Abstract
Hereditary hemochromatosis is a common autosomal recessive disorder of iron metabolism. Recent demonstration of an association between transferrin receptor (TfR) and HFE, a major histocompatibility complex class I-like molecule that has been implicated to play a role in hereditary hemochromatosis, further strengthens the notion that HFE is involved in iron metabolism. Herein we show that TfR is required for and controls the assembly and the intracellular transport and surface expression of HFE. Because surface-expressed HFE and TfR remain firmly associated physically, only the fraction of TfR that is associated with HFE during biosynthesis is affected functionally. Moreover, we show that HFE binding reduces the number of functional transferrin binding sites and impairs TfR internalization, thus reducing the uptake of transferrin-bound iron. Thus, iron homeostasis is indirectly regulated by HFE, a negative modulator of TfR.
Hereditary hemochromatosis (HH) is a common autosomal recessive disorder of iron metabolism characterized by progressive iron overload (1). In a study of HH patients using linkage desequilibrium and full haplotype analysis, a gene (originally HLA-H but since renamed Hfe) that encodes a major histocompatibility complex class I-like molecule was identified (2). The Hfe gene product was shown to be highly expressed in certain epithelial cells throughout the gastrointestinal tract and to have a unique localization in the cryptal cells of the small intestine (3), implying a role for HFE in iron absorption (for reviews, see refs. 4 and 5). Direct proof that Hfe is the gene that causes HH came from a study on Hfe-deficient mice (6) in which profound alterations in several parameters of iron homeostasis that recapitulate the biochemical abnormalities and histopathology found in human HH (1) were observed. However, the underlying mechanism by which HFE is involved in iron metabolism remained obscure until an association between the HFE protein and transferrin receptor (TfR) was demonstrated (7–9).
All dividing cells express TfR on their surface, and the synthesis rate of TfR is closely linked to the requirements of the cell for iron (for review, see refs. 10 and 11). TfR, a transmembrane glycoprotein composed of two identical disulfide-linked subunits (12–14), binds plasma transferrin (Tf) with a strong preference for diferric Tf. Ligand binding induces rapid endocytosis of the Tf–TfR complex that is mediated by a tyrosine-based internalization signal located in the N-terminal cytoplasmic portion of the receptor (15). Within the endosomes, the TfR–Tf complex releases its bound iron to the cytosol and the TfR-bound Tf recycles to the cell surface, where the apo-Tf rapidly dissociates (for review, see refs. 10 and 11). It has recently been reported that HFE binding may reduce the affinity of TfR for Tf (7, 16). However, it is difficult to imagine that a reduction in affinity of Tf to TfR would effectively result in a reduction of iron uptake, because TfR–Tf complexes undergo rapid recycling. Thus, the question of whether the alleged reduction in affinity directly correlates with a reduction in cellular iron uptake remained unanswered. Moreover, the nature of the molecular interaction between the HFE and TfR proteins remained unclear.
In this study we have analyzed the nature of the biochemical association between the HFE and TfR and have examined the effect of this interaction on TfR-mediated Tf-bound iron uptake. We present convincing evidence showing that HFE not only blocks Tf binding to the TfR on the extracellular side but also inhibits internalization of the TfR in the cytoplasmic side. Thus, HFE inhibition of TfR function is responsible for the diminished uptake of Tf-bound iron.
MATERIALS AND METHODS
DNA, Transfections, Expression, and Purification of the Soluble HFE Protein.
Full-length Hfe cDNA was generated by PCR amplification from a human liver cDNA library (CLONTECH) and the PCR product was sequenced and subcloned into the pBI (CLONTECH) or the pCDNA3 (Invitrogen) expression vector. Human TfR cDNA (17) was provided by Sandy Schmid (The Scripps Research Institute, La Jolla, CA). HeLa cells were transiently transfected by the calcium phosphate method as described (18). Stable transfectant cell lines were established by transfecting transactivator-expressing HeLa cells (19) with the pBI construct and with a plasmid conferring resistance against ouabain (20). The truncated HFE covering amino acid residues 1–275 was sequenced and subcloned into the pRMHa-3 Drosophila expression vector (21) and cotransfected into Drosophila melanogaster SC2 cells with pRMHa-3 containing the human β2-microglobulin (β2m) cDNA (22). Transfection, selection, generation of stable HFE-expressing SC2 cells, and protein purification of HFE–β2m heterodimers were performed essentially as described (23). Protein homogeneity was confirmed by SDS/PAGE and silver staining. Protein concentration was determined by BCA assay (Pierce). Typically, the yield of soluble HFE heterodimers was ≈1.5 mg/liter.
Antibodies, Metabolic Radiolabeling, Immunoprecipitation, and SDS/PAGE.
The rabbit anti-HFE antiserum HFE-C was raised against a peptide corresponding to the 20 C-terminal amino acids (RKRQGSRGAMGHYVLAERE). Anti-HFE monoclonal antibodies were raised by immunizing BALB/c mice with purified recombinant HFE–β2m heterodimers. Hybridomas were generated as described (24). Antibody specificity was determined by ELISA, flow cytometry (FACS) analysis, immunofluorescence staining, Western blotting, and immunoprecipitation of HeLa and SC2 cells transiently expressing HFE or HLA-A2.1. Among the antibodies used in this study, 10G4 is specific for HFE heavy chain and the polyclonal antiserum K355 recognizes β2m (25). Mouse anti-human TfR monoclonal antibodies were purchased from PharMingen. Metabolic radiolabeling, immunoprecipitations, SDS/PAGE, and fluorography were carried out as described by Yang et al. (18). When denaturing samples before immunoprecipitation was required, SDS was added to the Nonidet P-40 cell lysates to a final concentration of 0.1% (18).
