Abstract
The sequences of large insert clones containing genomic DNA that is orthologous to the maize adh1 region were obtained for sorghum, rice, and the adh1-homoeologous region of maize, a remnant of the tetraploid history of the Zea lineage. By using all four genomes, it was possible to describe the nature, timing, and lineages of most of the genic rearrangements that have differentiated this chromosome segment over the last 60 million years. The rice genome has been the most stable, sharing 11 orthologous genes with sorghum and exhibiting only one tandem duplication of a gene in this region. The lineage that gave rise to sorghum and maize acquired a two-gene insertion (containing the adh locus), whereas sorghum received two additional gene insertions after its divergence from a common ancestor with maize. The two homoeologous regions of maize have been particularly unstable, with complete or partial deletion of three genes from one segment and four genes from the other segment. As a result, the region now contains only one duplicated locus compared with the eight original loci that were present in each diploid progenitor. Deletion of these maize genes did not remove both copies of any locus. This study suggests that grass genomes are generally unstable in local genome organization and gene content, but that some lineages are much more unstable than others. Maize, probably because of its polyploid origin, has exhibited extensive gene loss so that it is now approaching a diploid state.
The grasses (Poaceae) are a monophyletic family of monocotyledonous flowering plants with ≈10,000 species (1), including the cereal crops, rice, maize, wheat, barley, and sorghum. The cereals differ considerably in haploid nuclear genome size, varying from ≈450 Mb in rice to ≈5,000 Mb in barley (2). Comparative genetic maps of sorghum and maize (3), and of more distantly related cereals (4–7), revealed significant genome conservation among the cereals despite their differences in nuclear DNA content. The discovery of conserved gene content and order (i.e., colinearity) in grasses has opened new frontiers for gene discovery, gene isolation, and the characterization of gene function in an evolutionary context (8).
Over the last several years, comparative genetics in the grasses has progressed from genetic map studies toward detailed assessments of microcolinearity at the sequence level. Comparative sequence analyses of related grass species (9–13) confirmed the general conservation of gene content and order, although some local sequence rearrangements such as deletions, tandem duplications, inversions, and translocations were revealed (14, 15). Although a lack of microcolinearity can complicate comparative genetic studies, such sequence rearrangements have become a valuable resource in characterizing local genome evolution. Unfortunately, these studies have lacked a multispecies perspective that can allow clear identification of the timing, nature, and lineages of the observed rearrangements.
Two closely related grasses of the tribe Andropogoneae, sorghum and maize, diverged from a common ancestor ≈20 million years ago (mya) (16). The small sorghum genome (750 Mb) can serve as a reference genome for the much larger and more complex genome of maize (2,400 Mb). Rice diverged from a common ancestor with sorghum and maize ≈50–70 mya (17). With the smallest genome among the cereals, rice has emerged as a model for all of the grasses (18), especially because the full genome sequence is determined (19, 20). Therefore, including rice in comparative studies of the orthologous regions in sorghum and maize can reveal details of local sequence evolution since the radiation of grasses, and may allow reconstruction of the genomic structure of their common ancestor (21).
Little is known about the origin of the modern maize genome. Comparisons of gene and DNA marker content/order in the maize genome has provided strong evidence that maize is derived from a tetraploid, and still retains chromosomal sets that can be viewed as at least partly homoeologous (i.e., pairs of chromosomes that are homologues of another pair of chromosomes in the same nucleus, as with the three homoeologous chromosome sets of allohexaploid wheat) (6, 22, 23). According to one hypothesis (16), the ancestral maize genome originated from a segmental allotetraploid event that took place ≈11 mya. Beyond this study, there is little evidence on the exact mode of tetraploid formation in maize, and especially on the events that led to the current diploid behavior of the maize genome.
Haldane (24) predicted, and numerous subsequent researchers have confirmed, that most duplicated genes are subsequently lost. Lynch and Force (25) proposed that most of the retained duplicated loci would become subfunctionalized, whereas only rarely would a duplicated gene evolve a new function. It is not known to what degree these processes have acted on the polypoloid maize genome.
