Abstract
We conducted an analysis of the topology of AtNHX1, an Arabidopsis thaliana vacuolar Na+/H+ antiporter. Several hydrophilic regions of the antiporter were tagged with a hemagglutinin epitope, and protease protection assays were conducted to determine the membrane topology of the antiporter by using yeast as a heterologous expression system. The overall structure of AtNHX1 is distinct from the human Na+/H+ antiporter NHE1 or any known Na+/H+ antiporter. It is comprised of nine transmembrane domains and a hydrophilic C-terminal domain. Three hydrophobic regions do not appear to span the tonoplast membrane, yet appear to be membrane associated. Our results also indicate that, whereas the N terminus of AtNHX1 is facing the cytosol, almost the entire C-terminal hydrophilic region resides in the vacuolar lumen. Deletion of the hydrophilic C terminus resulted in a dramatic increase in the relative rate of Na+/H+ transport. The ratio of Na+/K+ transport was twice that of the unmodified AtNHX1. This altered ratio resulted from a relatively small decrease in K+/H+ transport with a large increase in Na+/H+ transport. The vacuolar localization of the C terminus of the AtNHX1, taken together with the regulation of the antiporter selectivity by its C terminus, demonstrates the existence of luminal vacuolar regulatory mechanisms of the antiporter activity.
Na+/H+ antiporters play major roles in pH and Na+ homeostasis of cells throughout the biological kingdom (1–4). Na+/H+ antiporters are integral membrane proteins residing in the plasma membranes and endomembranes of cells. Mammalian Na+/H+ exchange relies on a sodium electrochemical gradient (ΔμNa+) established across the membrane by the action of Na+/K+-ATPases. Although their basic mode of action appears to be simple and invariant, they have been implicated in multiple diverse functions. In mammalian cells, these include the regulation of intracellular pH, regulation of cellular volume, elimination of excess acid from actively metabolizing cells, and the reabsorption of NaCl across renal, intestinal, and other epithelia (1). The NHE family of antiporters carries out this array of functions. Their activity is localized at the plasma membrane, mitochondrial inner membrane, and endoplasmic reticulum. A similar membrane topology is predicted for all isoforms on the basis of their primary structure with 10–12 membrane-spanning regions at the N terminus and a cytosolic region at the C terminus (2). In plants, vacuolar Na+/H+ antiporters (5) use the proton electrochemical gradient (ΔμH+) generated by the vacuolar H+-translocating enzymes, H+-ATPase, and H+-PPiase to couple the downhill movement of H+ with the uphill movement of Na+ (6). The CPA1 family of Arabidopsis cation/H+ antiporters comprises six members, At-NHX1–6 (7, 8), which have a significant similarity to the endosomal NHX1 from yeast (9, 10). These antiporters have functional Na+/H+ antiporter activity based on their ability to complement the endosomal yeast Nhx1p (9, 10). Immunological and GFP expression studies have shown that AtNHX1 (5, 11) and AtNHX2 (9) are localized to the vacuole. Plant vacuolar Na+/H+ antiporters have been shown to play important roles in cellular ion homeostasis, including the sequestration of Na+ ions into the vacuole (5, 12) and vacuolar pH regulation (13).
All six vacuolar AtNHX proteins share some basic structural similarities. They have a consensus amiloride-binding sequence and two short amino acid stretches that are similar to a putative cation-binding domain (14) and the last extracellular loop of the human NHE1 (15). Analysis of the AtNHX1–6 primary structure predicts 7–12 transmembrane domains. A number of secondary structure models can be derived exclusively from hydropathy analysis of primary sequences, but there is no additional biochemical evidence to support any proposed topology of the vacuolar transporters. In fact, the predicted secondary structure of transporters often varies depending on the programs used to analyze the hydropathy profiles. Clearly, biochemical and structural studies are needed to ascertain the topology of the vacuolar transporters. The Arabidopsis Genome Initiative (16) has identified at least 800 predicted plant membrane-bound transporters, and more than half are predicted to be H+-coupled transporters (7, 8). Despite all of the sequence information available, topology and structure/function studies of these transporters have not yet been initiated. This lack of fundamental knowledge poses a limitation for the analysis of ion transport mechanisms, ion homeostasis, and ultimately for the understanding of how plants cope with changing ionic environments.