Confocal Fluorescence Microscopy and FACS.
HeLa cells transiently transfected with Hfe were incubated in serum-free DMEM for 1 h at 37°C. For live cell-association experiments, cells were incubated with 100 nM FITC-Tf (Molecular Probes) for 1 h at 37°C, fixed with 3% formaldehyde, permeabilized with 0.1% Triton X-100, and then stained with the anti-HFE-C antiserum followed by Texas red-conjugated goat anti-rabbit IgG conjugate (Molecular Probes). Confocal fluorescence microscopy was performed with a Bio-Rad MRC 1024 microscope at a nominal magnification of ×100. FACS analysis was performed as described (24) with a Becton Dickinson LYSIS II instrument.
Tf Binding and Internalization Assays.
Saturation binding protocols, as described by Jing et al. (26), were used to estimate the number of cell-surface Tf binding sites and the affinity of the TfR for 125I-labeled Tf. The data obtained were further analyzed by Scatchard transformation. Briefly, cells were incubated at 4°C for 90 min with 2–100 nM 125I-labeled Tf (NEN). For each concentration of 125I-labeled Tf, nonspecific binding was determined by addition of a 200- fold excess of unlabeled human Tf. Cells were removed with 0.5 ml of 1 M NaOH for 15 min, and the radioactivity in the NaOH pellet was measured in a γ counter (Packard). Tf steady-state surface/internal distribution and internalization rates were determined as described (27). The internal pool of TfRs was determined by incubating the cells at 37°C with 125I-labeled Tf at 4 μg/ml. The surface-bound 125I-labeled Tf was removed by incubating the cells twice with 0.5 ml of ice-cold 0.2 M acetic acid/0.5 M NaCl, pH 2.4, for 3 min, and the cell-associated radioactivity was determined. In control plates, the number of surface TfR was measured by incubating for 90 min at 37°C with unlabeled diferric Tf at 4 μg/ml. After washing, cells were incubated at 4°C with 125I-labeled Tf at 4 μg/ml. Cells were solubilized in 1 M NaOH, and the radioactivity was measured. A 200-fold excess of unlabeled Tf was added to some wells to determine nonspecific binding, which was typically less than 10% of the total binding. The internalization rate constant was determined by using the in/sur plot (28) with some modifications as described (27). Briefly, cells were incubated for various times at 37°C with 125I-labeled Tf at 4 μg/ml. Surface-bound Tf was removed by acid wash and the cell-associated radioactivity was measured. The amount of surface 125I-labeled Tf was measured after incubating the cells for 2 h at 4°C with 125I-labeled Tf at 4 μg/ml. All data were corrected for nonspecific binding. The rate constant of internalization was determined as the slope of a plot of the ratio of internalized Tf to steady-state surface Tf binding versus time.
TfR Recycling Assay.
The rate at which internalized apo-Tf is released from cells into the medium is used as a measure of the externalization constant kext. Determination of the TfR recycling rates was performed by the method of Tanner and Lienhard (29). Briefly, cells were incubated for 1 h at 37°C with 125I-labeled Tf at 4 μg/ml, and the surface-bound Tf was removed by incubating the cells for 15 min at 4°C in 1 ml of 150 mM NaCl/2 mM CaCl2/20 mM NaOAc, pH 5, containing 50 μM desferoxamine (Sigma). After washing, cells were incubated for 20 min at 4°C in PBS containing 50 μM desferoxamine and 125 nM apo-Tf and then incubated at 37°C for various times in prewarmed PBS containing 0.1% BSA and unlabeled human Tf at 50 μg/ml. After each time point, the medium was collected and the cells were removed from the wells with 1 M NaOH. Radioactivity released into the medium and associated with the cells was measured. A fraction (8–10%) of the 125I-labeled Tf remained associated with the cells even after a 1-h incubation and was subtracted from the values of the cell-associated radioactivity at the various time points (29).
RESULTS
Physical Association and Cotransport of HFE and TfR.
To examine the overall intracellular transport of the HFE molecule and study its association with TfR during biosynthesis, we generated stable HeLa transfectants in which Hfe gene expression is controlled via a tetracycline-responsive promoter (19). One of these transfectants, LS10, was selected for further characterization, because it expresses high levels of HFE in the absence of tetracycline and its HFE expression is completely inhibited by tetracycline at a concentration of 1 μg/ml (data not shown). To rule out the possibility that tetracycline might affect TfR expression in HeLa cells, we examined the expression levels of TfR under various concentrations of tetracycline. We have found that under those conditions, TfR expression was not significantly altered. In addition, we generated an antiserum against a peptide corresponding to the 20 C-terminal amino acids of HFE (HFE-C) and a panel of monoclonal antibodies against purified recombinant HFE protein. The specificity of these antibodies was determined by ELISA, immunoprecipitation, FACS, and Western blot analysis. Epitope mapping by sandwich ELISA revealed that a monoclonal antibody, named 10G4, specifically recognizes an epitope on the HFE heavy chain (unpublished data) without disrupting the complexes of HFE–β2m heterodimers and HFE–β2m–TfR heterotrimers and, hence, was used to study the biosynthesis of HFE.