Orthologous adh regions in maize and sorghum were previously sequenced and analyzed in our laboratory (11), revealing a general conservation of gene content and order. However, the sequence of the rice adh1/adh2 region (26) indicated a complete lack of colinearity with the adh1-orthologous regions of maize and sorghum. Because all grass species outside the Andropogoneae contain adjacent adh1 and adh2 orthologues in syntenic locations with rice adh1/adh2, it appears that the maize and sorghum adh1-orthologous genes moved to their current locations in an ancestral Andropogoneae. In this paper, we report the sequence of a rice bacterial artificial chromosome (BAC) clone that is truly orthologous to the adh1 region of maize, and also the sequence of a maize BAC that represents the adh-homoeologous region. Comparative analysis of these segments revealed the specific natures of several genic rearrangements, demonstrating very different evolutionary dynamics of this region in maize, sorghum, and rice. Sequence comparison of homoeologous segments in the maize genome revealed the local genomic structure of the two diploid ancestors of maize, and also the nature and timing of numerous subsequent genomic rearrangements.
Materials and Methods
BAC Selection. Several genes flanking an adh gene in sorghum on BAC 110K05 (GenBank accession no. AF124045) (11) were used as probes to screen a HindIII BAC library from rice (Oryza sativa L., sp. japonica, cv. Nipponbare) and a HindIII BAC library from maize (Zea mays, L., sp. mays, cv. B73). Both libraries were obtained from Clemson University Genomics Institute (CUGI). The longest rice BAC that contained homologies to most of the genes from the sorghum adh region in a central location (OSJNBa0084L17) was chosen for sequencing. The maize BAC that hybridized to sorghum gene 2, the 5′ region of gene 4/5, and gene 7.5 was selected and sequenced (ZMMBBb0276N13). In addition, an MboI library of B73 maize DNA, made by Pieter de Jong at Children's Hospital Oakland Research Institute (www.chori.org/bacpac/home.htm), was screened with sorghum genes 4/5 and 10. The clone positive for these two probes (ZMMBBc0531D05) was further analyzed by restriction mapping to determine its overlap with CUGI BAC 276N13. Subsequently, another maize CUGI BAC (ZMMBBb0123C01) that hybridized to sorghum gene 10 was selected and analyzed as well. DNA manipulation, PCR amplification, and gel blot hybridizations were conducted according to standard procedures (27).
Sequencing. BAC DNA isolation, construction of sublibraries and shotgun DNA sequencing were as described previously (12). For details, see Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Sequence Analysis and Annotation. Sequence analysis and annotation were conducted as described in Supporting Text.
Gene Designations. Instead of using their numerical order of appearance on each BAC, the assigned numeric names of the genes in maize and rice were based on their homology to the candidate genes in the published sequence of sorghum BAC 110K05 (11). As a result, most of the genes from the previously annotated maize adh1 region (11) had changed names. For instance, gene 334B7.4 is now referred to as gene 4/5 (truncated), gene 334B7.5 is called gene 6, 334B7.6 is called gene 7, 334B7.7 is called gene 10, 334B7.8 is called gene 11, and 334B7.9 is now called gene 12.
Our current annotation corrects several apparent errors in the initial annotation (11). Two previously uncharacterized genes were found on the sorghum BAC and were annotated as genes 3.5 and 7.5. Also, two sorghum genes were reannotated as a single gene (4/5), based on homology to maize mRNA sequences (GenBank accession nos. AY107153 and AY109941) that joined predicted genes 4 and 5.
Expression Studies. Isolation of total plant RNA was performed by using TRIzol Reagent according to the manufacturer's recommendations (Invitrogen/Life Technologies). For details on RT-PCR and primer design, see Supporting Text.
Results
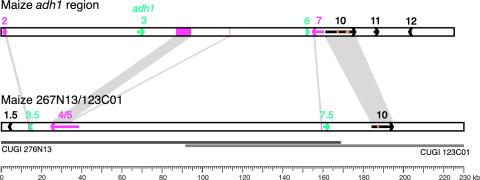
Selection and Sequence Analysis of Maize Contig 276N13/123C01. To select the maize segment that is homoeologous to the adh1 region in maize, we used genes 4/5, 8, and 9 from the orthologous sorghum region as hybridizational probes. From the BACs identified, no two probes hybridized to the same BAC clone. So, additional probes (sorghum genes 2, 7.5, and 10) were used to select the homoeologous segment. A BAC clone (276N13) that was positive for genes 2, 4/5, and 7.5 was selected for sequence analysis. To extend the contig further downstream, gene 10 was used to identify an additional BAC (123C01). The two overlapping clones provided a genomic segment of ≈230 kb long (Fig. 1).
Fig. 1.