Here we conducted an analysis of the topology of AtNHX1 by using yeast as a heterologous expression system. Our results show that AtNHX1 is comprised of nine transmembrane domains, with a hydrophilic C-terminal domain and three hydrophobic regions that do not appear to span the tonoplast membrane, yet they appear to be membrane associated. Whereas the N terminus faces the cytosol, the C terminus faces the vacuolar lumen. Moreover, the C terminus appears to be involved in the determination of the ion selectivity of the transporter.
Materials and Methods
Yeast Strains and Media. Saccharomyces cerevisiae vacuolar protease-deficient strain MM476 (MAT α, pep4–3, leu2, trp1, ura3–52, prb1–1122) was kindly provided by Morris Manolson (University of Toronto, Toronto). The strain TY001 [MAT α, pep4–3, leu2, trp1, ura3–52, prb1–1122, nhx1::TRP (TRP1)] was generated by replacing the NHX1 locus with the TRP1 gene of MM476 by the PCR-based gene deletion method (17).Yeast cells were grown in YPD (1% yeast extract/2% peptone/2% glucose; Sigma) or SD media (0.67% yeast nitrogen base/2% glucose; Sigma) with appropriate amino acid supplements as indicated.
Plasmid Construction and Transformation of Yeast. A 3xHA-epitope (hemagglutinin peptide epitope) was inserted into 14 different positions of the AtNHX1 ORF by fusion PCR. The primers used are listed in Table 3, which is published as supporting information on the PNAS web site, www.pnas.org. The 3xHA fragment and AtNHX1 were amplified from pKEB35 [provided by T. Stevens (University of Oregon); ref. 18] or pBSK-X1 (5), respectively. Each PCR was performed by using pfu-turbo DNA polymerase (Stratagene). The amplified fragments were ligated into the pCR-Blunt II TOPO vector by using the Zero Blunt TOPO cloning kit (Invitrogen). These plasmids were digested with EcoRI and SalI, and 1.6-kb fragments were gel purified and then ligated into the p426-TEF vector (obtained from American Type Culture Collection) (19). The resulting plasmids were designated as pHpl-1-pHpl-13.2. p426-X1 was constructed by ligating the BamHI-EcoRI fragment of pYPGE15-X1 (see below) containing the AtNHX1 ORF into p426-TEF. S. cerevisiae MM426. TY001 yeast cells were transformed with the pHpl vectors by the standard lithium acetate method. MM426 and TY001 were used for protease protection assays and functional complementation tests, respectively. For transport assays, the TY001 strain was transformed with pYPGE15 vectors (20) bearing either no insert (control) or truncated forms of the AtNHX1 gene (full length-amino acids 1–538, N terminus truncation amino acids 18–538, or C terminus truncation amino acids 1–456). The full length AtNHX1 ORF was subcloned into pYPGE15 by using the BamHI and EcoRI sites of pBSK-X1 (pYPGE15-X1). N and C terminus deletion constructs were generated by ligating PCR amplified fragments with appropriate primer sets (see Table 3) into the pCR-Blunt II TOPO vector, as described above. Each deletion fragment was subcloned into the BamHI and EcoRI sites of pYPGE15.
Isolation of Intact Vacuoles from Yeast. Intact vacuoles were isolated as described by Ohsumi and Anraku (21). The intact vacuoles were collected at the top of the gradient and resuspended in 10 mM Mes-Tris (pH 6.9). The vacuoles were almost free of contamination by unbroken spheroplasts and lipid granules (21). Protein concentrations were determined by the Bio-Rad DC Protein Assay according to the manufacturer's protocol.
Protease Protection Assay. Vacuoles (25 μg of protein) were resuspended in 100 μl of 10 mM Tris-Mes (pH 6.9), 50 μg of proteinase K, and 1% Triton X-100 (where indicated) and incubated on ice for 1 h. PMSF (to a final concentration of 20 mM) was added to terminate the reaction with incubation for 20 min on ice. Samples were denatured by adding 5× Laemmli's sample buffer. Twenty microliters (3.3 μg of vacuolar protein) was subjected to a 12% Tris-Tricine SDS/PAGE followed by Western blot analysis. Anti-HA-epitope monoclonal antibody 16B12 (Babco, Richmond, CA) and anti-mouse IgG antibody–horseradish peroxidase conjugate (Molecular Probes) were used at a dilution of 1/1,000 and 1/10,000, respectively. Vacuolar integrity was monitored by the susceptibility of pro-carboxypeptidase Y (CPY) to proteinase K according to Landolt-Marticorena et al. (22), with slight modifications. Samples were incubated with proteinase K in the presence or absence of Triton X-100 for the indicated times on ice, and PMSF and 5× sample buffer were added as above. The samples (1.65 μg of vacuolar protein) were resolved in 10% Tris-Glycine SDS/PAGE followed by Western blot. Anti-CPY monoclonal antibodies (Molecular Probes) were used at 1/5,000 dilution, and secondary antibodies were used as described above.
