Abstract
Understanding mammalian spermatozoan development and the events surrounding fertilization has grown slowly, in part because of uncertainty about the number and identity of the cellular components involved. Determination of those transcripts expressed specifically by germ cells should provide an inclusive list of probable critical proteins. Here, total mouse testis transcript profiles were trimmed of transcripts found in cultures enriched in Sertoli or interstitial cells to yield a germ cell-enriched transcript profile. Monitoring of changes of this profile in the developing testis identified 1,652 genes whose transcript abundance increased markedly coincident with the onset of meiosis. Remarkably, 351 of these genes (≈20%) appear to be expressed only in the male germline. Germ cell-specific transcripts are much less common earlier in testis development. Further analysis of the UniGene EST database coupled with quantitative PCR indicates that ≈4% of the mouse genome is dedicated to expression in postmeiotic male germ cells. Most or many of the protein products of these transcripts are probably retained in mature spermatozoa. Targeted disruption of 19 of these genes has indicated that a majority have roles critical for normal fertility. Thus, we find an astonishing number of genes expressed specifically by male germ cells late in development. This extensive group provides a plethora of potential targets for germ cell-directed contraception and a staggering number of candidate proteins that could be critical for fertilization.
The underlying basis of fertilization in mammals continues to be poorly understood. Contributing factors include: (i) paucity of eggs, (ii) heterogeneity of sperm cell populations, and (iii) lack of information on the number and nature of molecules that serve as potential players (1). Unlike many invertebrates and some lower vertebrates, where sperm cell behavior is highly synchronous, at any given moment, only a fraction of mammalian sperm cells seem to respond to components of the egg extracellular matrix, to chemoattractants, or to other signaling molecules (2, 3). Thus, sensitive functional bioassays have been difficult or impossible to establish. We have attempted to overcome this problem through the identification of all proteins potentially expressed in the spermatozoon. To make a list of potential molecules involved in fertilization more experimentally palatable, we also have restricted an initial list of potential players to proteins expressed exclusively by male germ cells. Additionally, because proteins involved specifically in fertilization are likely expressed late in spermatogenesis, we also have restricted our analysis to gene products expressed at meiosis or later. To this end, we initially generated signal peptide trap cDNA libraries from spermatid-enriched cell populations and successfully identified a number of important sperm proteins such as CatSper2 (1). On the basis of these screens, we estimated that >200 unique sperm proteins existed; even this was somewhat surprising, given that “leaky transcription” has long been attributed to the testis, suggesting that a large number of sperm transcripts might be shared with other cells (4).
Recent high-throughput efforts by other groups resulted in the identification of a few new testis-specific transcripts. For example, 19 novel premeiotic male germ cell-specific genes were identified through the use of cDNA subtraction (5), and use of a differential display reverse transcriptase PCR resulted in the identification of other genes expressed during spermatogenesis (6). Through the use of cDNA microarrays in mouse or human, four (7) and 42 (8) novel genes potentially involved in the regulation of spermatogenesis have been identified. Even though these approaches identified a number of new genes potentially important in spermatogenesis, the studies failed to determine the potential number or identity of genes specifically expressed in the male germline that serve as likely participants in germ cell development and fertilization.
We analyzed gene expression in the mouse testis from day 1 to adult by using the Affymetrix (Santa Clara, CA) Mouse U74v2 oligonucleotide array set, generating profiles for ≈20,000 genes expressed in the testis. The results provide an invaluable depository of gene transcripts that could be specifically involved in spermatogenesis and fertilization. In mining the microarray data, we identified 1,652 gene transcripts whose expression increased coincident with or after meiosis. Through further analysis (UniGene, www.ncbi.nlm.nih.gov/UniGene), we estimate that >2,300 genes (≈4% of the mouse genome) are dedicated to male germ cell-specific transcripts, >99% of which are first expressed during or after meiosis. That so many germ cell-specific genes are expressed late in development provides a vast number of candidates for contraceptive targeting as well as a large number of potential participants in the process of fertilization.