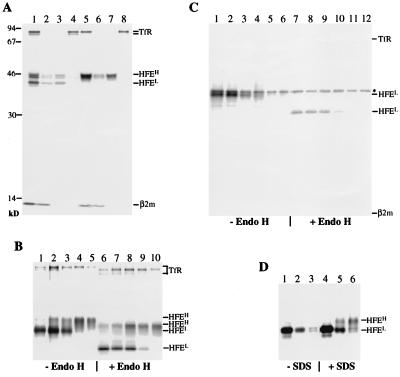
LS10 cells were labeled for 1 h with [35S]methionine followed by a 4-h chase, and cell lysates were immunoprecipitated with 10G4 (Fig. 1A, lanes 1 and 5). As visualized by SDS/PAGE and fluorography, 10G4 immunoprecipitates contained four major species of proteins with molecular masses of 12, 43, 46–48, and 75 kDa. The identities of these proteins were confirmed by reimmunoprecipitation with 10G4 and anti-β2m K355 (lanes 2 and 6), anti-HFE-C (lanes 3 and 7), and anti-TfR (lanes 4 and 8) antibodies. Immunoprecipitated HFE molecules consisted of two populations distinguishable by their sizes. A lower molecular mass form (HFEL) of 43 kDa, present at the start of the chase, was undetectable 4 h after synthesis, and a heterogeneous population of higher molecular mass forms (HFEH) of 46–48 kDa was detected in the chased samples. In addition, 10G4 coprecipitated TfR and β2m at both pulse and chase time points. These results suggested that HFE associates with TfR to form a trimeric complex composed of TfR, HFE heavy chain, and β2m.
Figure 1.
Biosynthesis, assembly, and intracellular transport of HFE and TfR. (A) Association of β2m and TfR with HFE. LS10 cells grown for 48 h in the absence of tetracycline were labeled with [35S]methionine for 1 h (lanes 1–4) followed by a 4-h chase (lanes 5–8). Nonidet P-40 lysates were first incubated with the monoclonal antibody 10G4 (lanes 1 and 5). Immunoprecipitates were then boiled in the presence of 0.1% SDS and reimmunoprecipitated with 10G4 and anti-β2m K355 (lanes 2 and 6), HFE-C (lanes 3 and 7), and anti-TfR (lanes 4 and 8) antibodies. Molecular mass markers are indicated in kDa on the left. (B and C) Pulse–chase analysis of the expression and intracellular transport of HFE in LS10 cells. Expression of HFE in LS10 cells was induced for 48 h. Cells were labeled for 30 min and then chased for various times: B, 0 min (lanes 1 and 6), 30 min (lanes 2 and 7), 1 h (lanes 3 and 8), 3 h (lanes 4 and 9), and 6 h (lanes 5 and 10); C, 0 min (lanes 1 and 7), 30 min (lanes 2 and 8), 1 h (lanes 3 and 9), 2 h (lanes 4 and 10), 4 h (lanes 5 and 11), and 6 h (lanes 6 and 12). The asterisk indicates nonspecific bands. Immunoprecipitated materials with 10G4 (B) or HFE-C (C) were divided into two portions and incubated at 37°C for 16 h with or without endo H. (D) Recognition of the C-terminal epitope of HFE by HFE-C. LS10 cells were labeled for 30 min and chased for 0 min (lanes 1 and 4), 2 h (lanes 2 and 5), and 4 h (lanes 3 and 6). Nonidet P-40 lysates were divided into two aliquots, one of which was boiled in the presence of 0.1% SDS. Samples were immunoprecipitated with HFE-C.
Because HFE association with TfR is detectable at pulse time points, we wanted to ascertain whether HFE displays different transport kinetics from that of the TfR. Therefore, we carried out a more detailed pulse–chase analysis followed by endoglycosidase H (endo H) digestion. These studies also enabled us to investigate the time of onset of the different HFE forms and to determine whether conformational changes and/or posttranslational modifications, such as glycosylation of HFE would affect their association with the TfR. LS10 cells were metabolically labeled for 1 h, chased for the indicated times, lysed, and immunoprecipitated with 10G4. Immunoprecipitated materials were divided into two equal aliquots and incubated overnight at 37°C in the absence or presence of endo H. As shown in Fig. 1B, analysis of the immunoprecipitates indicated that endogenously synthesized major histocompatibility complex class I molecules had a transport rate of ≈45 min (data not shown), whereas the transport rate of HFE was ≈150 min. Endo H digestion experiments showed that HFEL and HFEH forms correspond to HFE molecules that reside in the pre- and post-Golgi compartments, respectively. The high molecular mass form of the TfR, which is the surface-expressed mature form of TfR (30, 31), was also shown to be partially sensitive to endo H digestion (lanes 6–10 and refs. 30 and 31). Interestingly, partial endo H sensitivity was also observed for HFEH even after 8 h of chase (data not shown), suggesting that the HFEH and TfR molecules undergo similar posttranslational modifications during their intracellular transport. Fully endo H-sensitive TfR was shown to be coimmunoprecipitated with endo H-sensitive HFE at pulse time points (Fig. 1B, lane 1), suggesting that TfR first complexes with HFE in a pre-Golgi compartment. Thus, with the observation that both HFE and TfR molecules had similar transport kinetics, our data strongly suggest that upon association HFE and TfR are cotransported to the cell surface. Moreover, the stoichiometric ratios of HFE and TfR during the time courses of the pulse–chase experiments remained constant (data not shown and Fig. 2B), further suggesting that they remain associated during intracellular transport to the cell surface.
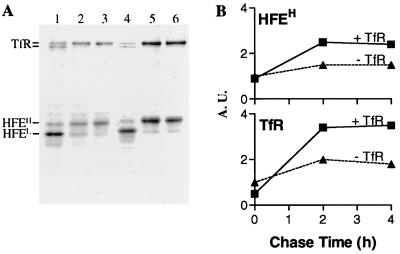
Figure 2.
