Abstract
Positive Darwinian selection on advantageous point substitutions has been demonstrated in many genes. We here provide empirical evidence, for the first time, that positive selection can also act on insertion/deletion (indel) substitutions in the evolution of a protein. CATSPER1 is a voltage-gated calcium channel found exclusively in the plasma membrane of the mammalian sperm tail and it is essential for sperm motility. We determined the DNA sequences of the first exon of the CATSPER1 gene from 15 primates, which encodes the intracellular N terminus region of ≈400 aa. These sequences exhibit an excessively high frequency of indels. However, all indels have lengths that are multiples of 3 nt (3n indels) and do not disrupt the ORF. The number of indel substitutions per site per year in CATSPER1 is five to eight times the corresponding rates calculated from two large-scale primate genomic comparisons, which represent the neutral rate of indel substitutions. Moreover, CATSPER1 indels are considerably longer than neutral indels. These observations strongly suggest that positive selection has been promoting the fixation of indel mutations in CATSPER1 exon 1. It has been shown in certain ion channels that the length of the N terminus region affects the rate of channel inactivation. This finding suggests that the selection detected may be related to the regulation of the CATSPER1 channel, which can affect sperm motility, an important determinant in sperm competition.
There have been dozens of reports on detection of positive Darwinian selection at the DNA sequence level (1–3) since the pioneering work by Hughes and Nei (4) on mammalian MHC genes. The majority of the positively selected genes are involved in host–pathogen interactions (4–8) or reproduction (9–18), although a small number of the genes are of other functions (19, 20). In all these cases, positive selection has been shown to promote nonsynonymous (amino acid-replacing) nucleotide substitutions that are presumably advantageous. In theory, certain insertion/deletion (indel) mutations in protein-coding regions may also be advantageous and subject to positive selection. Naturally occurring polymorphisms of indels that alter protein function have been reported (21). However, there has been no evidence for the operation of positive selection promoting fixations of indel mutations. This is probably because a large proportion of indel substitutions would disrupt the reading frame of a gene and thus be subject to strong purifying selection, which makes it difficult to detect positive selection. Nevertheless, here we provide evidence for the operation of positive selection on indel substitutions in the primate CATSPER1 gene, and demonstrate that positive selection plays a role in the evolutionary change of protein length.
CATSPER1 is a voltage-gated calcium ion channel that is exclusively found in the plasma membrane of the principal piece of the sperm tail (22). It is necessary for cAMP-induced Ca2+ influx, normal sperm motility, and penetration of the egg (22). Targeted disruption of the gene results in sperm immobility and male infertility in mice (22). The CATSPER1 protein contains an intracellular N terminus region, six transmembrane domains, a pore-forming domain, and an intracellular C terminus (22). In an alignment of the putative orthologous CATSPER1 sequences from the human and mouse, we noticed that the N terminus region (mostly encoded by exon 1) contains multiple gaps and a large number of amino acid differences, whereas the rest of the sequences are conserved (Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). Such a high frequency of gaps is unusual for orthologous mammalian proteins, which prompted us to examine this region in detail. Below, we describe the results from an analysis of 16 orthologous CATSPER1 sequences from primates and demonstrate the action of positive selection on indels in this gene.
Materials and Methods
PCR and Sequencing. Exon 1 (≈1,200 nucleotides) of the CATSPER1 gene was amplified by PCR from genomic DNAs of the common chimpanzee (Pan troglodytes), pygmy chimpanzee (Pan paniscus), gorilla (Gorilla gorilla), orangutan (Pongo pygmaeus), talapoin monkey (Miopithecus talapoin), rhesus monkey (Macaca mulatta), baboon (Papio hamadryas), African green monkey (Cercopithecus aethiops), colobus monkey (Colobus guereza), woolly monkey (Lagothrix lagotricha), owl monkey (Aotus trivirgatus), squirrel monkey (Saimiri sciureus), spider monkey (Ateles geoffroyi), cotton-top tamarin (Saguinus oedipus), and ring-tailed lemur (Lemur catta). The PCR products were purified and cloned into pCR4TOPO vector (Invitrogen) before being sequenced in both directions by using an automated DNA sequencer. In addition, we amplified and sequenced intron 1 (2,050 nucleotides) of the rhesus monkey CATSPER1 gene. The primer sequences are given in Fig. 8, which is published as supporting information on the PNAS web site.