Methods
Biological Material and RNA Isolation. Testes were individually collected from C57BL/6 mice (Harlan Breeders, Indianapolis) at days 1, 4, 8, 11, 14, 18, 21, 26, 29, and 60 (adult) postpartum and immediately frozen in liquid nitrogen. Total RNA was isolated from individual testis by using RNA Stat-60 (Tel-Test, Friendswood, TX), according to the manufacturer's protocol.
Microarray Processing. RNA labeling for subsequent microarray analysis was performed on three 1-day-old animals, two 4-day-olds, two 8-day-olds, two 11-day-olds, and one animal each for days 14, 18, 21, 26, 29, and adult. Five micrograms of total RNA was reverse transcribed into double-stranded cDNA containing a T7 promoter by using the RiboAmp RNA Amplification Kit (Arcturus, Mountain View, CA), in vitro transcribed, and biotin-labeled by using the BioArray HighYield RNA Transcript Labeling Kit (Enzo Life Sciences, Farmingdale, NY). RNA from the somatic cell samples was amplified and labeled from 50 ng of starting material by using two rounds of amplification. The amplified target RNA (aRNA) was purified by using an RNeasy Mini column (Qiagen, Valencia, CA). Twenty micrograms of aRNA of each sample were fragmented for hybridization to each microarray. Affymetrix Murine Genome U74v2 A, B, and C arrays were used for all samples. The arrays were hybridized and processed according to the manufacturer's specifications.
Data Analysis and Clustering. Results were analyzed by using the Affymetrix microarray suite (mas), Ver. 5.0. Signals on each chip were scaled to a mean intensity of 250. All sample comparisons were performed in genespring, Ver. 5.1 (Silicon Genetics, Redwood City, CA). Triplicates and duplicates were averaged, and all data were normalized to day 1. For subsequent data analysis, all genes were filtered as follows: the transcripts had to have a present call (assigned by mas, τ = 0.015) and a minimum signal intensity of 75 in at least one time point. Transcripts also had to be up- or down-regulated >3-fold in at least one time point compared with the day 1 samples. To enrich for germ cell genes, transcripts also had to be absent in a testicular Sertoli cell sample (9) and in an interstitial cell sample (10). Genes fulfilling these requirements were clustered in arrayminer 4.0 (Optimal Design, Brussels) by using the Gaussian clustering model, an algorithm that takes cluster variance into account and has the ability to recognize outliers (www.optimaldesign.com/Download/ArrayMiner/AM2whitepaper.pdf). Annotations of all filtered transcripts were updated by using Affymetrix netaffx (www.netaffx.com), based on the March 17, 2003 quarterly annotation update (11). All corresponding UniGene clusters were then screened for reported tissue expression by using the National Center for Biotechnology Information UniGene database (Build no. 122 Mus musculus, May 1, 2003).
Quantitative Real-Time PCR. Real-time PCR analysis was performed by using the abi prism 7900HT Sequence Detection System (Applied Biosystems). All primers were designed in primer express, Ver. 2.0 by using the mgb primer design method, and runs were analyzed by using sequence detection system, Ver. 2.1.
One microgram of total RNA of each sample was reverse transcribed in a 20-μl reaction by using 150 ng of random primers and the SuperScript II reagents (Invitrogen). Samples were diluted 1:10, and 0.5 μl was used for the PCR reaction. PCR was performed by using the SYBR Green PCR Master Mix (Applied Biosystems). For expression ratio verifications, all samples were normalized to the 18S signal. Ranges of fold changes were calculated according to the Applied Biosystems Comparative CT method by using standard deviations (n = 2).
In Situ Hybridization. Gene-specific PCR products of 200–350 bp were generated and linked to a T7-promoter-containing fragment by using the Lig'nScribe No-Cloning Promoter Addition Kit (Ambion, Austin, TX). Sense and antisense constructs were then generated by PCR by using a primer complementary to the T7 promoter fragment and the 3′ or 5′ gene primer, respectively. PCR products were gel purified and in vitro transcribed by using the MAXIscript T7 Kit (Ambion) incorporating S35-UTP (Amersham Pharmacia Biosciences). Probes were subsequently DNase treated and purified by using NucAway Spin Columns (Ambion). In situ hybridizations were performed by the Molecular Pathology Core Facility at the University of Texas Southwestern Medical Center, as described (12).