Effect of TfR on surface expression of HFE. (A) Effect of TfR supertransfection on the biosynthesis and intracellular transport of HFE. LS10 cells without (lanes 1–3) or with (lanes 4–6) expression of additional TfR were labeled for 30 min and chased for 0 min (lanes 1 and 4), 2 h (lanes 2 and 5), or 4 h (lanes 3 and 6). Immunoprecipitates with 10G4 were analyzed by SDS/PAGE and fluorography. (B) Densitometric analysis of intracellular transport of HFEH and TfR. Values were expressed relative to the level of HFEH or TfR in LS10 cells at zero chase time point, which was arbitrarily set at 1.0 arbitrary units (A.U.). Four experiments were performed.
TfR Masks the HFE-C Epitope.
When similar pulse–chase studies were performed with the HFE-C antiserum, only endo H-sensitive HFEL was recognized and neither β2m nor TfR were coimmunoprecipitated at any time point (Fig. 1C). Because endo H-resistant HFEH was detected by 10G4 (Fig. 1 A and B), these results suggest that the C-terminal portion of the HFEH molecule, which is associated with TfR, is not available for recognition by HFE-C. It seems likely that TfR induces conformational changes of HFE, which shield the HFE-C epitope from recognition by the HFE-C antiserum. Alternatively, TfR binding might sterically block the HFE-C epitope. To examine whether association with TfR makes the HFE-C- epitope inaccessible, we investigated the recognition of HFEH isoforms by HFE-C under dissociation conditions. LS10 cells were pulse-labeled for 1 h and chased for a period of 0, 2, or 4 h. At each time point, cells were lysed and the Nonidet P-40 lysates were divided into two aliquots. One aliquot was incubated directly with HFE-C, and the other was adjusted to 0.1% SDS before addition of the antiserum. Fig. 1D showed that HFE-C was able to recognize both endo H-sensitive HFEL and endo H-resistant HFEH, indicating that under dissociation conditions the C-terminal portion of HFE becomes available for recognition by HFE-C.
TfR Is Required for the Intracellular Transport and Surface Expression of HFE.
Because our data showed that the stoichiometric ratios of HFE and TfR remain constant during the course of intracellular transport and that endo H-sensitive HFEL associates with endo H-sensitive TfR, we reasoned that TfR might be required for HFE assembly and intracellular transport. If more TfR were available to associate with newly synthesized HFE, more HFE would be expressed on the cell surface. To test this hypothesis, we supertransfected LS10 cells with human TfR cDNA and reexamined the biosynthesis and surface expression of HFE and TfR under similar conditions. LS10 cells with and without transfected TfR were labeled for 1 h and chased for 0, 2, or 4 h. Materials immunoprecipitated by 10G4 were divided into two aliquots, one of which was digested with endo H (not shown), to distinguish endoplasmic reticulum (ER)-resident HFEL from surface-expressed HFEH. Although in untransfected LS10 cells, HFEL was still visible 2 h after biosynthesis (Fig. 2A, lane 2), almost all of the HFE molecules became endo H-resistant within the same period of time in the TfR supertransfected LS10 cells (lane 5). Densitometric analysis showed that even at the pulse time point, the amount of endo H-sensitive HFEL was decreased by 15% in the TfR supertransfectants (lane 4) compared with the nontransfectants (lane 1). Moreover, the amount of endo H-resistant HFEH in the supertransfected cells was approximately two times greater (Fig. 2B). Similarly, the amount of coimmunoprecipitated TfR was also increased ≈2-fold in the supertransfected cells (Fig. 2B). Because stoichiometric ratios of HFE and TfR remain constant in the TfR supertransfected LS10 cells, our data suggest that the level of HFE surface expression is determined by the level of TfR expression. In contrast, transfection of cells with Hfe had no apparent effect on the transport kinetics of TfR (data not shown). Thus, our results show that HFE depends on newly synthesized TfR for exit from the ER and transport through the Golgi reticulum network. Therefore, because HFE and TfR remain firmly associated, only those newly synthesized HFE-associated TfR are functionally affected by HFE.
HFE Diminishes the Level of Cell-Associated Tf.
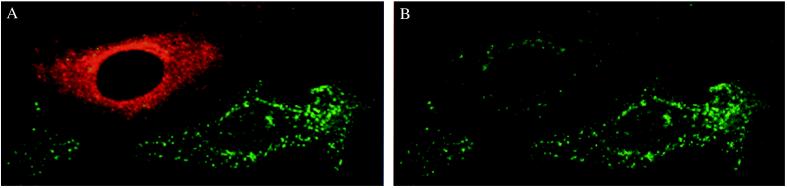
The biological function of cell surface-expressed TfR is to bind diferric Tf and deliver it to the endosomal system. Thus, any change in the number of functional surface-expressed receptors, their affinity for ligand, or internalization rate, will ultimately result in an alteration in the binding and/or uptake of Tf. To determine whether HFE has an effect on the binding of Tf to TfR, we used confocal fluorescence microscopy to examine the binding and uptake of FITC-labeled diferric Tf in HeLa cells transiently expressing HFE. Fig. 3 showed that internalized Tf was located in small punctate peripheral sorting endosomes and was mainly concentrated in a juxtanuclear region referred to as a recycling compartment (32) in both untransfected and HFE-transfected cells. However, a significant reduction of the internalized Tf was apparent in HFE-expressing cells (Fig. 3B, compare the cell on the upper left corner with the cell on the lower right corner), suggesting that HFE expression inhibits cellular Tf uptake.
Figure 3.
Effect of HFE on Tf uptake. HeLa cells transiently expressing HFE were incubated with FITC-Tf for 30 min at 37°C and examined by confocal fluorescence microscopy. (A) Cells expressing HFE (e.g., the cell on the upper left corner) were identified by incubation with the antiserum HFE-C and Texas red-conjugated goat anti rabbit IgG. (B) Differences in intensities of intracellular FITC-Tf between cells with (e.g., the cell on the upper left corner) and without (e.g., the cell on the lower right corner) HFE expression was apparent. The presence of surface-expressed HFE was also detected in an aliquot of the same cells with 10G4 (data not shown).