Sequence Analysis. The protein sequences for CATSPER1 exon 1 from the above 15 primates and the human (GenBank accession no. NM_053054) were aligned by using clustal x (23). The DNA sequences were then aligned following the protein alignment. To examine the robustness of the alignment, we used a variety of penalty parameters for gap opening (go) and gap extension (ge): go = 10, ge = 0.2; go = 20, ge = 0.2; go = 20, ge = 0.4; go = 40, ge = 0.8; go = 40, ge = 0.2; go = 10, ge = 0.8; go = 80, ge = 0.2; go = 60, ge = 0.2; go = 10, ge = 10; and go = 10, ge = 20. The default parameters in the program were go = 10 and ge = 0.2. The phylogeny of the 16 primate species studied here is relatively well established, especially for the major divisions (24–27), and use of alternative trees does not change our conclusion. We mapped the indels observed in the alignment of the CATSPER1 sequences to this phylogeny by using the parsimony principle and counted the number of indel substitutions in each branch of the tree.
To compare the rates of synonymous and nonsynonymous nucleotide substitutions, we implemented the modified Nei– Gojobori method (28) in a sliding window analysis using mega2 (29). The window size was set to be 20 codons. In addition, we used the likelihood (30) and parsimony (31) methods to identify codons that are under positive selection. Ancestral gene sequences were inferred by the Bayesian method (32, 33). Rates of conservative and radical nonsynonymous substitutions were estimated by the method of Zhang (34). winpep software (35) was used to identify intracellular N and C termini and transmembrane domains of CATSPER1 and other six-transmembrane voltage-gated channels.
Results
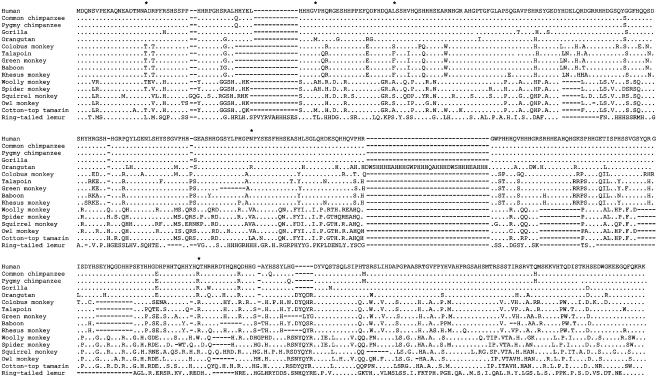
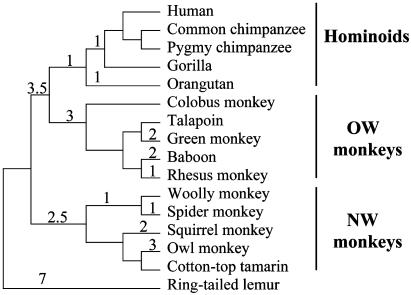
The Primate CATSPER1 Exon 1 Contains Many Indels. The exon 1 sequences of the CATSPER1 gene from 15 nonhuman primates were determined and compared with the sequence from the human. All sequences have an ORF throughout the exon as expected. But the sequence length of the exon varies among species, from 360 codons in the lemur to 443 codons in the orangutan. These sequences were conceptually translated and aligned by clustal x with the default parameters (Fig. 1), and the DNA sequences were subsequently aligned following the protein alignment. A gene tree of the 16 sequences was reconstructed by using the neighbor-joining method (36), which shows branching patterns (Fig. 9, which is published as supporting information on the PNAS web site) that are largely consistent with the known species tree (Fig. 2), indicating that the sequences obtained are orthologous to each other. Using the parsimony principle, we inferred events of indel substitutions in CATSPER1 exon 1 and mapped them onto the species tree. Note that parsimony makes our inference of the total number of indels conservative. Multiple parsimonious solutions are weighted equally. A total of 31 indel substitutions were found throughout the tree (Fig. 2). To investigate the robustness of this result, we used a wide variety of penalty parameters in alignment (see Materials and Methods). The resulting number of indels for the entire tree varied from 26 to 34. But, by our judgment, the alignment with 31 indels shown in Fig. 1, which was obtained by using the default parameters, appears most reasonable, and further analysis is based on this alignment. However, our conclusion is valid even when alignments with fewer indels are used (see below).
Fig. 1.
Amino acid sequence alignment of the exon 1 of CATSPER1 from 16 primates. This alignment is derived from clustal x with default parameters. The dots represent an amino acid identical to the first sequence, and dashes represent an alignment gap. Asterisks show five sites at which positive selection for amino acid substitutions is inferred by the likelihood method with >95% posterior probabilities (see text).
Fig. 2.
Phylogeny of the 16 primates studied here. The numbers on tree branches are parsimony-inferred numbers of indel substitutions in CATSPER1 exon 1, based on the alignment shown in Fig. 1.
