Abstract
Human cytomegalovirus has a complex double-stranded DNA genome of ≈240,000 bp that contains ≈150 ORFs likely to encode proteins, most of whose functions are not well understood. We have used an infectious bacterial artificial chromosome to introduce 413 defined insertion and substitution mutations into the human cytomegalovirus AD169 genome by random and site-directed transposon mutagenesis. Mutations were produced in all unique ORFs with a high probability of encoding proteins for which mutants have not been previously documented and in many previously characterized ORFs. The growth of selected mutants was assayed in cultured human fibroblasts, and we now recognize 41 essential, 88 nonessential, and 27 augmenting ORFs. Most essential and augmenting genes are located in the central region, and nonessential genes generally cluster near the ends of the viral genome.
Human cytomegalovirus (HCMV) is the prototypic β-herpes virus and a ubiquitous human pathogen. Although infections in healthy children and adults are generally asymptomatic, HCMV is a leading viral cause of birth defects and a major cause of morbidity and mortality in immunocompromised individuals (1).
HCMV contains a complex double-stranded DNA genome of ≈240,000 bp, the largest genome for a virus known to infect humans. The laboratory strain of HCMV, AD169, contains ≈150 ORFs likely to encode proteins (2-5). Most ORFs have not been well studied due to the limited host range and slow growth of HCMV in cultured cells and the lack of efficient tools to generate mutant viruses. Recently, the HCMV genome has been cloned as an infectious bacterial artificial chromosome (BAC) (6-9), greatly facilitating its genetic manipulation (8, 10).
We previously described an infectious BAC clone of HCMV AD169, termed pAD/Cre (6). The BAC vector is flanked by LoxP sites and contains a Cre-recombinase gene that is modified by the insertion of an intron into its coding sequence. Consequently, Cre is not expressed in bacterial cells, but it is expressed when its transcript is spliced in human cells and the BAC vector is excised from the virus. Now we report the use of both random and site-directed transposon mutagenesis to introduce 413 defined insertion and substitution mutations into the HCMV AD169 genome residing in pAD/Cre. Mutations were produced in all ORFs with a high probability of encoding proteins for which mutants have not been previously documented and in many previously characterized ORFs. We have begun to systematically delineate the functions of viral ORFs in HCMV-infected cells by analyzing the growth of HCMV mutants in cultured human fibroblasts. We now recognize 41 essential, 88 nonessential, and 27 augmenting ORFs. This work describes a functional map of the complete HCMV genome and provides a foundation for future genetic studies.
Methods
Cells, Viruses, and Plasmids. Primary human foreskin fibroblasts at passage 8-15 were propagated in medium supplemented with 10% FCS. The HCMV strain AD169 BAC, pAD/Cre (6), was the wild-type parent of all mutant viruses. AD169 and its mutant derivatives were propagated in fibroblasts. Virus titers were determined in duplicate by plaque assay in fibroblasts (6).
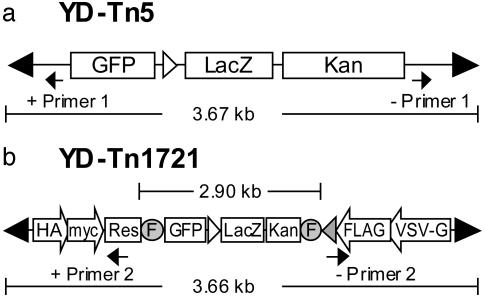
To construct YD-Tn5, a GFP cassette expressed under the control of the SV40 promoter and polyadenylation signals was amplified by PCR from pGET015 (11) and cloned into the NotI/NcoI sites of Tn5 (12). The LacZ coding region was amplified and cloned into the AflII site of the above construct to generate pC107. A part of pC107 was then amplified with primers containing priming sites for future use in sequencing the junctions of transposon insertions. The PCR-generated fragments were cut with restriction enzymes and used to replace the corresponding portions of pC107 to generate the final YD-Tn5 vector. To construct YD-Tn1721, the XhoI/FspI fragment of pTsTm8B (10) was cloned into the XhoI/EcoRV sites of pSP72 (Promega) to generate pC136. The hemagglutinin tag, the c-myc tag, and the sequencing primer (+Primer 2, Fig. 1) were amplified and cloned into the XhoI/BglII sites of pC136 to generate pC137. The bovine growth hormone poly(A) signal from cDNA3 (Invitrogen), the FLAG tag, and the VSV-G tag were amplified and cloned into the NotI site of pC137 to generate pC138. The LacZ/GFP cassette with an FRT site at one end was amplified and cloned into pC138 to generate pC144. A fragment containing a short version of the kanamycin resistance gene (Finnzymes, Helsinki) with an FRT site at one side was amplified and cloned into the pGEM-T-easy vector (Promega) to generate pC143. The SalI/BglII fragment of the kan+ cassette from pC143 was used to replace the XhoI/BamH I fragment containing the long version of the kanamycin resistance gene of pC138 to generate pC145. The BstE II/SpeI fragment of the kanamycin resistance gene from pC145 was used to replace the BstEII/SpeI fragment of pC144 to generate pC146. The ScaI/BstE II fragment of pC146 was inserted into TsTm8B to generate the final YD-Tn1721 construct.