Results and Discussion
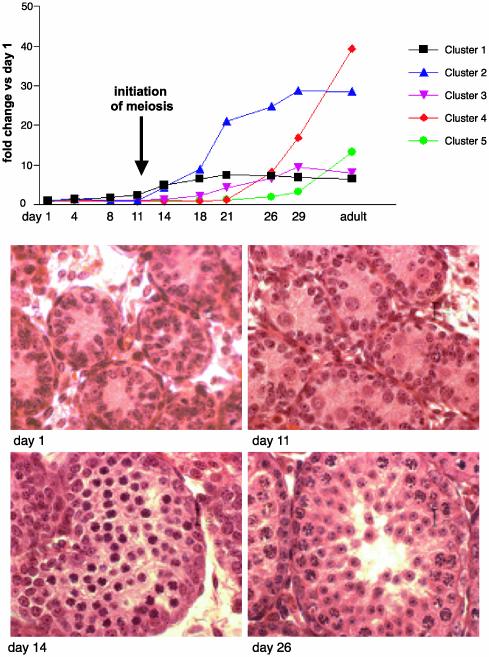
Testes from C57BL/6 mice were collected at various days ranging from day 1 postpartum to adult and the mRNA expression profiles determined (Affymetrix Mouse U74v2 oligonucleotide array set). The complete data set can be found as Data Set 1, which is published as supporting information on the PNAS web site, www.pnas.org. It can also be queried in the NCBI Gene Expression Omnibus (GEO) repository (www.ncbi.nlm.nih.gov/geo), GEO accession no. GSE640. See also Supporting Methods, which is published as supporting information on the PNAS web site. For the first few days after birth, the seminiferous tubules principally contain gonocytes, spermatogonia, and Sertoli cells. They are rapidly outnumbered by differentiating germ cells as the animal ages. Spermatocytes appear around days 11–14 and spermatids after about day 21 of age in the mouse.
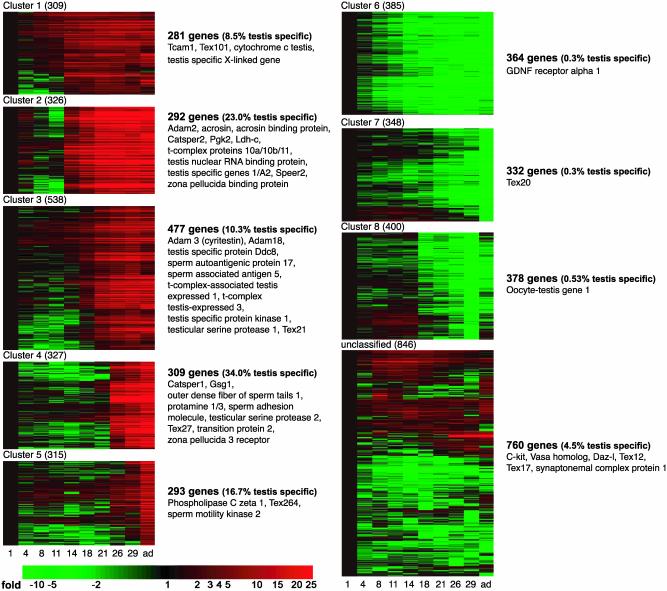
A total of 21,374 transcripts were identified as expressed in the developing mouse testis (a “present” call is as defined in Methods). To more clearly characterize a germ cell population of transcripts, we separately screened for genes found in a cultured testis somatic cell preparation containing principally Sertoli cells prepared as described (9). We also determined the genes expressed in an interstitial cell population prepared as described (10). The results from these two somatic cell populations were subtracted from those for whole testis to yield a germ cell-enriched population. Three thousand seven hundred ninety-four transcripts (3,486 different genes) were changed at least 3-fold compared with day 1 of age, and of these, 2,245 represented uncharacterized genes. Transcript expression profiles were clustered into eight distinct patterns; in five clusters, the expression of genes substantially increased coincident with the appearance of spermatocytes or spermatids. The pattern of gene expression in the other three clusters was highest during times when the spermatogonial population dominated the testis (Fig. 1). Although the transcript profiles of male germ cells early in development are critical to an understanding of male germline stem cells and the early steps of commitment to differentiation, we concentrated on the five clusters where transcript expression markedly increased late in development. If these transcripts were expressed by the germ cells, these would be the best candidate genes for a role in fertilization. The average patterns of expression for the genes found in clusters 1–5 are shown in Fig. 2. Meiosis begins in the mouse at about day 11, and elevations in transcript expression are evident starting on or after this day. It should be noted that the relative levels to which transcript expression increased were considerably different between each of the clusters, with genes in clusters 2 (expression starting in pachytene spermatocytes) and 4 (expression in spermatids) showing the highest degree of induction.