Yeast Complementation. All pHpl plasmids were introduced into TY001, as described above. Exponentially grown cells were harvested and adjusted to OD600 = 1.0 with double-distilled water. Serial dilutions of 1/10 and 1/100 were made, and 3 μl of each culture was spotted onto YPD and YPD/hygromicin (30 μg/ml) plates.
Transport Assays. The fluorescence quenching of acridine orange was used to monitor the establishment and dissipation of vacuolar-inside acidic pH gradients. Intact vacuoles (95 μg of vacuolar protein) were used for each assay. Vacuoles were added to a buffer containing 25 mM tetramethyl ammonium-Cl; 5 μM acridine orange; 5 mM Tris-Mes, pH 7.5; 1 mM DTT; and 3.125 mM MgSO4. The vacuolar ATPase was activated by the addition of 1.5 mM Tris-ATP, and time-dependent fluorescence changes were monitored on a Perkin–Elmer fluorescence spectrophotometer with excitation and emission wavelengths of 495 and 540 nm, respectively, and a slit width of 5 nm with a 1% transmittance filter. When a steady-state pH gradient was established, the vacuolar ATPase was stopped by addition of bafilomycin. After 3 min, Na+ or K+ was added as the gluconate salt (between 2.5 and 100 mM). Initial rates were measured as the slope of the relaxation of the quench over a period of ≈25 s. Rates were reported as percent quench per minute per milligram of protein. All curves were normalized to 100% quench before quantitation. Rates of transport in TY001 cells transformed with pYPGE15 only (as control) were small but were subtracted from the rates obtained for vacuoles containing the different AtNHX1 forms. Curves were fitted to the mean values of rates at each concentration measured by using KALEIDEGRAPH (Synergy Software, Reading, PA).
Results
HA Tagging of AtNHX1 Hydrophilic Regions. Hydropathy plot analyses (23) indicate that AtNHX1 contains 12 putative hydrophobic regions (Fig. 1). Different topology prediction programs (SOSUI, http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0.html; TOPPRED, http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html; and HMMTOP, www.enzim.hu/hmmtop) suggest that AtNHX1 is a protein that contains 9–12 membrane-spanning domains (not shown). We used 3xHA-epitope insertion mutagenesis to assess the topology of AtNHX1. AtNHX1 was tagged by 3xHA tag at different hydrophilic loop regions (Hpls) (Fig. 1) independently and expressed in S. cerevisiae under the control of the TEF promoter (24, 25). We used yeast as a heterologous expression system, taking advantage of the similarity between AtNHX1 and ScNhx1p (26) and the ability of AtNHX1 to functionally complement ScNhx1 (9, 10, 26).
Fig. 1.
HA-tagged constructs of AtNHX1. (A) Hydropathy plot of AtNHX1. (B) Schematic diagram indicating the 3xHA-tagged AtNHX1 constructs used in this study. Numerals above the boxes indicate the number of putative transmembrane domains. Blank boxes indicate putative transmembrane domains and shadowed box indicates 3xHA-tag.