Structures of two homoeologous genomic regions in maize. Arrows indicate the location, approximate size, and transcriptional orientation of predicted genes. Colored arrows represent the genes whose homoeologous copies are either completely (green) or partially (purple) deleted from one maize region. Purple boxes show the remaining fragments of partially deleted genes. Gray areas connect conserved sequences between the two maize regions. The only structurally intact candidate gene still present in both regions is gene 10. Orange arrowheads show long interspersed nuclear elements inserted in the intronic region(s) of two homoeologues. Two overlapping gray horizontal lines represent the two BACs (276N13 and 123C01) that were sequenced.
The two BACs, 276N13 (169 kb) and 123C01 (138 kb), had a 78-kb sequence overlap and formed two contigs: the main, 213,888-bp, contig (GenBank accession no. AY371488) and an additional short contig (≈15 kb), composed mainly of repetitive DNA. The two BACs are part of a BAC contig (including marker umc90) that maps to the short arm of maize chromosome 5 (E. Butler, Arizona Genomic Institute, personal communication). This map position is colinear/syntenic with adh1 of maize (located on chromosome 1) by the criteria of Gale and Devos (7), indicating that the 276N13/123C01 contig is homoeologous and orthologous to the adh1 region of maize.
Among the standard approaches that we used for gene identification in this maize region (see Supporting Text), we found that sequence comparison to the orthologous sorghum and rice regions was the most rapid, comprehensive, and reliable technique. These sequence analyses revealed five candidate genes: 1.5, 3.5, 4/5, 7.5, and 10 (Fig. 1). All gene candidates exhibited different degrees of homology to unknown proteins. Predicted gene 1.5 apparently encodes an unknown protein (258 aa) that has its highest homology to an unknown protein in rice (BAB92554, 2e-70) and maps to a nonsyntenic chromosomal location (rice chromosome 1). Candidate gene 3.5 was predicted on the basis of homology to a maize cDNA (BM382318, 6e-13) and an orthologous sorghum sequence located 500 bp down-stream of the adh gene (BAC110K05). The predicted protein has no significant homology to any entry in public protein databases. Furthermore, our RT-PCR studies did not detect expression of this gene in the maize and sorghum organs that we tested. The neighboring gene, renamed 4/5, was originally annotated as two genes in sorghum, 110K5.4 and 110K5.5 (11). However, homology to maize cDNA sequences (GenBank accession nos. AY107153 and AY109941) indicated that this was a single gene, occupying 13.3 kb of genomic DNA. Sequence identity to the orthologous genes in rice and sorghum was 90% and 89%, respectively (at the amino acid level). The putative protein of gene 4/5, with a predicted length of 754 aa, also has good homology to an Arabidopsis hypothetical protein (At4g02030.1, NP_192112, e-89). A 1.9-kb terminal-repeat retrotransposons in miniature (TRIM) element was detected in the intron 15 of candidate gene 4/5, with homology to eway_115G01-1 of Triticum monococcum (Genbank accession no. AF459639, 3e-15) (28). Candidate gene 7.5 apparently encodes a protein that is homologous to an Arabidopsis hypothetical protein (At2g14110.1, NP_179027, 4e-37) and a magnesium-dependent phosphatase-1 in Mus musculus (NM_023397, 4e-21). The last candidate gene in the contig, gene 10, with nine exons, is homologous to an Arabidopsis hypothetical protein (At2g39810.1, NP_181511, e-88).
There was only one case where genes were clustered together (genes 1.5 and 3.5) in the entire 230-kb maize contig. All other gene candidates are separated by large blocks of repetitive DNA (detailed description is provided in Supporting Text and Fig. 4, which is published as supporting information on the PNAS web site).
Comparison of Homoeologous Segments in Maize. Previous sequence analysis of a yeast artificial chromosome (YAC) containing the maize adh1 region DNA (YAC 334B7) yielded ≈225 kb of genomic sequence (Fig. 1) (11). YAC 334B7 and BACs 276N13/123C01 share four predicted genes or gene fragments, each in colinear order and orientation (shaded regions in Fig. 1). Hence, by this evidence and their syntenic positions on chromosomes 1 and 5, these appear to be homoeologous segments that have been retained since the paleotetraploid origin of maize ≈11–17 mya (16). Gene 4/5, previously 334B7.4 (11), appears to have been severely rearranged in the adh1 region compared with its homoeologous maize segment. Most of the 5′ region of the predicted gene is missing (first 10 exons). Instead, the region is occupied by a 35.3-kb block of nested retroelements, whereas the central portion of the truncated gene (separating exons 11 and 12) contains a nested 19-kb insertion of Huck-2 and Fourf (11).