Fig. 1.
Transposons used to construct the mutant BAC-HCMV library. The Tn5-derived transposon YD-Tn5 (a) and the Tn3-derived transposon YDTn1721 (b) both contain the kan+/LacZ cassette for identification of transposon-carrying BAC clones in E. coli. They also carry a GFP gene controlled by the SV40 early promoter and terminated by a bidirectional SV40 poly(A) signal (open arrow) to monitor mutant viruses in human cells. YD-Tn1721 contains four unique epitope tags. The HA (YPYDVPDYA) and c-myc (EQKLISEEDL) epitopes tag viral proteins encoded by two translation frames from one strand, whereas Flag (DYKDDDDK) and VSV-G (KGLRNMEIDTY) epitopes tag viral proteins encoded by two translation frames from the complementary strand. It carries the BGH poly(A) signal (gray arrow) and two FRT sites (shadowed circles), so that the 2.9-kb internal fragment of the transposon can be removed by FLP/FRT recombination. Many features built into YD-Tn1721 are not used in this report. Black arrows mark the locations of primers used for sequencing junctions of transposon insertion.
Transposon Mutagenesis. Random insertion mutagenesis with Tn5 was carried out as previously described (12). The procedure for mutagenesis with Tn1721 was slightly modified from earlier reports (10). YD-Tn1721 was electroporated into Escherichia coli DH10B cells harboring pAD/Cre and selected at 30°C overnight on medium containing ampicillin (50 μg/ml). The resulting colonies containing both pAD/Cre and YD-Tn1721 were grown in medium containing ampicillin (50 μg/ml) and chloramphenicol (15 μg/ml) at 30°C overnight. A series of dilutions of the culture were plated onto medium containing both antibiotics and selected for drug-resistant clones at 43°C overnight, which were replicated on a LB plate with ampicillin (50 μg/ml) and incubated at 30°C overnight to confirm that the YD-Tn1721 shuttle vector was lost.
YD-Tn1721 was introduced into specific locations within pAD/Cre by linear recombination (13, 14). To insert the transposon, a pair of 70-nt primers were designed so that the 5′-terminal 50 nucleotides were homologous to the viral gene to be disrupted, whereas the 3′-terminal 20 nucleotides corresponded to the ends of the transposon. Amplification with these primers by using YD-Tn1721 as the template generated a linear fragment in which the transposon was flanked on each end by 50 bp of the virus-specific DNA. The linear fragment was then electroporated into E. coli DY380 cells harboring pAD/Cre and the defective λ prophage. The λ functions mediate insertion of the electroporated linear DNA fragment into pAD/Cre by homologous recombination between the short 50-bp viral sequences residing at the ends of the PCR fragment and their corresponding alleles in the viral genome.
Analysis of the Mutant BAC Library. The gross integrity of BACHCMV clones carrying a transposon insertion was monitored by EcoRI digestion, and the sites of transposon insertions were determined by sequence and PCR analysis through both junctions.
BACs were judged to harbor mutations in essential viral genes when they generated GFP-positive cells 2-3 d after electroporation but failed to produce virus. For transfections in which viruses were produced, virus stocks were harvested ≈4 wk after electroporation, and virus yields were determined by plaque assay (6). BACs were judged to harbor mutations in genes that augment viral replication when they produced small plaques and/or generated ≥10-fold reduced yields of cell-free virus at days 5, 10, and 15 after infection at a multiplicity of 0.01 plaque-forming units per cell. In most cases, two independent mutations were evaluated, although one mutant plus a revertant were characterized in some cases for essential and augmenting genes. BACs were judged to harbor mutations in nonessential genes when they produced normal-sized plaques and a virus stock with a wild-type yield. The kan+/lacZ cassette in the transposons facilitated the efficient marker rescue of mutant BAC-HCMV clones by allelic exchange and blue-white screening (6).