Indels in CATSPER1 Are Under Positive Selection. To test whether the rate of indel substitutions in CATSPER1 exon 1 is significantly higher than the neutral expectation, it is necessary to first estimate the neutral rate of indel substitutions. For this, we used a recently published human–chimpanzee genomic comparison by Britten (37). In this comparison, 1,019 indels were found in an alignment of 779,142 nucleotides. Because only 1.1–1.4% of the human genome contains protein-coding sequences (38), this alignment is largely comprised of noncoding sequences and may thus be regarded as neutrally evolving regions. The earliest hominid fossil known to date has an age of 6–7 million years (MY) (39). We thus assume that the human and chimpanzee diverged ≈6.5 MY ago. The neutral indel substitution rate is then estimated to be 1,019/779,142/(6.5 × 106 × 2) = (1.01 ± 0.03) × 10–10 per site per year. Because we will compare the indel rates between noncoding and coding regions, it is more relevant to compute the neutral substitution rate of indels with sizes of multiples of three nucleotides (3n indels), as only 3n indels are potentially nondeleterious when they occur in protein-coding regions. In the above human–chimpanzee genomic data, there are 194 3n indels. Thus, the neutral substitution rate for 3n indels is 194/779,142/(6.5 × 106 × 2) = (1.92 ± 0.14) × 10–11 per site per year. The reason that the number of 3n indels is smaller than one third the number of all indels is that the frequency of indels declines quickly with the increase of the indel size and there are many more indels with one (or two) nucleotide(s) than with three nucleotides (37). In addition to the use of the Britten data, we also used the result from Silva and Kondrashov, who conducted a genomic comparison between human and baboon for 1,448,332 nt (40). They identified 5,883 indels, of which 1,001 were 3n indels. Assuming that humans and baboons diverged 23 MY ago (24, 41), we estimated from their data that the neutral substitution rate for indels is (8.83 ± 0.12) × 10–11 per site per year and that for 3n indels is (1.50 ± 0.05) × 10–11 per site per year.
With these estimates of genomic neutral substitution rates for 3n indels, we computed the expected number of 3n indels in CATSPER1 exon 1 under the assumption that all 3n indels are neutral. When the estimate from the human-chimpanzee genomic comparison is used, the expected number of 3n indels in exon 1 between hominoids and Old World (OW) monkeys is 1.92 × 10–11 × 1,329 × 23 × 106 × 2 = 1.17 (Table 1). Here, 1,329 is the number of nucleotides in the longest exon 1 sequence (orangutan) of the 16 primates. Use of this number makes our statistical test more conservative. The observed average number of 3n indels in exon 1 between the five hominoids and five OW monkeys is 6, which is significantly greater than the expected value of 1.17 under neutral evolution (P < 0.001, Poisson test, Table 1). Similarly, the comparison between hominoids and New World (NW) monkeys and that between OW and NW monkeys yielded the same conclusion (Table 1). Use of the neutral rate estimated from the human–baboon genomic data shows even higher statistical significance (Table 1). Use of the alignment with the smallest number (i.e., 26) of gaps gave similar results (Table 2, which is published as supporting information on the PNAS web site). These comparisons strongly suggest that 3n indels are positively selected for in the evolution of primate CATSPER1 exon 1. In the above tests, we assumed that the rate of indel mutations at the CATSPER1 locus is similar to the genomic average. To verify this assumption, we sequenced the rhesus monkey CATSPER1 intron 1, which is adjacent to exon 1, and compared it with the orthologous human sequence. We found nine indels in 2,102 aligned sites of intron 1, which translates into a rate of 9/2,102/(23 × 106 × 2) = (9.31 ± 0.96) × 10–11 indels per site per year. This rate is very close to the rates estimated from the two genomic comparisons (1.01 × 10–10 and 8.83 × 10–11), suggesting no elevation of the indel mutation rate at the CATSPER1 locus.
Table 1. Comparison of the substitution rates of 3n indels from genomic data and from primate CATSPER1 exon 1.