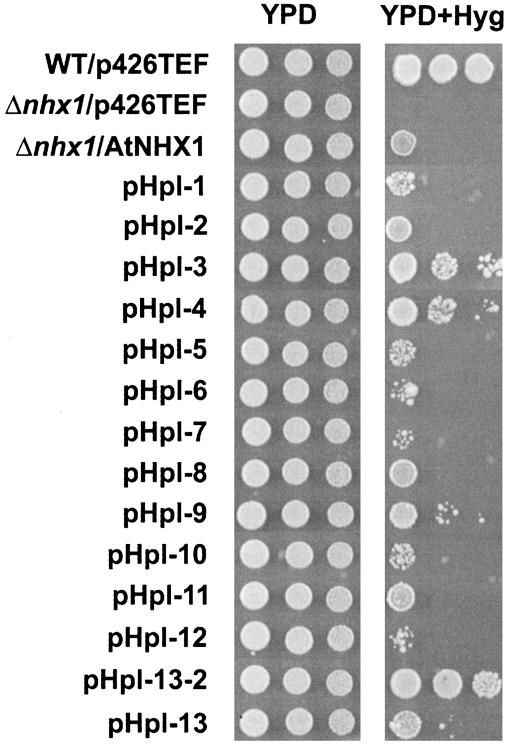
To test whether the tagged proteins were functional, we performed hygromycin-resistance tests on YPD media containing 30 μg/ml hygromycin (Fig. 2). Hygromycin is a toxic cation, and the hygromycin sensitivity of nhx1 mutants has been shown to correlate with the Na+ sensitivity of nhx1 mutants (9, 26, 27). The yeast strain TY001, with the endogenous nhx1 allele disrupted, showed hygromycin sensitivity relative to wild-type MM476, whereas yeast harboring the full length AtNHX1 cDNA displayed growth, emphasizing that AtNHX1 partially complemented the nhx1 mutation. All pHpl constructs were able to complement the hygromycin-sensitive phenotype of TY001, indicating that the HA-fusion proteins were functionally competent. In some cases, the complementation was weak (i.e., pHpl-6, -7, and -12), and in other cases, the complementation was stronger (i.e., pHpl-3, -4, and -13–2). These results would indicate that in some cases the insertion of the 3xHA tag modified AtNHX1 activity. Although the insertion of 3xHA tags could affect AtNHX1 activity, it is possible that they do not affect AtNHX1 localization and membrane topology, and therefore we performed the protease protection assays for all constructs.
Fig. 2.
Complementation test for hygromycin sensitivity of 3xHA-tagged constructs in S. cerevisiae Δnhx1 strain. The p426-TEF empty vector was introduced into MM476 (WT/p426TEF) and TY001 (Δnhx1/p426TEF). p426-X1 (Δnhx1/AtNHX1) and pHpl constructs (1–13.2) were transformed into TY001 (pHpl-1 to 13.2). Each strain was grown, and serial dilutions were made as described in Materials and Methods. Three microliters of each dilution (starting from 106 cells) were spotted onto YPD medium and incubated 2 days at 30°C, or onto YPD containing 30 μg/ml hygromycin (YPD+Hyg) and incubated 3 days at 30°C.
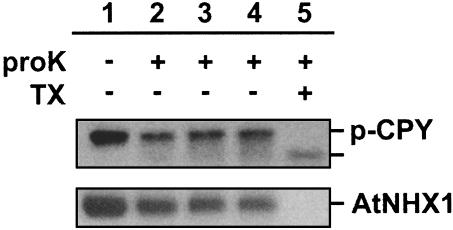
Protease Protection Assays. Vacuoles were subjected to protease protection assays. To monitor vacuolar integrity, we took advantage of the vacuolar protease-deficient strain MM476, which accumulates carboxypeptidase Y pro-form in the vacuolar lumen (22). Vacuoles were incubated with proteinase K in the presence or absence of 1% Triton X-100, and the vacuolar proteins were subjected to Western blot analysis by using an anti-CPY antibody (Fig. 3). When the vacuoles were incubated with proteinase K in the absence of Triton X-100, a 66-kDa band that represents pro-CPY was detected (Fig. 3, lanes 2–4), whereas in the presence of Triton X-100, a band corresponding to processed CPY was detected (Fig. 3, lane 5). A slight decrease in the relative amount of pro-CPY was seen after 30 min of incubation with proteinase K.
Fig. 3.
Vacuolar integrity tests. Intact vacuoles were isolated as described in Materials and Methods. Western blot was performed with anti-CPY antibodies (Upper) or anti-AtNHX1 (Lower). Vacuoles from yeast harboring the pHpl-2 plasmid were incubated with (lanes 2–5) or without (lane 1) 500 μg/ml proteinase K in the presence (lane 5) or absence (lanes 1–4) of Triton X-100. Untreated control vacuoles (lane 1) and vacuoles containing proteinase K and Triton X-100 (lane 5) were incubated for 90 min on ice. Vacuoles containing proteinase K only (lanes 2–4) were incubated for 30 min (lane 2), 60 min (lane 3), and 90 min (lane 4) on ice, respectively.