Predicted gene 10 in the maize adh1 region occupies 15 kb of space and contains two long interspersed nuclear elements (LINEs) inserted in introns 6 and 8 (11). Interestingly, gene 10 (occupying >10 kb) of the homoeologous maize segment contains two fragments (256 and 95 bp long) related to the same LINE (Colonist2, GenBank accession no. U90128) in the sixth intron. No homology to the LINE elements was found in intron 8 of this copy of gene 10, and consequently, this intron was considerably shorter (2,034 bp) than the corresponding intron of the gene 10 in the adh1 region (4,610 bp) (for details, see Supporting Text). Investigation by RT-PCR demonstrated that both copies of gene 10 were expressed in the maize organs that we tested. Because of the low gene density in both maize homoeologous segments, we could not determine whether candidates genes 11 and 12 from the maize adh1 region were present downstream of predicted gene 10 in the second homoeologous segment.
A few small fragments of residual gene sequence were detected based on their homology to homoeologous/orthologous genes in maize and sorghum adh regions. Two short DNA segments (163 and 147 bp) were found with homology to gene 2 in the maize adh1 region, (4e-28, 137/163 identities and 7e-26, 125/147 identities), corresponding to the terminal exon and 3′ untranslated region of gene 2 from the adh1 region (Fig. 1). In close proximity to predicted gene 7.5 (<600 bp upstream of the predicted translation initiation site), a 145-bp segment was detected that showed homology (1e-226, identities 121/145) to candidate gene 7 in the maize adh1 region. This short fragment was homologous to exon 11 (85 bp) and part of intron 11, whereas the rest of gene 7 was apparently deleted from both 5′ and 3′ ends. Thus, a total of 7 gene candidates were deleted (either partially or completely) from the two homoeologous regions: predicted genes 3.5, 4/5, and 7.5 from the adh1 region, and putative genes 2, 3, 6, and 7 from the homoeologous segment.
The nonsynonymous substitution rate (Ka) for the two maize genes 10 was 0.06 (± 0.006), whereas the synonymous substitution rate (Ks) was 0.112 (± 0.015), giving a Ka/Ks ratio of 0.54. Thus, these two genes appear to be under selection for a retained function, otherwise known as purifying selection (29). The RT-PCR gene expression analysis, using primers designed in the 3′ end of the coding region of gene 10, showed that both genes are expressed in 2-week-old seedlings, in roots and leaves of 4-week-old plants, and in reproductive organs of adult maize plants (data not shown). Hence, these duplicated genes appear to have not yet undergone subfunctionalization (25) or any other regulatory change.
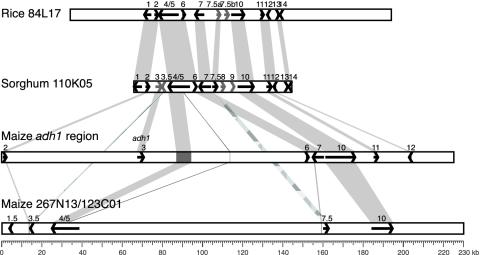
Sequence Analysis of Orthologous Regions from Rice and Sorghum. Screening of the rice BAC library filters was conducted by using the same sorghum probes from BAC 110K05 used to isolate maize BACs 276N13 and 123C01. Rice BAC 84L17 was selected, completely sequenced, and found to contain a 159,664-bp insert (GenBank accession no. AY387483). This BAC maps to the long arm of rice chromosome 3 (www.genome.arizona.edu/fpc/rice/WebAGCoL/WebFPC), in a syntenic position with all of the other BACs investigated in this study. BAC 84L17 contains a total of 21 candidate genes, or about one gene per 7.6 kb. The sequence colinear with sorghum BAC 110K05 is 67.8-kb long (from 37,275 to 105,075 bp) and contains 12 predicted genes (Fig. 2). With a few notable exceptions, the sorghum and rice regions show conserved gene content, order, and transcriptional orientation. This microcolinearity was interrupted in two regions where a total of four sorghum genes do not have homologues on the rice BAC. The longest noncolinear segment is a 10-kb stretch that contains the sorghum adh gene (also designated as gene 3) and adjacent gene 3.5.
Fig. 2.
Colinear genomic regions in rice, sorghum, and two homoeologous segments in maize. The shaded areas connecting the regions represent conserved sequence. Gene content of the central part of rice BAC 84L17 (68.7 kb) is depicted; candidate genes from the 37.2- and 54.6-kb rice segments flanking the compared region are not shown. Orthologous genes (shown as arrows) in all four regions have the same numerical designations. Gray arrows show a unique tandem gene duplication in rice. Also, four sorghum genes that are not shared with rice are shown as gray arrows. The blended-gray regions show orthologous genes (3.5 and 7.5) that are shared between sorghum and one homoeologous maize region (contig 276N13/123C01). Gene fragments and truncated genes in the two maize segments are shown as gray boxes and vertical lines.