| Indel rate (per site per 1011 year)
|
Number of indels in CATSPER1 exon 1
|
Probability†
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Comparisons | Divergence* (MY) | From ref. 37 | From ref. 40 | From CATSPER1 | Expectation from ref. 37 | Expectation from ref. 40 | Observation | Under ref. 37 | Under ref. 40 |
| Hominoids vs. OW monkeys | 23 × 2 | 1.92 | 1.50 | 9.81 | 1.17 | 0.92 | 6 | 1.3 × 10-3 | 3.9 × 10-4 |
| Hominoids vs. NW monkeys | 35 × 2 | 1.92 | 1.50 | 10.3 | 1.79 | 1.40 | 9.6 | 1.1 × 10-4 | 1.6 × 10-5 |
| OW vs. NW monkeys | 35 × 2 | 1.92 | 1.50 | 12.5 | 1.79 | 1.40 | 11.6 | 3.0 × 10-6 | 2.8 × 10-7 |
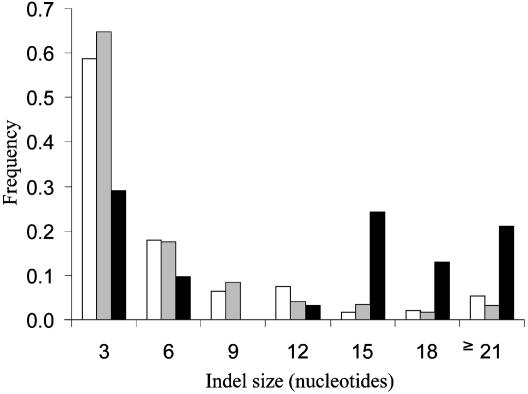
Longer Indels Are Selectively Favored. We further investigated whether indels of certain lengths are particularly favored in CATSPER1 by comparing the (3n) indel-size distributions for the CATSPER1 data and the two genomic data sets used above (Fig. 3). A significant distributional difference is detected between CATSPER1 and either of the two genomic data sets (P < 10–19, χ2 test). Longer indels are preferentially selected for in CATSPER1. For instance, the proportion of 3n indels with 15 or more nucleotides is 8–9% in the two genomic data, but 58% in the CATSPER1 data. However, even for indels of 3 nt, the number of observed indels in CATSPER1 is ≈2.5 times the expected number from the genomic data, and their difference is statistically significant (P < 0.02). This observation suggests that both short and long indels are selectively favored in CATSPER1, with longer ones being under stronger positive selection.
Fig. 3.
Size distribution of 3n indels. White and gray bars represent 3n indels of the genomic data from refs. 37 and 40, respectively, and black bars represent 3n indels from CATSPER1 exon 1 sequences of 16 primates. The frequencies are calculated by the number of indels of a particular size divided by the total number of 3n indels.
It should be noted that some sequence motifs such as simple nucleotide repeats are known to have relatively high rates of mutation (42). Most of these mutations are caused by slippage in DNA replication, which results in addition or deletion of one or occasionally a few repeats (42). The CATSPER1 exon1 sequences do not contain such repetitive sequences except that the sequences are rich in histidines, glycines, and serines. However, the indel-size distribution in Fig. 3 and the amino acid sequence alignment in Fig. 1 indicate that the majority of the indels in CATSPER1 were not caused by mutations of simple repetitive sequences.
Amino Acid Substitutions May Also Be Under Positive Selection. The alignment of the CATSPER1 exon 1 sequences from the 16 primates also reveals a relatively high number of amino acid substitutions (Fig. 1). To test whether the amino acid substitutions are under positive selection, we estimated the number of synonymous (dS) and nonsynonymous (dN) nucleotide substitutions per site between each pair of the sequences. We found that the average dN/dS ratio is 1.05 from a total of 120 pairwise comparisons, with 46 of these comparisons showing dN>dS (Fig. 10, which is published as supporting information on the PNAS web site). We further characterized the dN/dS ratio by a sliding-window analysis with a nonoverlapping window size of 60 nt. This analysis identified 9 of 17 windows that have an average dN greater than dS, one of them being marginally significant (Fig. 4). We also conducted a likelihood analysis, which may be more powerful in detecting positive selection for nonsynonymous substitutions at individual codons (30). Specifically, we compared a null model M7 with a more general model M8. M7 assumes that the dN/dS ratio across codon sites follows a β distribution between 0 and 1, whereas M8 adds to M7 an extra class of sites with any dN/dS. We found that M8 fits the data significantly better than M7 (χ2 = 37.3, df = 2, P < 10–8), with an additional class of sites of dN/dS = 2.98. Five codons were identified to be under positive selection with posterior probabilities >95% (Fig. 1). Similar results were obtained when models M1 and M2 were compared (see ref. 30 for details of the model description). Because the likelihood method is known to occasionally generate false positive results (43), we also used a more conservative test based on parsimony (31). With this test, none of the five aforementioned codons show significantly higher dN than dS. But when the five codons are tested together, the average dN is significantly greater than dS (P < 0.02). These tests thus are consistent in suggesting some sort of positive selection for amino acid substitutions. To further characterize the substitutions that are favored by selection, we compared the rates of conservative and radical nonsynonymous substitutions with regard to amino acid polarity (34). For this analysis, the ancestral sequences at all interior nodes of the tree (Fig. 2) were inferred by the Bayesian method, and the numbers of conservative and radical nonsynonymous substitutions were counted for each tree branch. For the entire tree, we observed 188 radical and 373 conservative nonsynonymous substitutions, respectively. The potential numbers of radical and conservative nonsynonymous sites are 195 and 478, respectively. Thus, the substitution rate at radical sites (188/195 = 0.96) is 1.23 times that at conservative sites (373/478 = 0.78), and their difference is statistically significant (P < 0.02, binomial test). This pattern is in contrast to that observed from a majority of mammalian genes (34), suggesting positive selection favoring changes of the polarity profile of CATSPER1. A similar analysis did not yield significant results when amino acid charge is considered.