The intensity of the pro-CPY band, however, did not decrease with incubation times of 60 and 90 min. Thus, we believe that the decrease in pro-CPY was caused by extravacuolar pro-CPY that probably leaked out during the preparation of the vacuoles. These results indicated that the vacuolar membrane was properly sealed for up to 90 min. AtNHX1 (≈48 kDa) was not digested completely either by incubation with up to 1 mg/ml proteinase K in the presence or absence of Ca2+ or with incubation times of up to 2 h (not shown). These results indicate that AtNHX1, at least partially, is protease resistant. Although the digestion was incomplete, incubation with proteinase K for 1 h digested approximately half the amount of AtNHX1 (Fig. 3). In addition, incubation for 1 h resulted in similar band patterns as those seen after 90 min (data not shown). Table 1 shows the expected molecular masses of the resulting HA-tagged peptides, which had been calculated on the basis of the hydropathy plot (Fig. 1).
Table 1. Predicted molecular masses of polypeptides released by protease protection assays.
| Construct | Predicted molecular mass, kDa | Region included |
|---|---|---|
| pHpl-1 (N terminus) | 7.4 | TM1 |
| pHpll-2 (42E)* | 8.4 | TM1—2 |
| pHpl-3 (70K)* | 8.5 | TM2—3 |
| pHpl-4 (99K)* | 9.3 | TM3—4 |
| pHpl-5 (138I)* | 9.2 (17.0)† | TM4—5 (TM4—7)† |
| pHpl-6 (169E)* | 7.8 (17.0)† | TM5—6 (TM4—7)† |
| pHpl-7 (202H)* | 10.0 (17.0)† | TM6—7 (TM4—7)† |
| pHpl-8 (246S)* | 10.3 | TM7—8 |
| pHpl-9 (291V)* | 10.7 | TM8—9 |
| pHpl-10 (328W)* | 9.4 | TM9—10 |
| pHpl-11 (375I)* | 9.1 (14.0)† | TM10—11 (TM10—12)† |
| pHpl-12 (402F)* | 9.4 (14.0)† | TM11—12 (TM10—12)† |
| pHpl-13.2 (441S)* | 16.7 | C terminus hydrophilic region |
| pHpl-13 (C terminus) | 16.7 | C terminus hydrophilic region |
All of the predicted molecular masses were calculated on the basis of the hydrophobicity plot (see Fig. 1). TM, putative transmembrane domain.
Position of amino acid just before the 3xHA tag is inserted
Molecular masses when the indicated regions face the vacuolar lumen
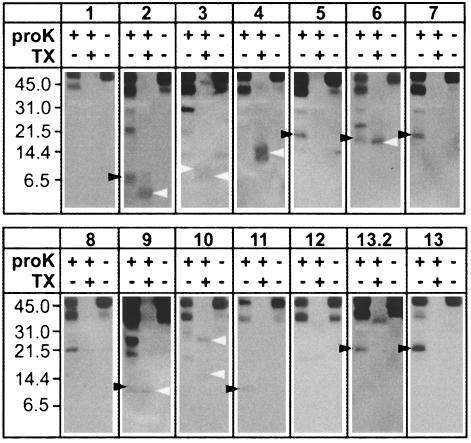
Membrane Topology of AtNHX1 in S. cerevisiae. Our experiments are based on the following rationale: if a particular hydrophilic region of the protein is facing the outside of the vacuole (i.e., the cytosol), then the HA tag will not be detected when the vacuoles are treated with proteinase K in the presence or absence of Triton X-100. On the other hand, if the protein region is facing the vacuolar lumen, the HA tag will be detected only when the vacuoles are treated with proteinase K in the absence of Triton X-100. Because the HA tag is highly hydrophilic, the detection of a tagged polypeptide in the presence of Triton X-100 would indicate that the polypeptide is protected from the protease, either by the formation of an intramolecular complex or on account of the interaction between AtNHX1 with another protein(s). If a band is detected both in the absence and presence of detergent, it would indicate nonspecific binding of the antibodies. Additionally, vacuoles from the yeast strain harboring p426-TEF vector did not show any band on Western blots (data not shown). Therefore, all the bands seen in Fig. 4 originated from 3xHA-tagged AtNHX1. When vacuoles containing 3xHA-tagged AtNHX1 were treated with proteinase K in the absence of detergent, polypeptides of molecular masses of ≈48 and ≈40 kDa were detected for all strains, representing the residual intact and partially digested AtNHX1, respectively (Fig. 4).