Results
Mutant BAC-HCMV Library Created by Transposon Mutagenesis. Of the ≈150 HCMV AD169 ORFs, five have been reported to be essential (5, 7, 9, 15), eight are known to augment viral replication (9, 11, 16-23), and 63 have been designated nonessential by studying spontaneous and site-directed mutations (5, 6, 24, 25). No mutants have been reported for the remaining ≈80 ORFs, and their roles during infection are largely unknown (5). To lay a foundation for the global genetic analysis of this complex viral genome, we constructed a BAC-HCMV library carrying mutations in the majority of viral ORFs, including all unique ORFs with a high probability of encoding proteins (3, 4) for which no mutants have been reported. We first generated random insertion mutations into HCMV ORFs by using transposon mutagenesis (9, 10). We modified the Tn5 and the Tn3-derived Tn1721 transposons to produce YD-Tn5 and YDTn1721 (Fig. 1), which contain markers that facilitate selection and screening within E. coli and fibroblasts. Standard transposon mutagenesis efficiently targeted most of the HCMV genome. To introduce transposons into spots where random insertions were absent, we used PCR-based site-directed mutagenesis (13, 14). YD-Tn1721 was amplified by PCR and introduced into BACHCMV by linear recombination, which was targeted by the sequence of the primers used for amplification. Sequence and PCR analysis located the insertions and demonstrated that unintentional DNA rearrangements were not introduced at the ends of the inserted transposon. The site of insertion or substitution was determined for 413 mutants (Table 2, which is published as supporting information on the PNAS web site, www.pnas.org); 81 resided between ORFs, and 332 were positioned within ORFs. Mutations were isolated in every putative functional ORF for which mutants have not been previously documented.
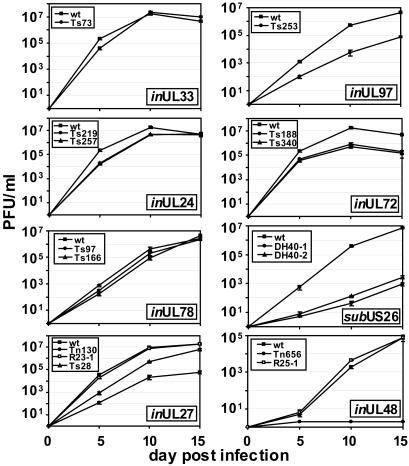
Growth of HCMV Mutants in Cultured Human Fibroblasts. We analyzed the growth phenotype of a subset of the mutants, favoring mutations within the N-terminal half of ORFs and categorizing them as essential, nonessential, or as ORFs whose products augment but are not absolutely essential for HCMV replication in fibroblasts (Fig. 2 and Table 1). ORFs were classified as essential if two different mutant BAC DNAs generated fluorescent cells after electroporation, indicating that the BAC DNA had successfully entered the fibroblasts and expressed the GFP marker in the transposon but failed to spread to additional cells. In some cases, only one mutant was tested, if its mutation was judged unlikely to affect expression of other ORFs and a marker-rescued derivative of the mutant BAC exhibited wild-type growth (e.g., UL32), or if a homologue had been characterized as essential in another herpes virus (e.g., UL46). ORFs were classified as nonessential if mutants produced plaques of normal size and generated a yield within 5-fold of that produced by wild-type virus when infected cultures were allowed to progress to full cytopathic effect. The growth of selected mutants with lesions in nonessential ORFs was confirmed by multistep growth analysis (representative results in Fig. 3). ORFs were considered to augment growth when mutants produced small plaques or a ≥10-fold-reduced yield of infectious progeny in the initial virus stock generated after delivery of the BAC DNA to fibroblasts. The growth behavior of all mutants with lesions in augmenting ORFs was verified by multistep growth analysis (representative results are shown in Fig. 3).