Fig. 1.
Heat maps showing the eight clusters of transcripts that change coincident with the formation of meiotic and postmeiotic male germ cells and the unclassified genes. Each row of the heat map represents a gene, and each column represents a time point in development (as labeled at the bottom). The color saturation represents differences in gene expression compared with day 1. Red indicates an increase in gene expression, whereas green indicates a decrease. The genes listed next to the heat maps are examples of known genes found in the respective clusters.
Fig. 2.
Testis developmental profiles for clusters 1–5. (Upper) The relative gene transcript abundance in testis from mice at ages days 4, 8, 11, 14, 18, 21, 26, and 29 and adult compared with testis at day 1 of age. The values are the mean relative abundance (compared with day 1) for all transcripts found in each of the clusters. Note the marked increase in relative transcript abundance for cluster 4 coincident with formation of spermatids. (Lower) Hematoxylin/eosin staining of sections of testis from the same mice at ages days 1, 11, 14, and 26 to show germ cell differentiation at each age.
The expression patterns of various transcripts are consistent with the behavior of these genes described in the literature (Fig. 1 lists several known genes for each cluster). Examples of well-characterized genes include lactate dehydrogenase C and phosphoglycerate kinase 2, which are known to be initially transcribed during meiosis; as expected then, they appear in cluster 2. Likewise, protamine 1 and transition protein 2 are initially expressed in spermatids and therefore as expected appear in cluster 4. An interesting aspect of the analysis is that transcription of CatSper 1 (cluster 4) and CatSper 2 (cluster 2), which may form subunits of the same channel, is not initiated at the same time during spermatogenesis.
Of the 1,652 genes found in the five clusters, where gene expression increased relative to day 1, ≈17.8% were represented by transcripts that may be testis-specific on the basis of the UniGene database (two or more sequence entries; no tissue other than testis) (Table 1; Tables 3 and 4, which are published as supporting information on the PNAS web site). We required at least two sequence entries in our analysis to help ensure that an EST sequence entered was real. The addition of transcripts present in one or two other tissues would increase the percentage to 28.9%. In contrast, in the other three clusters, only 0.4% of the genes appeared as testis-specific transcripts, suggesting that only a few of the genes in male germline stem cells or spermatogonia are unique to the testis. The known or named testis-specific genes are given in Table 3.
Table 1. The number of genes in each of the eight clusters that show apparent testis-specific expression.
| Cluster | Total | Named genes | Genes with unknown/unassigned function | Apparent testis specific* | Only one other tissue | Only two other tissues |
|---|---|---|---|---|---|---|
| 1 | 323 | 106 | 217 | 24 (7.4%) | 18 (5.6%) | 9 (2.8%) |
| 2 | 308 | 116 | 192 | 68 (22.1%) | 32 (10.4%) | 19 (6.4%) |
| 3 | 538 | 196 | 342 | 51 (9.5%) | 36 (6.0%) | 7 (1.3%) |
| 4 | 312 | 115 | 197 | 105 (33.7%) | 29 (9.3%) | 13 (4.2%) |
| 5 | 317 | 127 | 190 | 49 (15.5%) | 18 (5.7%) | 6 (1.9%) |
| 6 | 480 | 197 | 283 | 1 (0.2%) | 1 (0.4%) | 2 (0.4%) |
| 7 | 432 | 155 | 277 | 1 (0.2%) | 0 | 0 |
| 8 | 571 | 161 | 410 | 3 (0.5%) | 2 (0.4%) | 1 (0.2%) |
| Unclassified | 910† | 328 | 582 | 36 (4.1%) | 15 (1.7%) | 3 (0.3%) |
Expression data are based on tissue distribution information in the UniGene database.