HFE Reduces the Number of Cell Surface Tf Binding Sites.
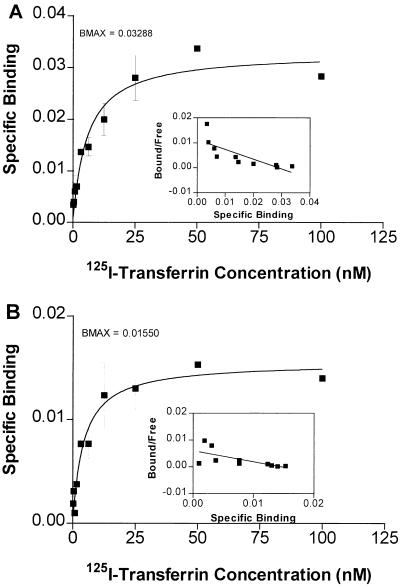
To confirm the observations obtained with confocal fluorescence microscopy, we next examined whether HFE expression alters the cell surface binding of Tf. Cell surface binding studies were undertaken with LS10 cells, in which the expression levels of HFE can be regulated by the concentration of tetracycline used (19). Thus, we were able to directly evaluate the dosage effect of increasing surface-expressed HFE on the function of TfR in LS10 cells. The amounts of cell surface HFE under the different tetracycline concentrations were determined by FACS analysis and immunoprecipitation (data not shown). At a tetracycline concentration of 1 μg/ml, there was no detectable expression of transfected HFE in LS10 cells. Low level of HFE was expressed at a tetracycline concentration of 0.01 μg/ml, whereas the expression level of HFE was further increased 10-fold in the absence of tetracycline. The degree of cell-surface Tf binding was determined by incubating the LS10 cells that were grown for 48 h in the presence of tetracycline at 1, 0.01, or 0 μg/ml for 90 min at 4°C in the presence of increasing concentrations of diferric 125I-labeled Tf. These experimental conditions allowed for Tf cell-surface binding to approach equilibrium without significant internalization. The saturation curves shown in Fig. 4 indicate that HFE expression was able to reduce the cell-surface binding of Tf. These results are in agreement with the findings of Feder et al. (7). Scatchard analysis of Tf binding was carried out to determine whether the decrease in Tf binding at the cell surface was due to decreased affinity of TfR for Tf (Fig. 4 Insets). In the absence of HFE, the apparent KD for TfR-bound Tf was 4.9 ± 1.0 nM (mean ± SEM), whereas in the presence of HFE, the value was 4.9 ± 1.1 nM. These values were highly reproducible in four experiments and are similar to the values reported previously for the affinity between TfR and Tf (26), suggesting strongly that HFE binding did not change the affinity of TfR for Tf.
Figure 4.
Effect of HFE on number of cell-surface Tf-binding sites. LS10 cells were grown for 48 h in medium containing tetracycline at concentrations of 1.0 μg/ml (A) or 0 μg/ml (B). Saturation binding curves were determined after incubating cells for 90 min at 4°C in the presence of various concentrations of 125I-labeled Tf. Results are expressed as means ± SEM of triplicate measurements. Scatchard analyses were performed with the prizm software program (GraphPad, San Diego). It was calculated that in the absence of HFE the apparent KD for TfR-bound Tf was 4.9 ± 1.0 nM, whereas in the presence of HFE the KD was 4.9 ± 1.1 nM. Four experiments were performed.
Under our experimental conditions, the number of surface-expressed TfRs available for Tf binding was estimated to be 2 × 105 molecules per cell in LS10 cells whose HFE expression was completely inhibited by tetracycline (Fig. 4A Inset); this is similar to the previously reported values for HeLa cells (33). In contrast, the number of surface-expressed TfRs available for Tf binding decreased to 0.9 × 105 molecules per cell in cells expressing high levels of HFE (Fig. 4B Inset). Cells expressing a low level of HFE (i.e., cultured in the presence of tetracycline at 0.01 μg/ml) had 1.6 × 105 available receptors per cell (Table 1). These results were highly reproducible and were also obtained when using a different tetracycline-regulated stable clone (data not shown). Because HFE expression reduced the number of functional Tf binding sites available at the cell surface, our data strongly suggest that the effect of HFE on surface Tf binding is the result of changes in number of available functional receptors rather than ligand affinity. Our findings are in contrast to previously published results that reported a 10-fold increase in apparent KD values in the presence of HFE (16). A likely explanation for this disparity lies with the different methodologies and experimental systems used. Although wild-type HFE was used in our study, mutant HFE with a C-terminal Flag epitope was used by others (16). In light of our finding that the cytoplasmic portion of HFE is very important for its association with TfR, it is likely that even though this C-terminal modified molecule can still associate with TfR, its behavior differs significantly from its wild-type counterpart.
Table 1.
Internalization and externalization rate constants
| Relative levels of HFE expression | kint, min−1 | kext, min−1 | kext/kint | S/I | Functional TfR, no. |
|---|---|---|---|---|---|
| No | 0.16 ± 0.01 | 0.06 ± 0.00 | 0.38 | 0.35 | 2.0 × 105 |
| Low | 0.13 ± 0.01 | 0.05 ± 0.00 | 0.35 | 0.39 | 1.6 × 105 |
| High | 0.06 ± 0.00 | 0.05 ± 0.00 | 0.86 | 0.75 | 0.9 × 105 |
S/I ratios, internalization, and externalization rate constants were calculated. The relative levels of functional TfRs on the surface of LS10 cells grown in the presence of various concentrations of tetracycline are expressed in molecules per cell. Values are means ± SEM. Triplicate measurements from three experiments (e.g., Fig. 5) were performed.