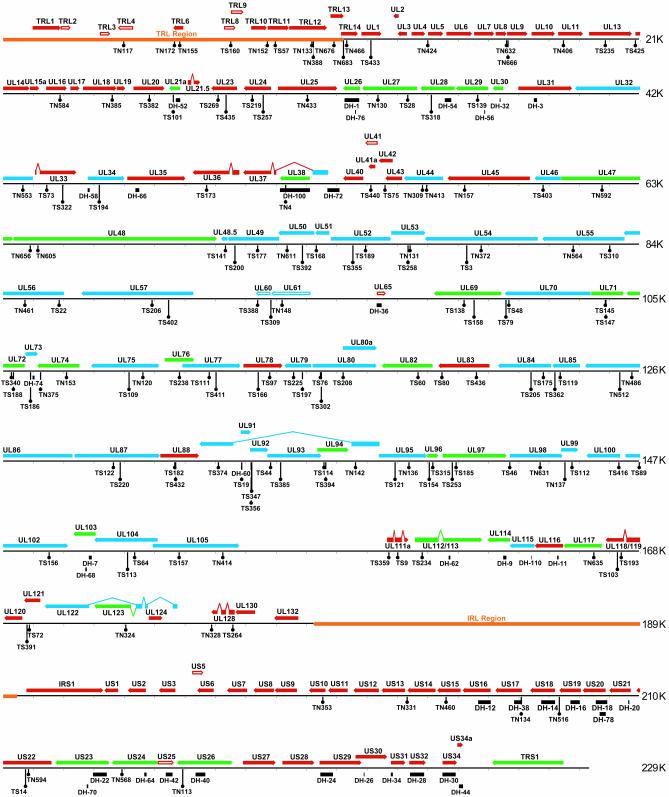
Fig. 2.
Functional map of the HCMV genome. The genome of HCMV strain AD169 is presented as a series of lines with approximate nucleotide sequence positions indicated to the right of each line. Above the viral genome, solid arrows represent putative ORFs predicted by Davison et al. (3), and open arrows represent additional ORFs predicted by the original annotation of Chee et al. (2) and the recent reannotation of Murphy et al. (4). The subset of mutants from the BAC library whose growth was analyzed in fibroblasts are positioned on the map below the viral genome, and the nature of each mutation is indicated: Tn5 insertions (TN), Tn1721 insertions (TS), and mutations made by linear recombination (DH). ORFs are color coded according to whether they are essential (blue) or nonessential (red), or augment (green) replication in fibroblasts. ORFs without an insertion or substitution notation were classified on the basis of earlier published reports. The terminal repeat long (TRL) sequence and the internal repeat long (IRL) sequence that is identical to the TRL sequence but in the inverted orientation are indicated as orange arrows on the viral genome. Only the set of ORFs in the TRL region is displayed because the set of ORFs in IRL are identical, and we did not distinguish which repeat contained the mutations.
Table 1. Analysis of mutants in previously uncharacterized HCMV ORFs.
| ORF | Function* | Position | Plaque† | Growth‡ |
|---|---|---|---|---|
| UL21a | A | 26626, 26696-26842§ | Small | ≥50 |
| UL23 | NE | 28101, 28372 | Normal | Normal |
| UL24 | NE | 29987, 29234 | Normal | Normal |
| UL25 | NE | 31051 | Normal | Normal |
| UL26 | A | 32260-32746§, 32608-32639§ | Small | ≥50 |
| UL27 | A¶ | 33373∥, | Small | ≥50 |
| 34360 | Normal | <50 | ||
| UL28 | A | 35138, 35568-35780§ | Small | <50 |
| Small | ||||
| UL29 | A | 36662, 36866-36896§ | Small | ≥50 |
| UL30 | A | 37369-37400§ | Small | ≥50 |
| UL31 | NE | 38502-38603§ | Normal | ND |
| UL32 | E | 42665∥ | None | ND |
| UL34 | E | 45188, 44791-44857§ | None | ND |
| UL35 | NE | 46366-46490§ | Normal | Normal |
| UL37 | E | 52698-53091§ | None | ND |
| UL38 | A | 51331∥, 51131-52126§ | Small | ≥50 |
| UL44 | E | 55839∥, 55982 | None | ND |
| UL46 | E | 59823 | None | ND |
| UL48 | A | 63917∥, 64165 | V. small | >104 |
| UL48.5 | E | 70346 | None | ND |
| UL49 | E | 71412, 70658 | None | ND |
| UL50 | E | 72376∥, 72886 | None | ND |
| UL51 | E | 73343 | None | ND |
| UL52 | E | 74965, 74547 | None | ND |
| UL53 | E | 76432∥, 76371 | None | ND |
| UL54 | E | 78776∥, 78300 | None | ND |
| UL56 | E | 84734∥, 85868 | None | ND |
| UL57 | E | 88931, 89476 | None | ND |
| UL60 | E | 92405 | None | ND |
| UL61 | E | 93220, 92848 | None | ND |
| UL65 | NE | 96316-96479§ | Normal | ND |
| UL70 | E | 100690, 100598 | None | ND |