The 910 gene transcripts in the unclassified category did not show a pattern of expression that fit the other eight clusters.
To determine whether transcripts reported only for the testis actually reflected testis-specific expression, 50 genes with unassigned function (principally from cluster 4) were examined for expression in spleen, liver, testis, brain, ovary, thymus, kidney, heart, embryo, and lung. Of the 50 transcripts, 46 yielded signals in the testis only (Table 4). Thus, it is predicted that >90% of the transcripts now entered in the UniGene database as testis-only (at least two sequence entries), in fact, are testis-specific.
To then determine whether all such genes were also germ cell-specific (likely given the subtraction against testicular somatic cells), we performed in situ hybridization on 11 (randomly chosen) genes with unassigned function from clusters 1–5. All transcripts were expressed specifically by spermatocytes or spermatids, strongly suggesting that all of the testis-specific genes identified above are germ cell-specific (Table 2).
Table 2. Meiotic and postmeiotic germ cells express the transcripts found in clusters 1-5.
| Gene identifier | Probe identification | Cluster | Germ cell stage of expression* |
|---|---|---|---|
| Mm.56002 | 163837_at | 1 | Spermatocytes/spermatids |
| Mm.87419 | 168262_at | 3 | Round spermatids |
| Mm.59283 | 129740_at | 3 | Spermatids (steps 1-14) |
| Mm.45611 | 164171_at | 4 | Spermatids (steps 1-12) |
| Mm.84522 | 167943_f_at | 4 | Spermatids (steps 1-16) |
| Mm.46121 | 165889_f_at | 4 | Spermatids (steps 7-14) |
| Mm.45306 | 165824_at | 4 | Spermatids (steps 5-14) |
| Mm.46151 | 165900_f_at | 4 | Spermatids (steps 5-14) |
| Mm.45302 | 165822_f_at | 4 | Spermatids (steps 7-12) |
| Mm.56501 | 166073_at | 4 | Spermatids (steps 7-14) |
| Mm.72970 | 166193_at | 5 | Spermatids (steps 7-16) |
All 11 genes have no assigned function.
In situ hybridization was performed as defined in Methods.
Intrigued by the large number of testis-specific genes in clusters 1–5, we then examined the UniGene database to determine whether a significant number of other transcripts not represented on the arrays would also be entered as testis-specific. As of May 2003, there were 62,692 entries with a cluster size of at least two sequence entries. Again, we limited our survey to at least two entries to help ensure that a given EST entry represented a real transcript sequence. Of these, 2,931 gene transcripts were reported only for the testis (7% with known functions, 68% with a predicted ORF but unknown or unassigned function, and 25% as ESTs). The question then arose whether these apparent testis-specific gene transcripts were also elevated during or after meiosis. Therefore, we arbitrarily chose 12 of the 2,931 (the 12 were genes not represented on the microarrays). Quantitative PCR showed that for 11 of 12, expression was significantly elevated after day 11, again suggesting expression in spermatocytes or spermatids (Table 5, which is published as supporting information on the PNAS web site). Additionally, a screen of testis-specific genes subtracted by using the somatic cell populations suggests that <10% of all testis-specific genes on the microarray (independent of cluster) are found in Sertoli or interstitial cells. Thus, a vast majority of the 2,931 gene sequences appear to represent meiotic or postmeiotic germ cell-specific transcripts. Subtraction of the low numbers of testis-specific genes that appear early in spermatogenesis, those confined to Sertoli or interstitial cells, and the ≈10% of genes expressed in tissues outside the testis, leads to the conclusion that ≈2,375 of the 62,692 genes represented in UniGene (at least two sequence entries) are expressed specifically in meiotic and postmeiotic male germ cells. This represents ≈3.8% of the genome. The number of entries in the mouse UniGene database appears to adequately reflect the actual number of mouse genes, given that the RIKEN Genome Exploration Research Group predicts ≈70,000 mouse genes (13). Although some EST entries will represent identical full-length transcripts, there is no apparent reason that the testis would have more such redundant entries than other tissues.