HFE Impairs the Internalization of Tf-Bound Receptor.
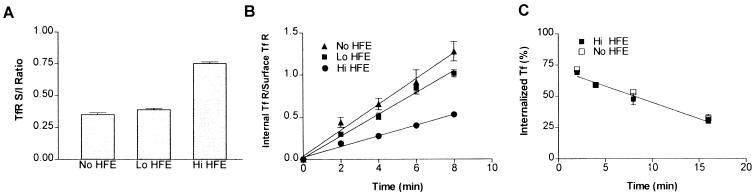
To examine the effect of HFE on the steady-state distribution of cell surface and intracellular Tf-bound TfR, we investigated the efficiency of endocytosis and recycling of the Tf–TfR complexes in LS10 cells. The internalization efficiency of the Tf–TfR complexes was estimated from measurements of the steady-state distribution of the complexes at 37°C and from their rate of recycling back to the cell surface (29). The distribution of the TfR between the cell surface and intracellular compartments reflects the ratio of the externalization rate constant, kext (which relates to the return to the cell surface of the endocytosed receptor) to the internalization rate constant, kint. In other words, at steady-state the internalization rate of the diferric Tf–TfR complexes (TR)sur equals the rate of externalization of the internal pool of apo-Tf–TfR complexes, (TR)int as follows: kint(TR)sur = kext(TR)int or (TR)sur/(TR)int = kext/kint, if we assume an insignificant rate of intracellular degradation (26, 27). Indeed, degradation appears to be minimal in our system, because the TfR–HFE complexes were stable for at least 12 h in LS10 cells (data not shown). The ratio of surface TfR to internal TfR (S/I) was determined. In the absence of HFE the S/I ratio was ≈0.35, indicating that the internalization rate constant is ≈3 times greater than that of the externalization rate constant (Fig. 5A). In HFE-expressing cells, the S/I ratio was ≈0.75. These data are in agreement with our immunofluorescence microscopic observations that suggest that, at steady state, cells expressing HFE consistently showed a significantly lower amount of internalized Tf than non-HFE-expressing cells. Direct measurement of the internalization rate constants, which represent the fraction of surface TfR internalized per minute, was performed by using the protocol described by McGraw and Maxfield (27). We found that the LS10 cells in the presence of tetracycline at 1 μg/ml had an internalization rate of 0.16 min−1, whereas a value of 0.13 min−1 was obtained in LS10 cells cultured in the presence of tetracycline at 0.1 μg/ml. Moreover, LS10 cells in the absence of tetracycline had an internalization rate of ≈0.06 min−1 (Fig. 5B), strongly suggesting that HFE expression impairs Tf–TfR internalization and that the degree of inhibition depends on the expression levels of HFE.
Figure 5.
Effect of HFE on TfR internalization. LS10 cells were grown for 48 h in the presence of tetracycline at 1.0 μg/ml (No HFE), 0.01 μg/ml (Lo HFE), or 0 μg/ml (Hi HFE). (A) The S/I ratio represents the ratio of the endocytic externalization to internalization rate constants. (B) Determination of the internalization constants. Cells were incubated for 2, 4, 6, or 8 min at 37°C with 125I-labeled Tf, and an acid wash was used to distinguish between membrane-bound and internalized Tf. A plot of the ratio of internalized to steady-state surface Tf binding versus time yields a straight line whose slope is the internalization rate constant. All measured kint values are presented in Table 1. (C) Determination of the externalization constants. After cell surface-bound 125I-labeled Tf was removed by the deferoxamine wash, cells were then incubated at 37°C for 2, 4, 8, or 16 min, and the radioactivity that remained associated with the cells as well as that was released into the media was determined. Values were corrected for nonspecific binding and the values for kext are presented in Table 1. The results presented are the means ± SEM from triplicate determinations. Three experiments were performed.
Recycling rates were determined by removing cell-surface-bound Tf under conditions that do not affect the recycling efficiency of endocytosed Tf–TfR complexes (26). Fig. 5C showed that the internal pool of 125I-labeled Tf was externalized at the same rate regardless of the expression levels of HFE in cells. Because kext was the same for LS10 cells with and without HFE expression, kint is proportional to (TR)int/(TR)sur. Thus, comparing kint values in cells with and without HFE expression would reveal the relative internalization efficiency of their Tf–TfR complexes. From the steady-state distribution of TfR, the relative efficiency of receptor internalization in LS10 cells with and without HFE expression was calculated according to the following formula: (kint TfR of HFE-expressing cells/kint TfR of non-HFE-expressing cells) × 100%. These values were determined to be ≈85% and ≈40% for the LS10 cells cultured in the presence of tetracycline at 0.01 and 0 μg/ml, respectively. Furthermore, comparison of the Tf endocytic rates in LS10 cells with different levels of HFE expression revealed that the kext/kint ratios were similar to the S/I distributions of 0.35, 0.39, and 0.75 (Table 1), which are in agreement with the previous steady-state analysis. Thus, our data strongly suggest that the changes in the steady-state S/I ratio were the result of the changes in internalization rates. We have estimated the efficiency of TfR internalization by determining the steady-state distribution of receptors and their internalization and externalization rates, revealing that TfRs in cells expressing HFE were internalized ≈40–50% as efficiently as in cells without HFE expression. Because efficiency of internalization and recycling of the Tf–TfR complexes correlates well with the uptake of Tf-bound iron by cells and its intracellular iron accumulation, it can be concluded that HFE expression inhibits Tf–TfR internalization, thus reducing iron uptake and accumulation.