| UL71 | A | 103867, 103891 | V. small | >104 |
| UL72 | A | 105358, 105294 | Normal | <50 |
| UL73 | E | 105923, 105974-106038§ | None | ND |
| UL76 | A | 110832 | Small | ≥50 |
| UL77 | E | 111817, 112039 | None | ND |
| UL78 | NE | 113804, 113432 | Normal | Normal |
| UL79 | E | 114597, 114885 | None | ND |
| UL80 | E | 115482, 115511 | None | ND |
| UL80/UL80a | E | 116225 | None | ND |
| UL84 | E | 122829, 122419 | None | ND |
| UL85 | E | 123381, 123207 | None | ND |
| UL86 | E | 125741∥, 125349 | None | ND |
| UL87 | E | 129657, 129882 | None | ND |
| UL88 | NE | 131675, 131710 | Normal | Normal |
| UL89 | E | 137636∥, 133118 | None | ND |
| UL91 | E | 133888, 133848-133863§ | None | ND |
| UL92 | E | 134210, 134193 | None | ND |
| UL93 | E | 134841, 134779 | None | ND |
| UL94 | A | 136593, 136650 | V. small | >104 |
| UL95 | E | 139394∥, 138943 | None | ND |
| UL96 | A | 140067, 140182 | V. small | >104 |
| UL98 | E | 142723, 143720∥ | None | ND |
| UL99 | E | 144775**, 144537 | None | ND |
| UL102 | E | 146992, 148532 | None | ND |
| UL103 | A | 149828-149929§, 149717-149759§ | V. small | >104 |
| UL104 | E | 151356, 151117 | None | ND |
| UL105 | E | 154266∥, 152821 | None | ND |
| UL111a | NE | 159730, 160030 | Normal | Normal |
| UL112/113 | A | 160832, 161691-161748§ | V. small | >104 |
| UL116 | NE | 165263-165313§ | Normal | ND |
| UL117 | A | 166495∥ | Small | ≥50 |
| UL121 | NE | 168888, 168803 | Normal | ND |
| US16 | NE | 204664-205080§ | Normal | Normal |
| US17 | NE | 206104, 205845-206106§ | Normal | ND |
| US18 | NE | 206740-207198§ | Normal | Normal |
| US19 | NE | 207357, 207708-208018§ | Normal | ND |
| US20 | NE | 208546-208924§, 208665-208870§ | Normal | Normal |
| US21 | NE | 209606-309645§ | Normal | Normal |
| US22 | NE | 210759, 210852 | Normal | Normal |
| US23 | A | 212967-213419§, 212763-212805§ | Normal | <50 |
| US24 | A | 213931∥, 214655-214733§ | Small | ≥50 |
| US25 | NE | 215362-215585§ | Normal | Normal |
| US26 | A | 215941, 216346-216665§ | Small | ≥50 |
| US29 | NE | 220459-220878§ | Normal | ND |
| US30 | NE | 221894-221923§ | Normal | ND |
| US31 | NE | 222784-222854§ | Normal | ND |
| US32 | NE | 223418-223885§ | Normal | ND |
| US34 | NE | 224483-224920§ | Normal | ND |
| US34a | NE | 225027-225162§ | Normal | ND |
E, essential; NE, nonessential; A, augmenting
Plaque size was measured. None, no plaques found after BAC transfection; V. small, very small
Multistep growth analysis was performed if a mutant produced small plaques or produced yields at least 10-fold lower than wild type. Numbers indicate folds of growth reduction compared to wild type. ND, not determined
Mutation constructed by site-directed mutagenesis
A, tentative assignment
Revertant made that grows like wild-type virus
The transposon insertion truncates one-third of the C terminus of UL99, and the mutant still replicates efficiently
Fig. 3.