Fig. 4.
Sliding-window analysis of CATSPER1 exon 1 sequences from 15 primates. The lemur sequence was not used here because of the presence of a large number of indels. The nonoverlapping window size is 20 codons. Average numbers of synonymous (dS) and nonsynonymous (dN) substitutions per site among the 15 sequences are shown by open and filled bars, respectively, with the error bars representing one standard error. One window has a marginally significant dN > dS (Z test, P = 0.053), and it is indicated by an asterisk. The nucleotide positions are from an alignment of the 15 sequences with all of the indels removed and thus do not directly correspond to the positions in Fig. 1.
Discussion
By analyzing DNA sequences from 16 primates, we discovered exceptionally frequent incidences of indel substitutions in the evolution of the first exon of the CATSPER1 gene. In all likelihood, the first exon is a functional part of CATSPER1, as no frame-shifting indels or nonsense substitutions are found in any of the species examined. We found that the indel substitution rate in exon 1 is five to eight times that of the genomic average, which represents the neutral rate. Furthermore, larger indels (≥15 nt) are significantly more prevalent in exon 1 than in the neutral genomic regions. These observations provide strong evidence that indel substitutions, particularly those with greater sizes, are positively selected for in the evolution of primate CATSPER1.
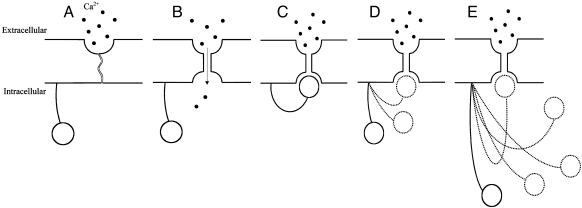
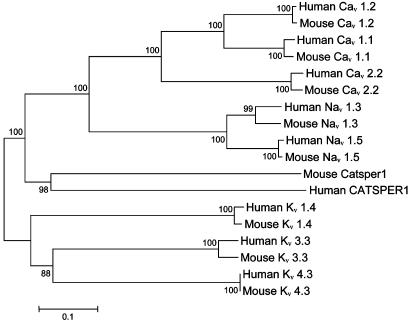
Why would indel substitutions be beneficial in CATSPER1 exon 1, which encodes the intracellular N terminus region of the ion channel? To address this question, we turn to the structure and function of ion channels. Ion channels are transmembrane proteins that form pores through which ions can pass. A voltage-gated ion channel such as CATSPER1 is activated by depolarization (reduction in electric potential) of the cell membrane, which causes a conformational change of the channel and allows ions to pass through it (Fig. 5). Within 1 ms of activation, the channel is inactivated and is impermeable to the ions, even though the membrane is still depolarized. The membrane must be repolarized or hyperpolarized to remove the channel from the inactive state and return it to the closed state where it is prepared for subsequent activation. Inactivation prevents the channel from remaining open, and is also responsible for the unidirectional propagation of action potential. The “ball-and-chain” model of ion channel inactivation (Fig. 5), proposed by Bezanilla and Armstrong (44) and demonstrated by Aldrich and colleagues (45, 46), offers a possible scenario where the length of the N terminus region plays an important functional role. Specifically, Aldrich and colleagues showed that the N terminus of a Drosophila voltage-gated potassium (KV) channel named Shaker acts to inactivate the channel (45, 46). Here, a “ball on a chain” structure is located at the N terminus of the channel and acts as a tethered plug, which is able to physically block the intracellular end of the ion channel pore region and cause inactivation of the channel (45, 46) (Fig. 5). The first ≈20 residues of the N terminus of Shaker channel form the intracellular “plug” and the next ≈60 residues represent the “tether” (45, 46). It was found that the length of this tethered plug controls the rate of channel inactivation (45). That is, lengthening or shortening of the tether resulted in slow or rapid channel inactivation, respectively. This is probably because a shorter tether restricts the space in which the “plug” wanders, making it easier for the “plug” to find the pore. Although this ball-and-chain model has only been demonstrated in KV channels, it is possible that CATSPER1 has a similar mechanism of regulating its inactivation. In fact, structurally, CATSPER1 resembles KV channels more than voltage-gated Ca (CaV) or Na (NaV) channels, because CATSPER1 and KV channels are each formed by four identical peptides, each having a single, six-transmembrane-spanning repeat, whereas CaV and NaV channels are made of a single peptide with four repeats of six-transmembrane-spaning regions. The amino acid sequence of the pore-forming region, however, is more similar between CATSPER1 and CaV, presumably reflecting the identical ion selectivity. The hydropathy profile shows a greater similarity of CATSPER1 to KV than to CaV or NaV channels (Fig. 11, which is published as supporting information on the PNAS web site). Evolutionarily, it is generally believed that KV channels originated before CaV