The other exception to colinearity is the segment that contains candidate sorghum genes 8 and 9. These two genes are also missing from both orthologous regions in maize (Fig. 1). However, screening the Clemson University Genomics Institute HindIII library with probes derived from predicted sorghum genes 8 and 9 indicated that there are at least two copies of both genes in maize. A database search confirmed that two copies of gene 8 existed in the rice genome, in two nonsyntenic chromosomal locations (on chromosome 12). Only one rice homologue of gene 9 was found (GenBank accession no. AP002844), also in a nonsyntenic location (chromosome 1).
In rice, we discovered a tandem duplication of gene 7.5 (7.5A and 7.5B, Fig. 2). Based on the synonymous nucleotide substitution rate (Ks), the estimated time of this gene duplication was ≈44 mya (see Supporting Text).
Eleven predicted genes in the comparable rice region have colinear homologues in sorghum. Only one of these candidate genes has high identity to a protein with a predicted function. This locus, gene 7, apparently encodes a cyclin H-1, based on 100% identity to the cyclin H-1 protein in rice (GenBank accession no. BAB11694). All other gene candidates exhibited varying degrees of homology to unknown or hypothetical proteins in Arabidopsis and/or rice (see Supporting Text).
Sequence Comparison of Rice, Sorghum, and Maize Segments. General conservation of gene content and order in the four regions analyzed in this study was evident. However, several gene rearrangements were observed.
Genes 8 and 9, found in the sorghum adh region, were absent from both segments in maize and in rice. This is a strong indication that these two genes were inserted in the sorghum adh region after the divergence of maize and sorghum. However, these two genes are not linked in rice, because two homologues of gene 8 are located on chromosome 12, whereas the single homologue of gene 9 was found on chromosome 1.
Predicted genes 4/5 and 7.5 are either present as truncated genes or completely deleted in the adh1 region of maize. They were found in sorghum, rice, and the other maize region. These data suggest that at least two separate deletion events took place in the adh1 region of maize.
Genes 2, 3, 6, and 7 are present in the adh regions of maize and sorghum and in the orthologous rice region, but intact copies of the genes are missing from the other maize homoeologous segment. Thus, these four genes were deleted from the region in several separate events, probably after the two diploid genomes merged to form the tetraploid maize ancestor. Considering the location of the remaining gene fragments, particularly a short fragment of gene 7, it is clear that deletion of this gene occurred from both 5′ and 3′ ends. Most of gene 7 (the first 10 exons, ≈3.5 kb) was deleted, whereas neighboring gene 7.5 (only 600 bp away) remained intact and expressed in maize. Although gene 2 appeared to be deleted only from the 5′ end, a complete deletion of neighboring gene 3 indicates two separate deletion sites.
Candidate gene 3.5 was conserved between sorghum and one maize segment. Interestingly, a 45-bp fragment in maize and a 37-bp fragment in sorghum (part of the terminal exon of gene 3.5) had homology (9e-08 and 3e-05, respectively) to putative gene RZ53 in the adh1/adh2 region of rice (26). RZ53 is located ≈1.7 kb downstream of the rice adh1 gene. This short fragment associates both sorghum and maize regions to the nonsyntenic adh1 location in rice, indicating that adh1 moved from its original adh1/adh2 position to its current location in the Andropogonae lineage before sorghum and the two maize progenitors diverged (see Supporting Text). Gene 3.5 probably moved to its current location together with the adh gene. The movement of genes 3 and 3.5 into this region in a common Andropogoneae ancestor of maize and sorghum is much more likely than the deletion of these genes in the rice lineage (from BAC 84L17) because recombinational mapping studies have shown that adh1 (gene 3) orthologs from all grasses except the Andropogoneae are in syntenic locations with the adh1 gene of rice and not with the maize adh1 gene (7). The simplest model suggests that the other copy of gene 3.5 was subsequently deleted from the adh1 region in maize, probably after the origin of paleopolyploid maize. Gene RZ53 has the same transcriptional direction as the adh1 gene in rice (26), in contrast to gene 3.5 in sorghum, indicating a single gene inversion either in the rice lineage or the sorghum/maize lineage.