Representative multistep growth curves of mutants with alterations in ORFs that are nonessential or augment replication of HCMV in fibroblasts. Also shown are the growth curves of the marker-rescued viruses R25-1 and R23-1 for the UL48 mutant Tn656 and the UL27 mutant Tn130, respectively. UL33 (30) and UL97 (22) have been reported previously to be nonessential and augmenting ORFs, respectively. UL24 and UL78 are newly defined nonessential genes, for which the growth kinetics of mutants is within an order of magnitude of that of the wild type, whereas UL72, US26, and UL48 are augmenting ORFs. Fibroblasts were infected at a multiplicity of 0.01 plaque-forming units per cell, and infectious progeny were quantified by plaque assay on fibroblasts at the indicated times. All growth analyses were performed in duplicate. The standard deviation for each data point is indicated.
Three Functional Categories of Viral ORFs. We now recognize 41 essential, 88 nonessential, and 27 ORFs whose products augment viral growth in fibroblasts (Fig. 2). The locations and properties of mutants used to assign ≈80 previously uncharacterized ORFs to a functional category are detailed in Table 1.
A total of 41 ORFs are essential for HCMV replication in fibroblasts. Our results confirmed earlier reports for five essential ORFs (5, 7, 9, 15) and placed 36 additional ORFs into this category. Most essential ORFs are homologous to genes in other herpes viruses encoding proteins required for DNA replication and virion assembly. However, 11 of the newly identified essential ORFs (UL32, -34, -49, -60, -61, -79, -84, -87, -91, -92, and -99) do not have well studied counterparts in other herpes viruses (3, 4). These gene products might identify novel aspects of the HCMV replication strategy as compared with other herpes viruses. Alternatively, they could be functionally homologous to genes with diverged sequences in their relatives. It is conceivable that some of these ORFs score as essential, because they are located in regions of the genome that encode critical cis-acting sequences. One case in which it is likely that mutations have altered a cis-acting elements is in the UL60-61 region, which contains the lytic origin of viral DNA replication (26). It is important to note that some of the ORFs assigned to this category by our analysis might prove to be required for HCMV replication in fibroblasts only in a multiplicity-dependent manner. Multiplicity dependence could prevent the initial recovery of mutant virus from transfected BAC DNA in normal fibroblasts. There is precedence for such a multiplicity-dependent effect. A mutant virus with a lesion in UL123, which we define as an augmenting ORF, was initially recovered in complementing fibroblasts but subsequently found to replicate well in normal fibroblasts infected at a high-input multiplicity (23).
The majority of HCMV ORFs, 88, encode products that are nonessential for efficient replication in fibroblasts. Among them, one copy of RL1∼RL9 as well as both copies of RL10∼RL13 have been reported to be nonessential for HCMV infection in fibroblasts (5, 6, 25). We have verified a portion, 26, of the previously reported classifications (5, 6, 24, 25) and placed 25 new ORFs into this category. A pseudo-ORF would score as nonessential. However, all but three ORFs (RL2, RL8, and UL65) newly assigned in this category have orthologues in the distantly related chimpanzee cytomegalovirus (3, 4), arguing that they likely encode functional products. A substantial portion of ORFs, 86/88, that are nonessential in fibroblasts do not have an identifiable counterpart in other herpes viruses (5), raising the possibility that some contribute to unique aspects of HCMV biology. Many ORFs that are nonessential in fibroblasts support replication and spread with an infected host, and most known nonessential functions involve viral strategies to antagonize the immune system. Some might encode redundant functions, and others will likely be required for growth in cell types other than fibroblasts or for HCMV latency.
In all, 27 ORFs augment the production of HCMV in fibroblasts by a factor of ≥10. Our results confirm earlier reports for seven ORFs (9, 16-23) and assign an additional 19 ORFs to this category. Augmenting ORFs might encode partially redundant functions or produce proteins, which accelerate reactions that nevertheless proceed slowly in their absence. It is also possible that some genes in this group are essential but the mutants replicated to a limited extent due to the accumulation of a truncated protein, although we generally analyzed mutants with insertions in the N-terminal region of ORFs.