DISCUSSION
HFE, a class Ib molecule encoded in the major histocompatibility complex region, does not have an obvious role in the immune response against pathogens (for review, see ref. 34). In this study we show that HFE associates with TfR in the ER and that the HFE–TfR complex is then cotransported through the Golgi reticulum network. During the course of intracellular transport to the cell surface, the stoichiometry of the TfR–HFE complexes remains unchanged, suggesting that HFE and TfR are in firm association on reaching the cell surface. Our finding that HFE was unable to exit the ER without TfR, although surprising, is not unique. In a recent report, McLatchie et al. (35) have shown that the members of a family of proteins called receptor-activity-modifying proteins are required for, and control, the intracellular transport of the calcitonin-receptor-like receptor. A consequence of this cotransport is that, depending on its expression level, HFE can only affect the fraction of TfR with which it associates. This limitation might explain why it takes years to develop HH in the absence of functional HFE. This is an important point in understanding the underlying mechanism of HH, especially in light of our findings that HFE is expressed in all cell types examined and that the expression levels of HFE in different cell types vary (data not shown). Therefore, it is important to study the factors that influence HFE expression to understand how TfR function is regulated. In this regard, we failed to find any obvious regulatory sites for interleukin- or iron-mediated transcriptional factor binding by sequence analysis of the HFE promoter and untranslated regions. Experimentally, we found no induction of HFE expression in HEK 293 or HeLa cells by various cytokines (data not shown), even though almost all the major histocompatibility complex molecules are up-regulated by interferon.
Our data further showed that HFE expression does not change the affinity of Tf to TfR, but rather it reduces the number of Tf-binding competent TfR molecules on the cell surface. More importantly, our data further showed that, on HFE binding, internalization of TfR-bound Tf is completely inhibited. Thus, Tf-bound iron uptake by TfR is negatively modulated by HFE. It has recently been reported that in the presence of HFE, the affinity of TfR for Tf was reduced by 2- to10-fold (7, 16), which is in sharp contrast to our findings that the effect of HFE on surface binding of Tf was the result of changes in functional receptor number rather than ligand affinity. Because interaction between HFE and TfR induces conformational changes on the cytoplasmic portions of HFE and/or TfR, the use of the C-terminal Flag epitope-tagged HFE and/or the soluble HFE without its cytoplasmic portion could severely affect its interaction with the TfR and that might, in turn, reduce the affinity of TfR for Tf. With respect to the finding that HFE does not occlude both Tf-binding sites of the TfR homodimer (9), it is difficult to imagine what evolutionary selective pressures would work to inactivate only one-half of the TfR homodimer. One would argue that it is more evolutionarily feasible to achieve the same effect by down-regulating the expression of TfR. Thus, it is difficult to conceptualize that HFE influences TfR activity primarily by reducing Tf affinity to TfR (7, 16). Most likely, the reduction of Tf binding to TfR is just the result of the physical blockage of Tf binding sites on TfR by the HFE molecule. Moreover, our study shows that HFE-associated TfR is not capable of delivering Tf-bound iron into the cells, due to the inhibition of TfR internalization by HFE. Thus, the extent of the functional regulation of TfR by HFE is dependent upon the expression levels of HFE, which vary among different types of cells.
Cellular iron homeostasis seems to be regulated at the level of iron uptake, which is dependent on TfR activity (11, 36, 37). In this study we show that HFE plays an essential role in modulating TfR activity, thus indirectly regulating the level of cellular iron uptake. Because TfR activity is closely linked to the requirements of the cell for iron, it is conceivable that complete inhibition of TfR function by HFE would ultimately lead to cell death. In fact, we have observed that a significant fraction of LS10 cells died after a 6-day induction of HFE overexpression and that almost all cells died after a 8-day induction (unpublished data). On the other hand, it can be concluded that excessive cellular iron uptake is the result of an Hfe deficiency due to the lack of negative TfR regulation. In animals, either an excess or a deficiency of iron leads to diseases. In HH patients as well as in Hfe-deficient mice, high levels of iron can be found in most tissues (1, 6), implying that HFE is normally expressed in these tissues. Indeed, it has been found that HFE is constitutively expressed in every tissue examined and in many cell types, as demonstrated by Northern blot (ref. 2 and unpublished data) and in situ hybridization analysis (unpublished data). In conclusion, cellular iron uptake by TfR appears to be a delicate process that is negatively regulated by HFE.
Acknowledgments
We thank Jonathan Blevitt, Lars Karlsson, Veronica Moreno, and Ola Winqvist for technical assistance and/or discussions and Anne M. Fourie, Charlie Glass, and Gary Schoenhals for critical reading. The technical assistance of the DNA and peptide synthesis facilities of the R. W. Johnson Pharmaceutical Research Institute is gratefully acknowledged.