Do genes in the five clusters identified here serve as potential targets for male germ cell-directed contraception? It is reasonable to assume that some or most are intimately involved in fertilization, but are they critical components? A search of the literature coupled with our own studies using targeted disruption of germ cell-specific genes suggests a high number of these genes will be required for fertility. We have disrupted three genes expressed late in germ cell development, a sperm-specific apparent sodium hydrogen exchanger (NHE; D. Wang and D.L.G., unpublished results), a cation channel (CatSper2; T. A. Quill and D.L.G., unpublished results; ref. 1) and an aminophospholipid transporter (L. Wang and D.L.G, unpublished results); in all cases normal sperm numbers were found in the epididymis, and sperm morphology was indistinguishable from that of wild-type spermatozoa, yet two of the three genes (CatSper 2; NHE) appeared absolutely required for fertility. These results, coupled with a search of the literature for other genes in the five clusters, show that 17 of 19 germ cell-specific genes eliminated by homologous recombination are essential for normal male fertility (Table 6, which is published as supporting information on the PNAS web site). The genes include proteases or proteins that interact with proteases (14–17), protein kinases, or proteins that interact with protein kinases (18), transcription factors (19), proteins associated with chromatin (20–22), channels or transporters (1, 23), mitochondrial-associated proteins (24, 25), adhesion proteins (26), RNA polymerases (27), and genes involved in the interaction between spermatids and Sertoli cells (28). More than 50% of these genes result in a complete loss of fertility when disrupted.
The apparent asynchronous behavior of mammalian spermatozoa coupled with the paucity of female gametes (eggs) prompted many to use invertebrate animal models to understand the molecular basis of fertilization (29, 30). Functional bioassays allowed identification of important signaling components in the invertebrate sperm cells, and the subsequent identification of homologous genes in the mammal seemed a strong approach for unraveling fertilization mechanisms in the mammal. However, in many or most cases, homologues of the signaling components specifically important for fertilization in the invertebrate were not found in mammalian germ cells (29). In retrospect, this is not surprising, given the rapid evolution of fertilization-specific proteins (31). These negative findings indicated that direct experimental approaches in the mammal were likely required. However, the continued use of functional bioassays to understand mammalian fertilization also appeared an antiquated and generally unsuccessful means of approach. Of various more global and unbiased approaches, genetic screens for fertilization-defective mutations could provide the strongest method. However, the implementation of a saturation-mutagenesis screen in the mouse to search for such mutations seems untenable at this point in time, given the number of personnel and the costs associated with such a labor-intensive screen, at least within individual laboratories. Among alternatives are the use of proteomics, in silico subtraction, microarrays and signal peptide trapping to identify candidate gene targets. Such targets could then be mutated (disrupted) to determine whether they are critical for fertilization.
A number of underlying basic assumptions would further reduce the list of gene candidates to palatable numbers. First, it is a reasonable assumption that sperm-specific gene products play an important role in male germ cell function. Second, transcripts that markedly increase in the haploid cell are more likely to play a significant role in the fertilization process. Third, proteins found on the cell surface of gametes are more likely to play a role in germ cell interactions with their environment and thus in fertilization. The use of the yeast-based signal peptide trap method (32) offers the opportunity to produce spermatid (haploid cell)-enriched cDNA libraries from which mRNA-encoded cell surface proteins can be identified. Through the use of such libraries, we estimated that the number of sperm-specific membrane proteins is >200 (1). This seemingly large number of sperm-specific proteins led us to enlarge our searches for sperm-specific transcripts through the use of microarrays to examine testis expression profiles after subtraction of Sertoli and interstitial cell culture transcripts.
Although tissues such as heart contain transcripts common to many other tissues (33), a previous analysis of 49 tissues demonstrated that testis contained more outliers than other tissues, supportive of a conclusion that testis might contain a higher number of cell-specific transcripts than found in most other tissues (34). Olfactory tissue, like testis, also contains a large number of specifically expressed transcripts, in this case a large family of odorant receptors, where it has been suggested there are >1,500 such receptors in the mouse (35).