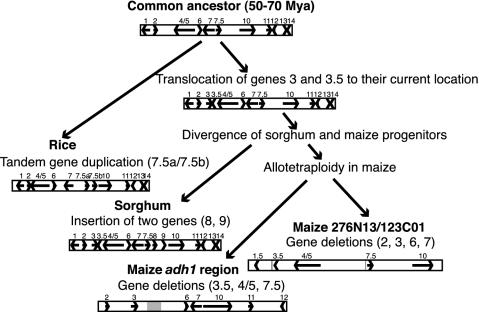
These analyses not only provide insights into the natures and frequencies of such small chromosomal rearrangements, but also indicate their lineages and approximate timing. A cladistic analysis of the origin of these orthologous segments from an ancestral region is depicted in Fig. 3.
Fig. 3.
Schematic diagram of the simplest evolutionary model for events that took place in this orthologous region of the rice, sorghum, and maize genomes after their divergence from a common ancestor. Arrows indicate genes, whereas gray bars in the two maize segments show truncated gene fragments. The genic composition of the common ancestor of the three species is proposed, with 11 genes in this region, and is structurally very similar to the rice region.
Discussion
Genomic Stability in Rice and Instability in Maize. This study provides a detailed comparison of homoeologous loci in maize at the sequence level. The analysis was enriched by comparisons to the orthologous regions in sorghum (a close relative of maize) and rice (a distant relative of sorghum and maize), providing numerous insights into the modes of local sequence evolution over the last 60 million years.
The extent of gene deletion seen in this region of the maize genome is higher than previously estimated for the complete maize genome. Over 40% of the total genes from each homoeologous region were deleted. These gene removals or fragmentations would have required several separate deletion events, either sited with one end within a gene or with both ends flanking the deleted gene. In the studied homoeologous regions of maize, only one of eight homoeologous gene pairs (12.5%) remain in a structurally intact and active state. This is in contrast to an earlier study by Ahn and Tanksley (4) indicating that 72% of maize genes, when compared with rice, were still duplicated by hybridizational criteria. In this regard, we observed that some genes in the maize adh1-homoeologous regions were deleted completely, whereas three genes had remnants of partially deleted genes. Such residual gene sequences could cause misleading hybridizational results. If one also takes into account the number of highly conserved paralogs, both linked and unlinked to an ortholog, that would be detected by hybridization, then it is easy to explain the 72% figure calculated by Ahn and Tanksley (4).
Considering the high frequency of gene deletion in the two maize segments that we studied, it is reasonable to expect that residues of deleted genes will be scattered throughout the maize genome (see Supporting Text). These may not have been noted earlier because short sequence homologies and fragments of genes can be overlooked by homology-searching and gene-finding programs. In fact, we were only able to identify and define several of these gene fragments in the studied homoeologous regions because we could do fine-scale comparative sequence analyses across four orthologous genomes.
Compared with its homoeologous region in maize, the adh1 region contains more genes, with four of its predicted genes grouped within a span of 39 kb. However, transposable element insertions and deletions were widespread in both maize segments. We found no evidence for preferential elimination of the genes from one maize subgenome, a possibility suggested by Gaut and Doebley (16), and reported for other polyploids (30). Also, we found no instance where both homoeologous genes had been deleted from maize. Therefore, selective gene deletions occurred in a nondiscriminative manner, affecting only one of the two syntenic copies, and were limited either to an individual gene or to two neighboring genes. These results suggest that natural selection acts against loss of all copies of any of these genes, also suggesting that most or all of these deletions took place some time after the formation of an allotetraploid maize. Rapid and directional elimination of low-copy-number sequences immediately after polyploid formation has been reported for synthetic polyploids of Brassica (30) and in the studies of synthetic and natural allopolyploids of the Aegelops–Triticum group (31–33).
Subsequent to the submission of this manuscript, we learned that extensive gene loss has also been observed from homoeologous lg2 and lrs1 regions of the maize genome. Langham and coworkers (34) found that a predicted 13 homoeologous gene pairs in ancient tetraploid maize had been “fractionated” to a current 12 single genes, with only one homoeologous pair (lg2/lrs1) retained. Taken together with our results, these observations indicate that maize is approaching a diploid genic state.
In contrast to maize, the orthologous region in rice appears to be very stable. At most, one genic change, a duplication of gene 7.5, appears to have occurred. In this region, sorghum represents a lineage with an intermediate instability between stable rice and unstable maize, exhibiting at least two genic rearrangements relative to rice. In other multispecies sequence comparisons to rice, the data also support a highly conserved structure of the rice genome relative to other grasses (35).