ABBREVIATIONS
- Tf
transferrin
- TfR
Tf receptor
- HH
hereditary hemochromatosis
- β2m
β2-microglobulin
- FACS
flow cytometry
- ER
endoplasmic reticulum
- S/I
ratio of surface TfR to internal TfR
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Bothwell T H, Charlton R W, Motulski A G. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 2237–2269. [Google Scholar]
- 2.Feder J N, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy D A, Basava A, Dormishian F, Domingo R, Ellis M C, Fullan A, et al. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 3.Parkkila S, Waheed A, Britton R S, Feder J N, Tsuchihashi Z, Schatzman R C, Bacon B R, Sly W S. Proc Natl Acad Sci USA. 1997;94:2534–2539. doi: 10.1073/pnas.94.6.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuthbert J A. J Invest Med. 1997;45:518–529. [PubMed] [Google Scholar]
- 5.Cox T M, Kelly A L. Curr Opin Genet Dev. 1998;8:274–281. doi: 10.1016/s0959-437x(98)80081-6. [DOI] [PubMed] [Google Scholar]
- 6.Zhou X Y, Tomatsu S, Fleming R E, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt E M, Ruddy D A, Prass C E, et al. Proc Natl Acad Sci USA. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feder J N, Penny D M, Irrinki A, Lee V K, Lebron J A, Watson N, Tsuchihashi Z, Sigal E, Bjorkman P J, Schatzman R C. Proc Natl Acad Sci USA. 1998;95:1472–1477. doi: 10.1073/pnas.95.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkkila S, Waheed A, Britton R S, Bacon B R, Zhou X Y, Tomatsu S, Fleming R E, Sly W S. Proc Natl Acad Sci USA. 1997;94:13198–13202. doi: 10.1073/pnas.94.24.13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lebron J A, Bennett M J, Vaughn D E, Chirino A J, Snow P M, Mintier G A, Feder J N, Bjorkman P J. Cell. 1998;93:111–123. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- 10.Harford J B, Rouault T A, Huebers H A, Klausner R D. In: Molecular Basis of Blood Diseases. Stamatoyannopoulos G, Nienhhuis A, Majerus P M, Varmus H, editors. Philadelphia: Saunders; 1994. p. 351. [Google Scholar]
- 11.Richardson D R, Ponka P. Biochim Biophys Acta. 1997;1331:1–40. doi: 10.1016/s0304-4157(96)00014-7. [DOI] [PubMed] [Google Scholar]
- 12.Schneider C, Owen M J, Banville D, Williams J G. Nature (London) 1984;311:675–679. doi: 10.1038/311675b0. [DOI] [PubMed] [Google Scholar]
- 13.Seligman P A, Schleicher R B, Allen R H. J Biol Chem. 1979;254:9943–9946. [PubMed] [Google Scholar]
- 14.Jing S Q, Trowbridge I S. EMBO J. 1987;6:327–331. doi: 10.1002/j.1460-2075.1987.tb04758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collawn J F, Stangel M, Kuhn L A, Esekogwu V, Jing S, Trowbridge I S, Tainer J A. Cell. 1990;63:1061–1072. doi: 10.1016/0092-8674(90)90509-d. [DOI] [PubMed] [Google Scholar]
- 16.Gross C N, Irrinki A, Feder J N, Enns C A. J Biol Chem. 1998;273:22068–22074. doi: 10.1074/jbc.273.34.22068. [DOI] [PubMed] [Google Scholar]
- 17.McClelland A, Kuehn L C, Ruddle F H. Cell. 1984;39:267–274. doi: 10.1016/0092-8674(84)90004-7. [DOI] [PubMed] [Google Scholar]
- 18.Yang Y, Waters J B, Frueh K, Peterson P A. Proc Natl Acad Sci USA. 1992;89:4928–4932. doi: 10.1073/pnas.89.11.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Frueh K, Ahn K, Peterson P A. J Biol Chem. 1995;270:27687–27694. doi: 10.1074/jbc.270.46.27687. [DOI] [PubMed] [Google Scholar]
- 21.Bunch T A, Grinblat Y, Goldstein L S B. Nucleic Acids Res. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson M R, Song E S, Yang Y, Peterson P A. Proc Natl Acad Sci USA. 1992;89:12117–12121. doi: 10.1073/pnas.89.24.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sykulev Y, Brunmark A, Jackson M, Cohen R J, Peterson P A, Eisen H N. Immunity. 1994;1:15–22. doi: 10.1016/1074-7613(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 24.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 25.Ahn K, Angulo A, Ghazal P, Peterson P A, Yang Y, Fruh K. Proc Natl Acad Sci USA. 1996;93:10990–10995. doi: 10.1073/pnas.93.20.10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jing S, Spencer T, Miller K, Hopkins C, Trowbridge I S. J Cell Biol. 1990;110:283–294. doi: 10.1083/jcb.110.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGraw T E, Maxfield F R. Cell Regul. 1990;1:369–377. doi: 10.1091/mbc.1.4.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiley H S, Cunningham D D. J Biol Chem. 1982;257:4222–4229. [PubMed] [Google Scholar]
- 29.Tanner L I, Lienhard G E. J Biol Chem. 1987;262:8975–8980. [PubMed] [Google Scholar]
- 30.Omary M B, Trowbridge I S. J Biol Chem. 1981;256:12888–12892. [PubMed] [Google Scholar]
- 31.Do S I, Enns C, Cummings R D. J Biol Chem. 1990;265:114–125. [PubMed] [Google Scholar]
- 32.Dunn K W, McGraw T E, Maxfield F R. J Cell Biol. 1989;109:3303–3314. doi: 10.1083/jcb.109.6.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beauchamp J R, Woodman P G. Biochem J. 1994;303:647–655. doi: 10.1042/bj3030647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shawar S M, Vyas J M, Rodgers J R, Rich R R. Annu Rev Immunol. 1994;12:839–880. doi: 10.1146/annurev.iy.12.040194.004203. [DOI] [PubMed] [Google Scholar]
- 35.McLatchie L M, Fraser N J, Main M J, Wise A, Brown J, Thompson N, Solari R, Lee M G, Foord S M. Nature (London) 1998;393:333–339. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- 36.Sayers M H, English G, Finch C. Am J Hematol. 1994;47:194–200. doi: 10.1002/ajh.2830470309. [DOI] [PubMed] [Google Scholar]
- 37.Huebers H A, Finch C A. Physiol Rev. 1987;67:520–582. doi: 10.1152/physrev.1987.67.2.520. [DOI] [PubMed] [Google Scholar]