 3105–3117. [DOI] [PubMed] [Google Scholar]
- 31.Kim, S. A., Vacratsis, P. O., Firestein, R., Cleary, M. L. & Dixon, J. E. (2003) Proc. Natl. Acad. Sci. USA 100, 4492–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sbrissa, D., Ikonomov, O. C., Deeb, R. & Shisheva, A. (2002) J. Biol. Chem. 277, 47276–47284. [DOI] [PubMed] [Google Scholar]
- 33.Jones, D. R., Gonzalez-Garcia, A., Diez, E., Martinez, A. C., Carrera, A. C. & Merida, I. (1999) J. Biol. Chem. 274, 18407–18413. [DOI] [PubMed] [Google Scholar]
- 34.Wolf, E., Kim, P. S. & Berger, B. (1997) Protein Sci. 6, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hurley, J. H. & Misra, S. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 49–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemmon, M. A. (2003) Traffic 4, 201–213. [DOI] [PubMed] [Google Scholar]
- 37.Sbrissa, D., Ikonomov, O. C. & Shisheva, A. (2002) J. Biol. Chem. 277, 6073–6079. [DOI] [PubMed] [Google Scholar]
- 38.Cui, X., De Vivo, I., Slany, R., Miyamoto, A., Firestein, R. & Cleary, M. L. (1998) Nat. Genet. 18, 331–337. [DOI] [PubMed] [Google Scholar]
- 39.Previtali, S. C., Zerega, B., Sherman, D. L., Brophy, P. J., Dina, G., King, R. H., Salih, M. M., Feltri, L., Quattrini, A., Ravazzolo, R., et al.. (2003) Hum. Mol. Genet. 12, 1713–1723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.