Our analysis indicates that a rather staggering number of germ cell-specific genes exist as possibly important participants in fertilization. These genes are potential targets for male-directed contraception and are candidates to explain many cases of male infertility. On the basis of the genes already disrupted, the inhibition of many of these genes seems likely to lead to male infertility. Because the genes are seemingly expressed exclusively in germ cells, inhibitory drugs would be expected to have few side effects. That a vast majority of germ cell-specific transcripts are expressed only late in development (during or after meiosis) also suggests that most of these protein products are likely present in mature spermatozoa.
Supplementary Material
Acknowledgments
We thank Dr. James Richardson for assistance with in situ hybridization and Karen M. Chapman for technical assistance, and we acknowledge the contributions of the University of Texas Southwestern Medical Center Microarray Facility. This research was supported by the Howard Hughes Medical Institute, the Cecil H. and Ida Green Center for Reproductive Biology Sciences, and National Institutes of Health Grant HD 36022.
References
- 1.Quill, T. A., Ren, D., Clapham, D. E. & Garbers, D. L. (2001) Proc. Natl. Acad. Sci. USA 98, 12527–12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee, M. A., Kopf, G. S. & Storey, B. T. (1987) Biol. Reprod. 36, 617–627. [DOI] [PubMed] [Google Scholar]
- 3.Eisenbach, M. (1999) Dev. Genet. 25, 87–94. [DOI] [PubMed] [Google Scholar]
- 4.Garbers, D. & Quill, T. (2000) in The Testis: From Stem Cell to Sperm Function, ed. Goldberg, E. (Springer, New York), pp. 186–197.
- 5.Wang, P. J., McCarrey, J. R., Yang, F. & Page, D. C. (2001) Nat. Genet. 27, 422–426. [DOI] [PubMed] [Google Scholar]
- 6.Anway, M. D., Li, Y., Ravindranath, N., Dym, M. & Griswold, M. D. (2003) J. Androl. 24, 173–184. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka, K., Tamura, H., Tanaka, H., Katoh, M., Futamata, Y., Seki, N., Nishimune, Y. & Hara, T. (2002) Dev. Biol. 246, 466–479. [DOI] [PubMed] [Google Scholar]
- 8.Sha, J., Zhou, Z., Li, J., Yin, L., Yang, H., Hu, G., Luo, M., Chan, H. C. & Zhou, K. (2002) Mol. Hum. Reprod. 8, 511–517. [DOI] [PubMed] [Google Scholar]
- 9.Hamra, F. K., Gatlin, J., Chapman, K. M., Grellhesl, D. M., Garcia, J. V., Hammer, R. E. & Garbers, D. L. (2002) Proc. Natl. Acad. Sci. USA 99, 14931–14936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mather, J. P., Saez, J. M. & Haour, F. (1981) Steroids 38, 35–44. [DOI] [PubMed] [Google Scholar]
- 11.Liu, G., Loraine, A. E., Shigeta, R., Cline, M., Cheng, J., Valmeekam, V., Sun, S., Kulp, D. & Siani-Rose, M. A. (2003) Nucleic Acids Res. 31, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shelton, J. M., Lee, M. H., Richardson, J. A. & Patel, S. B. (2000) J. Lipid Res. 41, 532–537. [PubMed] [Google Scholar]
- 13.Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. (2002) Nature 420, 563–573.12466851 [Google Scholar]
- 14.Adham, I. M., Nayernia, K. & Engel, W. (1997) Mol. Reprod. Dev. 46, 370–376. [DOI] [PubMed] [Google Scholar]
- 15.Cho, C., Bunch, D. O., Faure, J. E., Goulding, E. H., Eddy, E. M., Primakoff, P. & Myles, D. G. (1998) Science 281, 1857–1859. [DOI] [PubMed] [Google Scholar]