Fig. 4.
Protease protection assays. S. cerevisiae MM476 cells were transformed with the indicated plasmids (see Fig. 1), and purified vacuoles were treated with proteinase K in the presence or absence of Triton X-100. Western blot was probed with anti-HA antibodies as described in Materials and Methods. Relative molecular masses are indicated (Left). The molecular masses of different polypeptides (indicated by arrowheads) are discussed in the text. Results are representative of at least three independent experiments.
The vacuoles from the strains containing pHpl-1, 4, 8, 10, and 12 did not show bands with predicted molecular masses after the treatment with proteinase K (Fig. 4 and Table 1). These results indicated that these regions are facing the outside of the vacuole (i.e., cytosol). The vacuoles from strains containing pHpl-3 showed a 10-kDa band in agreement with the prediction (9.3 kDa); the same band, however, was also observed for the untreated control. This result suggested that Hpl-3 is facing the cytosol (Fig. 4). Treatment with protease in the absence of Triton X-100 resulted in bands with similar mass to the prediction (Table 1) in the strain harboring pHpl-2, 5, 7, and 11 (Fig. 4 and Table 1). These observations indicated that Hpl-2, 5, 7, and 11 are facing the vacuolar lumen. Interestingly, in vacuoles from the strain containing pHpl-2, an ≈4-kDa band was observed in the presence of detergent. The molecular mass observed (4 kDa) was close to the sum of Hpl-2 (1 kDa) and 3xHA tag (3 kDa). These observations might indicate that Hpl-2 is protected from proteolysis probably via binding with other vacuolar protein(s). The treatment of vacuoles harboring pHpl-3 and -4 with proteinase K in 1% Triton X-100 also resulted in the appearance of bands (Fig. 4). These results would suggest that these regions were protected from proteolysis either through binding to other molecules or by not being completely exposed to the cytosol. Vacuoles from the strain containing pHpl-6 displayed bands of molecular masses of ≈16 and 15 kDa in the absence and presence of Triton X-100, respectively. Because the predicted molecular mass from the polypeptide comprising the region between 107F and 237I (region spanning TM4–TM7) is 16.7 kDa (Table 1), these results indicated that this region is vacuolar luminal. The different band pattern observed for the strain containing pHpl-6 with pHpl-5 and -7 could be the result of the formation of an intramolecular complex. Vacuoles from the strain containing pHpl-9 showed two bands (9 and 10 kDa, respectively), similar to the predicted molecular mass (9.4 kDa) after proteinase K treatment. Although a 10-kDa band also appeared in the presence of Triton X-100, no band corresponding to the upper band (indicated by a filled arrowhead in Fig. 4) was observed. This indicated that Hpl-9 is facing the vacuolar lumen.
Vacuoles containing pHpl-13 displayed an ≈22-kDa band when treated with proteinase K, and no bands were detected in the presence of detergent. Because Hpl-12 is facing the cytosol, and the C terminus portion of the AtNHX1 is highly hydrophilic, it is likely that Hpl-13 is facing the vacuolar lumen. To confirm the orientation of the C terminus, we introduced a 3xHA tag next to the predicted TM12 (pHpl-13.2). The treatment of vacuoles harboring pHpl-13.2 with proteinase K displayed the same pattern observed with Hpl-13, indicating that the entire C terminus region of AtNHX1 resides in the vacuolar lumen. The difference between the molecular mass of the observed band (≈22 kDa) and the predicted band (≈16.7 kDa) is probably due to changes in the mobility of the HA-tagged polypeptides in the gel, because the mobility of the AtNHX1 tagged at the C (Hpl-13) or N terminus (Hpl-1) was slower than AtNHX-1 tagged internally in SDS/PAGE (Fig. 4). It has been reported that Nhx1 of S. cerevisiae is glycosylated on the C terminus region at 515N and 550N (28). Although no apparent similarity between AtNHX1 and ScNhx1 has been observed in these regions, it is possible that the larger molecular masses detected in Hpl-13 and -13.2 are due to glycosylation in the C-terminal hydrophilic region.