and NaV channels (47) and that metazoan CaV and NaV channels each form a monophyletic group in exclusion of KV channels (48). To investigate the phylogenetic position of CATSPER1 in relation to other six-transmembrane voltage-gated channels, we reconstructed a phylogeny by using the human and mouse sequences of N termini regions of several KV, NaV, and CaV channels (Fig. 6). The tree is consistent with the current understanding of the evolution of KV, NaV, and CaV channels (47–49). Surprisingly, CATSPER1 does not cluster with other CaV channels, but resides outside the monophyletic group of CaV and NaV channels (Fig. 6). This finding suggests that CATSPER1 is one of the earliest branches splitting from KV channels, originating before the divergence of other CaV channels and NaV channels. This finding would further suggest that the emergence of the structure of a single peptide with four repeats that is seen in non-CATSPER1 CaV channels and NaV channels postdated the origin of CATSPER1. It is unlikely that the branching pattern in Fig. 6 is caused by long-branch attraction between CATSPER1 and KV channels, because CATSPER1 does not cluster with the longest branch, the outgroups (legend to Fig. 6). And the same branching patterns are also observed when the non-N-terminal sequences, which are conserved in CATSPER1, are used in tree-making. Taken together, the evolutionary and structural analyses suggest similarity of CATSPER1 to KV channels, which makes the “ball- and-chain” a more plausible model of channel inactivation for CATSPER1. If this model indeed works in CATSPER1, the indels in the N terminus region can potentially affect the inactivation rate of the channel, as in the Drosophila KV channel Shaker. Because CATSPER1 determines sperm motility by regulating the cellular Ca2+ concentration (22), it is likely that the rate of channel inactivation influences sperm motility. Because sperm motility is one of the most important factors in sperm competition (50), the exceptionally high rate of indel substitutions in CATSPER1 may be a signature of intense sperm competition. A population genetic study will be useful to further test this hypothesis. Our preliminary data from mice (Mus musculus) show intraspecific indel polymorphisms in the first exon of Catsper1 (J.Z. and David Webb, unpublished data).
Fig. 5.
Schematics of the “ball-and-chain” model of channel inactivation. (A) Closed state. (B) Open state. (C) Inactivated state. According to this model, the N terminus represents a tethered plug that can physically block the intracellular side of the ion channel pore and cause inactivation. Different lengths of the N terminus region result in different rates of channel inactivation (45, 46), where a shorter N terminus (D) causes a more rapid inactivation in comparison to a longer N terminus (E).
Fig. 6.
The evolutionary relationship of CATSPER1 with mammalian Kv, Nav, and Cav channels. The tree was reconstructed with the N terminus region of each ion channel, which was determined by hydropathy analysis. The neighbor-joining method with protein p distance (29) was used. Numbers at interior nodes are bootstrap percentages from 1,000 replications. Branch lengths are drawn to scale (number of amino acid substitutions per site). The root of the tree was determined to be on the deepest branch shown here, by using Drosophila and vertebrate inward-rectifier K channels as outgroups (49). When the entire sequences of the ion channels are used, the tree topology remains the same with the exception of the interrelationships among KV1.4, KV3.3, and KV4.3. The GenBank accession numbers are: human KV1.4, A39922; mouse KV1.4, NP_067250.1; human KV3.3, NP_004968.1; mouse KV3.3, Q63959; human KV4.3, NP_004971.1; mouse KV4.3, NP_064315.1; human CaV2.2, Q00975; mouse CaV2.2, O55017; human CaV1.2, NP_000710.1; mouse CaV1.2, A44467; human CaV1.1, A55645; mouse CaV1.1, NP_055008.1; human NaV1.3, Q9NY46; mouse NaV1.3, NP_035453.1; human NaV1.5, Q14524; mouse NaV1.5, NP_067519.1; human CATSPER1, AF407333_1; mouse Catsper1, AF407332_1.
In addition to indel substitutions, our results indicate that amino acid substitutions in the N terminus region of CATSPER1, particularly those that alter amino acid polarity, are probably under positive selection as well. It is possible that such changes in hydrophobicity affect the folding of the N terminus region and influence the rate of channel inactivation. In the future, it would be interesting to use in vitro and in vivo assays to investigate the functional consequences of the indel and amino acid substitutions in the N terminus of CATSPER1.