- 16.Shamsadin, R., Adham, I. M., Nayernia, K., Heinlein, U. A., Oberwinkler, H. & Engel, W. (1999) Biol. Reprod. 61, 1445–1451. [DOI] [PubMed] [Google Scholar]
- 17.Uhrin, P., Dewerchin, M., Hilpert, M., Chrenek, P., Schofer, C., Zechmeister-Machhart, M., Kronke, G., Vales, A., Carmeliet, P., Binder, B. R., et al. (2000) J. Clin. Invest. 106, 1531–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miki, K., Willis, W. D., Brown, P. R., Goulding, E. H., Fulcher, K. D. & Eddy, E. M. (2002) Dev. Biol. 248, 331–342. [DOI] [PubMed] [Google Scholar]
- 19.Pearse, R. V., II, Drolet, D. W., Kalla, K. A., Hooshmand, F., Bermingham, J. R., Jr., & Rosenfeld, M. G. (1997) Proc. Natl. Acad. Sci. USA 94, 7555–7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adham, I. M., Nayernia, K., Burkhardt-Gottges, E., Topaloglu, O., Dixkens, C., Holstein, A. F. & Engel, W. (2001) Mol. Hum. Reprod. 7, 513–520. [DOI] [PubMed] [Google Scholar]
- 21.Cho, C., Willis, W. D., Goulding, E. H., Jung-Ha, H., Choi, Y. C., Hecht, N. B. & Eddy, E. M. (2001) Nat. Genet. 28, 82–86. [DOI] [PubMed] [Google Scholar]
- 22.Yu, Y. E., Zhang, Y., Unni, E., Shirley, C. R., Deng, J. M., Russell, L. D., Weil, M. M., Behringer, R. R. & Meistrich, M. L. (2000) Proc. Natl. Acad. Sci. USA 97, 4683–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren, D., Navarro, B., Perez, G., Jackson, A. C., Hsu, S., Shi, Q., Tilly, J. L. & Clapham, D. E. (2001) Nature 413, 603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nayernia, K., Adham, I. M., Burkhardt-Gottges, E., Neesen, J., Rieche, M., Wolf, S., Sancken, U., Kleene, K. & Engel, W. (2002) Mol. Cell. Biol. 22, 3046–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narisawa, S., Hecht, N. B., Goldberg, E., Boatright, K. M., Reed, J. C. & Millan, J. L. (2002) Mol. Cell. Biol. 22, 5554–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baba, D., Kashiwabara, S., Honda, A., Yamagata, K., Wu, Q., Ikawa, M., Okabe, M. & Baba, T. (2002) J. Biol. Chem. 277, 30310–30314. [DOI] [PubMed] [Google Scholar]
- 27.Kashiwabara, S., Noguchi, J., Zhuang, T., Ohmura, K., Honda, A., Sugiura, S., Miyamoto, K., Takahashi, S., Inoue, K., Ogura, A., et al. (2002) Science 298, 1999–2002. [DOI] [PubMed] [Google Scholar]
- 28.Mannan, A. U., Nayernia, K., Mueller, C., Burfeind, P., Adham, I. M. & Engel, W. (2003) Biol. Reprod. 69, 788–796. [DOI] [PubMed] [Google Scholar]
- 29.Garbers, D. L. (1992) Cell 71, 1–4. [DOI] [PubMed] [Google Scholar]
- 30.Mengerink, K. J., Moy, G. W. & Vacquier, V. D. (2002) J. Biol. Chem. 277, 943–948. [DOI] [PubMed] [Google Scholar]
- 31.Swanson, W. J. & Vacquier, V. D. (2002) Nat. Rev. Genet. 3, 137–144. [DOI] [PubMed] [Google Scholar]
- 32.Klein, R. D., Gu, Q., Goddard, A. & Rosenthal, A. (1996) Proc. Natl. Acad. Sci. USA 93, 7108–7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bortoluzzi, S., d'Alessi, F. & Danieli, G. A. (2000) J. Mol. Cell Cardiol. 32, 1931–1938. [DOI] [PubMed] [Google Scholar]
- 34.Kadota, K., Nishimura, S., Bono, H., Nakamura, S., Hayashizaki, Y., Okazaki, Y. & Takahashi, K. (2003) Physiol. Genomics 12, 251–259. [DOI] [PubMed] [Google Scholar]
- 35.Young, J. M. & Trask, B. J. (2002) Hum. Mol. Genet. 11, 1153–1160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.