Some of the mutants in this category successfully spread from cell to cell but produced little infectious progeny (>104-fold reduction): UL48 (Fig. 3), -71, -94, -96, -103, and -112/113. The ability to isolate a series of mutant viruses that mark augmenting ORFs and exhibit profound growth defects argues that the mutants with lesions in ORFs we have classified as essential would grow exceedingly poorly, if at all, in normal fibroblasts. Although we have tentatively classified UL27 as an augmenting ORF, it is equally possible that the defects observed for the mutants in this ORF result from their detrimental effects on UL26 expression. Tn130 carries a transposon insertion at the C terminus of UL27 (Fig. 2), which overlaps the UL26 transcript (27). The mutant produces small plaques and has ≈300-fold reduced growth compared with wild-type virus and the marker-rescued virus, R23-1 (Fig. 3). Ts28 contains an insertion at the N terminus of UL27, produces plaques of near wild-type size, and exhibits only a slight delay in growth kinetics.
Discussion
We have created mutations in all HCMV strain AD169 ORFs that are likely to encode proteins but have not been previously studied, and we have assigned the entire set of HCMV ORFs into three classes on the basis of their growth properties. AD169 contains 41 essential, 88 nonessential, and 27 ORFs whose products augment viral growth in fibroblasts. It is possible that some ORFs are categorized incorrectly due to effects of mutations on neighboring ORFs. However, our analysis correctly verified the growth behavior of 38 ORFs previously placed into these categories, providing strong validation for our conclusions. In general, the essential and augmenting ORFs are located in the central region and the nonessential ORFs cluster at the ends of the viral genome (Fig. 2).
Seven clusters of conserved herpes virus ORFs have been identified (28) by comparing the genomes of the distantly related α-herpes viruses, such as herpes simplex virus, and γ-herpes viruses, such as Epstein-Barr virus, to that of HCMV. Interestingly, 43/46 of the HCMV ORFs contained within these conserved clusters are essential or augmenting ORFs, consistent with the view that these genes represent a core set of genes required for herpes virus replication and assembly. Only 3/88 nonessential HCMV ORFs fall within these conserved clusters, arguing that most nonessential ORFs are specific to HCMV and its β-herpes virus relatives.
The YD-Tn1721 transposon contains several features (Fig. 1b) that were not used in this study but will likely prove useful in future studies. It contains markers that allow further manipulation of the locus in which it resides. For example, a substitution mutation can readily be converted to a point mutation by blue-white screening by using the β-galactosidase marker in E. coli. The transposon contains two FRT sites that bracket its GFP, β-galactosidase, and kanamycin markers, allowing them to be removed from recombinant viruses by using the yeast Flp recombinase (29). The markers can be removed to reduce the size of the transposon insert from 3,660 to 760 bp and to facilitate reuse of the markers for modification of the viral genome at additional sites. YD-Tn1721 also contains elements encoding epitopes that tag disrupted proteins in four of six possible ORFs. This makes it possible to search for expressed proteins produced by ORFs with an insert.
The library of 413 defined insertion mutants should prove useful in future genetic studies of the AD169 strain of HCMV, providing the starting materials for analysis of phenotypes or further manipulation of selected genomic regions. Indeed, investigation of the physiological basis for the poor growth of mutants carrying inserts within the 19 newly identified augmenting mutants should rapidly provide new insights to HCMV gene functions.
Supplementary Material
Acknowledgments
We thank W. Brune (University of Würzburg, Würzburg, Germany) for the TsTm8B plasmid; W. Bresnahan for participation in designing TD-Tn5; D. Court (National Cancer Institute) for the DY380 bacterial strain; S. J. Flint for critical reading of the manuscript, and J. P. Cong for excellent technical assistance. This work was supported by grants from the National Institutes of Health to T.S. (CA82396, CA85786, and 87661). D.Y. is a Fellow of the Leukemia and Lymphoma Society. M.S. was supported by a grant from the Brazilian government [(Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq)].
Abbreviations: HCMV, human cytomegalovirus; BAC, bacterial artificial chromosome.
References
- 1.Britt, W. J. & Alford, C. A. (1996) in Fields Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott-Raven, Philadelphia), pp. 2493-2523.