Approaching a Diploid Genetic State in the Maize Genome. Our data indicate that the ancestral allotetraploid maize has progressed quite a way toward functional diploidy. In the segment that we analyzed, the two homoeologous maize regions combined have about the same number of genes as the orthologous rice and sorghum segments. A similar high level of deletion in another segment of the maize genome, around the bronze1 (bz1) locus, was recently reported by Fu and Dooner (36). Interestingly, analysis of this same region in several maize inbreds indicated polymorphism for these deletions. That is, some haplotypes still contained genes (at least at the fragment hybridization level) that were deleted from other haplotypes. By hybridizational criteria, all of the genes that were sometimes missing from a bz1 region still had at least one copy elsewhere in the maize genome. These results suggest that the bz1 region is undergoing the same gene-loss events that we detected in adh1-orthologous segments, but that the deleted state is not yet fixed near bz1. Our study is limited to one maize haplotype, so we might see variability for gene deletions in the adh1 region as well if we investigated additional genetic backgrounds of maize. However, the bz1 study (36) lacks an outgroup (like the rice and sorghum genomes analyzed in our investigation), so one cannot determine whether the differences in gene content in the bz1 region are caused by gene deletion or gene insertion.
Extensive Sequence Conservation Between Rice and Sorghum Allows Prediction of an Ancestral Grass Genome Segment. Analysis of gene content and order in orthologous rice and sorghum regions, with 11 genes shared between the two, revealed a high degree of sequence conservation. As mentioned earlier, our data and earlier studies indicate that rice has a very stable genome (9, 12). The only rearrangement detected in the current study, the possible tandem duplication of gene 7.5, could actually have existed in an ancient grass genome, with one copy being deleted from a common ancestor of maize and sorghum. The apparent timing of the gene 7.5 duplication (≈44 mya) is more consistent with duplication in rice, although this could be a misleading date if frequent unequal conversions caused many million years of concerted evolution for these two genes or because the two genes appear to be evolving at different rates. In any case, it is clear that gene content and order in this region of the ancestral grass genome were very similar to that currently observed in rice. This observation provides further evidence that rice can serve as an excellent model for grass genome studies and as a surrogate for map-based applications relative to large genome grasses (37).
Further sequence comparisons between rice and other grass genomic segments will be needed to confirm the validity of rice as a genomic model. These comparisons will be most useful if they contain at least three orthologous segments from different grasses, because pairwise comparisons do not permit assignment of the lineage in which an identified rearrangement occurred. Additional value comes from multiple sequence comparisons, as demonstrated in this study by the number of cases where only the presence of a fourth segment allowed clear lineage attribution.
Our studies shed a new light on local sequence evolution in the maize, sorghum, and rice genomes, revealing very different evolutionary dynamics in this region in these three lineages. Additional investigations are needed of a larger number of orthologous gene segments within maize. Particular emphasis should be placed on functional analyses of the orthologous genes, with the goal of relating changes in evolved genome structure to conserved or altered functions of orthologous loci in maize, sorghum, and rice.
Supplementary Material
Acknowledgments
We are grateful to Wusirika Ramakrishna and Katrien Devos for help with sequence analysis and many useful suggestions. We are also thankful to Ed Butler from the Arizona Genomics Institute (Tucson, AZ) for providing information on chromosomal locations for maize BACs. This work was supported by U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service National Research Initiative Competitive Grants Program Award 00-35300-9412.
Abbreviations: BAC, bacterial artificial chromosome; mya, million years ago.