The phylogenetic tree in Fig. 6 also shows that the level of sequence divergence in the N terminus region between the human and mouse orthologous CATSPER1 proteins is exceptionally high in comparison to that in other voltage-gated ion channels. All of these channels are expressed in somatic tissues with the exception of CATSPER1, which is solely expressed in sperm. Recently, a second gene encoding a sperm calcium channel has been cloned, and it is named CATSPER2 (51). Our preliminary analysis suggests a remote evolutionary relationship between CATSPER1 and CATSPER2 (data not shown). Furthermore, the N terminus region of CATSPER2 appears conserved between the human and mouse, with only a few amino acid substitutions and virtually no indels. Thus, the physiological functions of CATSPER1 and CATSPER2 might be different.
Positive selection for nonsynonymous nucleotide substitutions has been documented in many genes (1–3). To our knowledge, CATSPER1 represents the first case in which positive selection for indel substitutions is detected. This success largely relies on the availability of genomic sequence data from closely related species, from which a neutral rate of indel substitution can be reliably estimated. From the present study, it seems that the statistical test for detecting selection on indels is relatively powerful. For instance, the number of expected 3n indels is ≈1 for the CATSPER1 exon 1 sequences between hominoids and OW monkeys (Table 1). Under the assumption that the number of indels follows a Poisson distribution, an observation of 4 indels would lead to the rejection of the null hypothesis of neutral evolution, with a statistical confidence of 98%. Because protein-length variation among orthologs and paralogs is quite common and indels are often seen in protein sequence alignments, we hypothesize that positive selection for indels is not rare. With the establishment of the basic methodology here and the estimation of neutral rates of indel substitutions from many more species, this hypothesis can be tested in the near future.
Supplementary Material
Acknowledgments
We thank Priscilla Tucker, Xiaoxia Wang, David Webb, and the anonymous reviewers for valuable comments. This work was supported by a start-up fund and a Rackham fellowship from the University of Michigan and by National Institutes of Health Grant GM67030 (to J.Z.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: OW, Old World; NW, New World; MY, million years; indel, insertion/deletion; CaV, voltage-gated Ca channel; NaV, voltage-gated Na channel; KV, voltage-gated K channel.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY382294–AY382305, AY382307–AY382309, and AY380795).
References
- 1.Hughes, A. L. (1999) Adaptive Evolution of Genes and Genomes (Oxford Univ. Press, New York).
- 2.Yang, Z. & Bielawski, J. P. (2000) Trends Ecol. Evol. 15, 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfe, K. H. & Li, W. H. (2003) Nat. Genet. 33, Suppl., 255–265. [DOI] [PubMed] [Google Scholar]
- 4.Hughes, A. L. & Nei, M. (1988) Nature 335, 167–170. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka, T. & Nei, M. (1989) Mol. Biol. Evol. 6, 447–459. [DOI] [PubMed] [Google Scholar]
- 6.Fitch, W. M., Leiter, J. M., Li, X. Q. & Palese, P. (1991) Proc. Natl. Acad. Sci. USA 88, 4270–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mindell, D. P. (1996) Proc. Natl. Acad. Sci. USA 93, 3284–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang, J., Dyer, K. D. & Rosenberg, H. F. (2000) Proc. Natl. Acad. Sci. USA 97, 4701–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, Y. H., Ota, T. & Vacquier, V. D. (1995) Mol. Biol. Evol. 12, 231–238. [DOI] [PubMed] [Google Scholar]
- 10.Swanson, W. J. & Vacquier, V. D. (1995) Proc. Natl. Acad. Sci. USA 92, 4957–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metz, E. C. & Palumbi, S. R. (1996) Mol. Biol. Evol. 13, 397–406. [DOI] [PubMed] [Google Scholar]
- 12.Tsaur, S. C. & Wu, C. I. (1997) Mol. Biol. Evol. 14, 544–549. [DOI] [PubMed] [Google Scholar]
- 13.Rooney, A. P. & Zhang, J. (1999) Mol. Biol. Evol. 16, 706–710. [DOI] [PubMed] [Google Scholar]
- 14.Wyckoff, G. J., Wang, W. & Wu, C. I. (2000) Nature 403, 304–039. [DOI] [PubMed] [Google Scholar]