- 2.Chee, M. S., Bankier, A. T., Beck, S., Bohni, R., Brown, C. M., Cerny, R., Horsnell, T., Hutchison, C. A., III, Kouzarides, T., Martignetti, J. A., et al. (1990) Curr. Top. Microbiol. Immunol. 154, 125-169. [DOI] [PubMed] [Google Scholar]
- 3.Davison, A. J., Dolan, A., Akter, P., Addison, C., Dargan, D. J., Alcendor, D. J., McGeoch, D. J. & Hayward, G. S. (2003) J. Gen. Virol. 84, 17-28. [DOI] [PubMed] [Google Scholar]
- 4.Murphy, E., Rigoutsos, I. & Shenk, T. (2003) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 5.Mocarski, E. S. & Courcelle, C. T. (2001) in Fields Virology, eds. Knipe, D. M., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B. & Straus, S. E. (Lippincott-Raven, Philadelphia), Vol. 2, pp. 2629-2673. [Google Scholar]
- 6.Yu, D., Smith, G. A., Enquist, L. W. & Shenk, T. (2002) J. Virol. 76, 2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchini, A., Liu, H. & Zhu, H. (2001) J. Virol. 75, 1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borst, E. M., Hahn, G., Koszinowski, U. H. & Messerle, M. (1999) J. Virol. 73, 8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobom, U., Brune, W., Messerle, M., Hahn, G. & Koszinowski, U. H. (2000) J. Virol. 74, 7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brune, W., Menard, C., Hobom, U., Odenbreit, S., Messerle, M. & Koszinowski, U. H. (1999) Nat. Biotechnol. 17, 360-364. [DOI] [PubMed] [Google Scholar]
- 11.Blankenship, C. A. & Shenk, T. (2002) J. Virol. 76, 12290-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith, G. A. & Enquist, L. W. (1999) J. Virol. 73, 6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu, D., Ellis, H. M., Lee, E. C., Jenkins, N. A., Copeland, N. G. & Court, D. L. (2000) Proc. Natl. Acad. Sci. USA 97, 5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, E. C., Yu, D., Martinez de Velasco, J., Tessarollo, L., Swing, D. A., Court, D. L., Jenkins, N. A. & Copeland, N. G. (2001) Genomics 73, 56-65. [DOI] [PubMed] [Google Scholar]
- 15.Heider, J. A., Bresnahan, W. A. & Shenk, T. E. (2002) Proc. Natl. Acad. Sci. USA 99, 3141-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bechtel, J. T. & Shenk, T. (2002) J. Virol. 76, 1043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bresnahan, W. A. & Shenk, T. E. (2000) Proc. Natl. Acad. Sci. USA 97, 14506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayashi, M. L., Blankenship, C. & Shenk, T. (2000) Proc. Natl. Acad. Sci. USA 97, 2692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Courcelle, C. T., Courcelle, J., Prichard, M. N. & Mocarski, E. S. (2001) J. Virol. 75, 7592-7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prichard, M. N., Duke, G. M. & Mocarski, E. S. (1996) J. Virol. 70, 3018-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf, D. G., Courcelle, C. T., Prichard, M. N. & Mocarski, E. S. (2001) Proc. Natl. Acad. Sci. USA 98, 1895-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prichard, M. N., Gao, N., Jairath, S., Mulamba, G., Krosky, P., Coen, D. M., Parker, B. O. & Pari, G. S. (1999) J. Virol. 73, 5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mocarski, E. S., Kemble, G. W., Lyle, J. M. & Greaves, R. F. (1996) Proc. Natl. Acad. Sci. USA 93, 11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bresnahan, W. A. & Shenk, T. (2000) Science 288, 2373-2376. [DOI] [PubMed] [Google Scholar]
- 25.Atalay, R., Zimmermann, A., Wagner, M., Borst, E., Benz, C., Messerle, M. & Hengel, H. (2002) J. Virol. 76, 8596-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masse, M. J., Karlin, S., Schachtel, G. A. & Mocarski, E. S. (1992) Proc. Natl. Acad. Sci. USA 89, 5246-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamminger, T., Gstaiger, M., Weinzierl, K., Lorz, K., Winkler, M. & Schaffner, W. (2002) J. Virol. 76, 4836-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roizman, B. & Pellett, P. E. (2001) in Fields Virology, eds. Knipe, D. M., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B. & Straus, S. E. (Lippincott-Raven, Philadelphia), Vol. 2, pp. 2381-2397. [Google Scholar]
- 29.Cox, M. (1988) in Genetic Recombination, eds. Kucherlapati, R. & Smith, G. R. (Am. Soc. Microbiol., Washington, DC), pp. 429-443.
- 30.Margulies, B. J., Browne, H. & Gibson, W. (1996) Virology 225, 111-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.