References
- 1.Clark, L. G., Zhang, W. & Wendel, J. F. (1995) Syst. Bot. 20, 436–460. [Google Scholar]
- 2.Bennett, M. D. & Leitch, I. J. (1995) Ann. Bot. 76, 113–176. [Google Scholar]
- 3.Hulbert, S. H., Richter, T. E., Axtell, J. D. & Bennetzen, J. L. (1990) Proc. Natl. Acad. Sci. USA 87, 4251–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn, S. & Tanksley, S. D. (1993) Proc. Natl. Acad. Sci. USA 90, 7980–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn, S., Anderson, J. A., Sorrells, M. E. & Tanksley, S. D. (1993) Mol. Gen. Genet. 241, 483–490. [DOI] [PubMed] [Google Scholar]
- 6.Moore, G., Devos, K. M., Wang, Z. & Gale, M. D. (1995) Curr. Biol. 5, 737–739. [DOI] [PubMed] [Google Scholar]
- 7.Gale, M. D. & Devos, K. M. (1998) Science 282, 656–659. [DOI] [PubMed] [Google Scholar]
- 8.Bennetzen, J. L. & Freeling, M. (1993) Trends Genet. 9, 259–261. [DOI] [PubMed] [Google Scholar]
- 9.Chen, M., SanMiguel, P. J., de Oliveira, A. C., Woo, S. S., Zhang, H., Wing, R. A. & Bennetzen, J. L. (1997) Proc. Natl. Acad. Sci. USA 94, 3431–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feuillet, C. & Keller, B. (1999) Proc. Natl. Acad. Sci. USA 96, 8265–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tikhonov, A. P., SanMiguel, P. J., Nakajima, Y., Gorenstein, N. M., Bennetzen, J. L. & Avramova, Z. (1999) Proc. Natl. Acad. Sci. USA 96, 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubcovsky, J., Ramakrishna, W., SanMiguel, P. J., Busso, C. S., Yan, L., Shiloff, B. A. & Bennetzen, J. L. (2001) Plant Physiol. 125, 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song, R., Llaca, V. & Messing, J. (2002) Genome Res. 12, 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennetzen, J. L. & Ramakrishna, W. (2002) Plant Mol. Biol. 48, 821–827. [DOI] [PubMed] [Google Scholar]
- 15.Feuillet, C. & Keller, B. (2002) Ann. Bot. 89, 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaut, B. S. & Doebley, J. F. (1997) Proc. Natl. Acad. Sci. USA 94, 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfe, K. H., Gouy, M., Yang, Y. W., Sharp, P. M. & Li, W. H. (1989) Proc. Natl. Acad. Sci. USA 86, 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izawa, T. & Shimamoto, K. (1996) Trends Plant Sci. 1, 95–99. [Google Scholar]
- 19.Sasaki, T. & Burr, B. (2000) Curr. Opin. Plant Biol. 3, 138–141. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki, T., Matsumoto, T., Yamamoto, K., Sakata, K., Baba, T., Katayose, Y., Wu, J., Niimura, Y., Cheng, Z., Nagamura, Y., et al. (2002) Nature 420, 312–316. [DOI] [PubMed] [Google Scholar]
- 21.Bennetzen, J. L. & Ma, J. (2003) Curr. Opin. Plant Biol. 6, 128–133. [DOI] [PubMed] [Google Scholar]
- 22.Helentjaris, T., Weber, D. & Wright, S. (1988) Genetics 118, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaut, B. S. (2002) New Phytol. 154, 15–28. [Google Scholar]
- 24.Haldane, J. B. S. (1933) Am. Nat. 67, 5–19. [Google Scholar]
- 25.Lynch, M. & Force, A. (2000) Genetics 154, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarchini, R., Biddle, P., Wineland, R., Tingey S. & Rafalski, A. (2000) Plant Cell 12, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., Maniatis, T. & Fritsch, E. F. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 28.SanMiguel, P. J., Ramakrishna, W., Bennetzen, J. L., Busso, C. S. & Dubcovsky, J. (2002) Funct. Integr. Genomics 2, 70–80. [DOI] [PubMed] [Google Scholar]
- 29.Ophir, R., Itoh, T., Graur, D. & Gojobori, T. (1999) Mol. Biol. Evol. 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 30.Song, K., Lu, P., Tang, K. & Osborn, T. C. (1995) Proc. Natl. Acad. Sci. USA 92, 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feldman, M., Liu, B., Segal, G., Abbo, S., Levy, A. A. & Vega, J. M. (1997) Genetics 147, 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, B., Vega, J. M. & Feldman, M. (1998) Genome 41, 535–542. [DOI] [PubMed] [Google Scholar]
- 33.Shaked, H., Kashkush, K., Ozkan, H., Feldman, M. & Levy, A. A. (2001) Plant Cell 13, 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langham, R. J., Walsh, J., Dunn, M., Ko, C., Goff, S. A. & Freeling, M. (2003) Genetics, in press. [DOI] [PMC free article] [PubMed]
- 35.Ramakrishna, W., Dubcovsky, J., Park, Y. J., Busso, C., Emberton, J., San-Miguel, P. & Bennetzen, J. L. (2002) Genetics 162, 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu, H. & Dooner, H. K. (2002) Proc. Natl. Acad. Sci. USA 99, 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson, W. A., Harrington, S. E., Woodman, W. L., Lee, M., Sorrells, M. E. & McCouch, S. R. (1999) Genetics 153, 453–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.