- 15.Yang, Z., Swanson, W. J. & Vacquier, V. D. (2000) Mol. Biol. Evol. 17, 1446–1455. [DOI] [PubMed] [Google Scholar]
- 16.Swanson, W. J., Yang, Z., Wolfner, M. F. & Aquadro, C. F. (2001) Proc. Natl. Acad. Sci. USA 98, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galindo, B. E., Vacquier, V. D. & Swanson, W. J. (2003) Proc. Natl. Acad. Sci. USA 100, 4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swanson, W. J. & Vacquier, V. D. (2002) Nat. Rev. Genet. 3, 137–144. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, J., Zhang, Y. P. & Rosenberg, H. F. (2002) Nat. Genet. 30, 411–415. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg, A., Wildman, D. E., Schmidt, T. R., Huttemann, M., Goodman, M., Weiss, M. L. & Grossman, L. I. (2003) Proc. Natl. Acad. Sci. USA 100, 5873–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osterberg, M. K., Shavorskaya, O., Lascoux, M. & Lagercrantz, U. (2002) Genetics 161, 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren, D., Navarro, B., Perez, G., Jackson, A. C., Hsu, S., Shi, Q., Tilly, J. L. & Clapham, D. E. (2001) Nature 413, 603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodman, M., Porter, C. A., Czelusniak, J., Page, S. L., Schneider, H., Shoshani, J., Gunnell, G. & Groves, C. P. (1998) Mol. Phylogenet. Evol. 9, 585–598. [DOI] [PubMed] [Google Scholar]
- 25.Page, S. L. & Goodman, M. (2001) Mol. Phylogenet. Evol. 18, 14–25. [DOI] [PubMed] [Google Scholar]
- 26.Singer, S. S., Schmitz, J., Schwiegk, C. & Zischler, H. (2003) Mol. Phylogenet. Evol. 26, 490–501. [DOI] [PubMed] [Google Scholar]
- 27.Steiper, M. E. & Ruvolo, M. (2003) Mol. Phylogenet. Evol. 27, 121–130. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, J., Rosenberg, H. F. & Nei, M. (1998) Proc. Natl. Acad. Sci. USA 95, 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 30.Yang, Z., Nielsen, R., Goldman, N. & Pedersen, A. M. (2000) Genetics 155, 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki, Y. & Gojobori, T. (1999) Mol. Biol. Evol. 16, 1315–1328. [DOI] [PubMed] [Google Scholar]
- 32.Yang, Z., Kumar, S. & Nei, M. (1995) Genetics 141, 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, J. & Nei, M. (1997) J. Mol. Evol. 44, Suppl. 1, S139–S146. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, J. (2000) J. Mol. Evol. 50, 56–68. [DOI] [PubMed] [Google Scholar]
- 35.Hennig, L. (1999) BioTechniques 26, 1170–1172. [DOI] [PubMed] [Google Scholar]
- 36.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 37.Britten, R. J. (2002) Proc. Natl. Acad. Sci. USA 99, 13633–13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 39.Brunet, M., Guy, F., Pilbeam, D., Mackaye, H. T., Likius, A., Ahounta, D., Beauvilain, A., Blondel, C., Bocherens, H., Boisserie, J. R., et al. (2002) Nature 418, 145–151. [DOI] [PubMed] [Google Scholar]
- 40.Silva, J. C. & Kondrashov, A. S. (2002) Trends Genet. 18, 544–547. [DOI] [PubMed] [Google Scholar]
- 41.Glazko, G. V. & Nei, M. (2003) Mol. Biol. Evol. 20, 424–434. [DOI] [PubMed] [Google Scholar]
- 42.Hancook, J. M. (1999) in Microsatellites, Evolution and Applications, eds. Goldstein, D. B. & Schlotterer, C. (Oxford Univ. Press, New York), pp. 1–9.
- 43.Suzuki, Y. & Nei, M. (2002) Mol. Biol. Evol. 19, 1865–1869. [DOI] [PubMed] [Google Scholar]
- 44.Bezanilla, F. & Armstrong, C. M. (1977) J. Gen. Physiol. 70, 549–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoshi, T., Zagotta, W. N. & Aldrich, R. W. (1990) Science 250, 533–538. [DOI] [PubMed] [Google Scholar]
- 46.Zagotta, W. N., Hoshi, T. & Aldrich, R. W. (1990) Science 250, 568–571. [DOI] [PubMed] [Google Scholar]
- 47.Lodish, H., Berk, A., Zipursky, S. L., Matsudaira, P., Baltimore, D. & Darnell, J. (2000) Molecular Cell Biology (Freeman, New York).
- 48.Piontkivska, H. & Hughes, A. L. (2003) J. Mol. Evol. 56, 277–285. [DOI] [PubMed] [Google Scholar]
- 49.Hille, B. (2001) Ion Channels of Excitable Membranes (Sinauer, Sunderland, MA).
- 50.Gomendio, M., Harcourt, A. H. & Roldan, E. R. S. (1998) in Sperm Competition and Sexual Selection, eds. Birkhead, T. R. & Moller, A. P. (Academic, London), pp. 667–751.
- 51.Quill, T. A., Ren, D., Clapham, D. E. & Garbers, D. L. (2002) Proc. Natl. Acad. Sci. USA 98, 12527–12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.