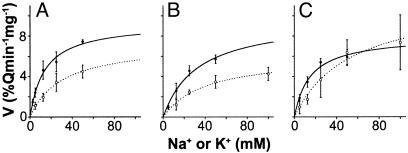
Transport Assays. It has been shown that the activity of H+-coupled antiporters can be affected by modifications at the C or N terminus (29–32). To test whether the C or N terminus of AtNHX1 plays a role in regulating antiporter activity, we measured the H+-coupled cation transport activity in vacuoles containing the full length AtNHX1 and the N or C terminus truncated forms of AtNHX1 (Fig. 5 and Table 2). The three forms of AtNHX1 displayed both Na+- and K+-coupled H+ transport activity. In all cases, this transport showed Michaelis–Menten-type saturation kinetics. Although differences in expression efficiency leading to different amounts of AtNHX1 in the yeast strain preclude direct comparisons of either Na+/H+ or K+/H+ transport of each form to the others, the comparison of the ratio of Na+ to K+ transport relieves this limitation. The deletion of the N terminus of AtNHX1 did not significantly alter the apparent Vmax of the Na+/H+ and the K+/H+ transport (Fig. 5 and Table 2). On the other hand, the deletion of the C terminus of AtNHX1 resulted in significantly higher Na+/H+ and lower K+/H+ transport rates, with a 2-fold increase in the ratio of Na+ to K+ transport.
Fig. 5.
Transport assays. Initial rates of cation-dependent H+ movement were assayed by measuring the initial rates of fluorescence quench recovery after addition of Na+- or K+-gluconate (see Materials and Methods). Solid lines with filled circles are fitted curves for rates of K+ transport, whereas dashed lines with open circles are fitted curves for Na+ transport. Curves were fitted for each of unmodified AtNHX1 (A), N terminus truncation of AtNHX1, Ndl (B), and C terminus truncation of AtNHX1, Cdl (C). Each data point is the mean ± SD (n ≥ 3).
Table 2. Kinetics Na+/H+ and K+/H+ exchange of vacuoles harboring unmodified AtNHX1, N terminus truncation of AtNHX1 (Ndl), and C terminus truncation of AtNHX1 (Cdl).
| AtNHX1 | Ndl | Cdl | |
|---|---|---|---|
| Vmax (Na+) | 7.3 ± 0.8 | 6.1 ± 0.6 | 11.7 ± 1.9 |
| Vmax (K+) | 9.3 ± 0.7 | 9.7 ± 1.6 | 7.9 ± 0.7 |
| Na+/K+ (Vmax) | 0.78 | 0.63 | 1.48 |
| Km (Na+) | 30 ± 6.6 | 40 ± 9.4 | 53 ± 1.9 |
| Km (K+) | 14 ± 2.8 | 33 ± 12.1 | 15 ± 4.3 |
Apparent Vmax and Km values were calculated from data in Fig. 5 by using KALEIDEGRAPH. Values are mean ± SD (n ≥ 3).
Discussion
We conducted an analysis of the topology of AtNHX1, an Arabidopsis thaliana vacuolar Na+/H+ antiporter by using heterologous expression in yeast. The expression of AtNHX1 cDNA is able to complement yeast nhx1 null mutants (9, 10, 26), suggesting that the AtNHX1 protein is localized in endosomal compartments and that it folds properly in the yeast vacuoles. Previous studies have shown that the ScNhx1p::GFP fusion protein is sequestered in the prevacuolar compartment in yeast (33). Although there is no additional evidence on the specific localization of AtNHX1 in yeast, the colocalization of AtNHX1 and Vph1, the 100-kDa subunit of the vacuolar H+-ATPase strongly suggests that AtNHX1 is a vacuolar protein. There is a possibility that the V–ATPase complex is also localized in prevacuolar compartments, in view of reports of the comigration in sucrose gradients of the Vph1 with the prevacuolar marker protein Pep12 (33, 34). Although we have used intact vacuoles for our protease protection assays, we cannot rule out the presence of prevacuoles in our preparations. All the HA-tagged proteins localized in the vacuolar fractions, indicating that all tagged proteins were present in the yeast vacuoles and the prevacuolar compartment, or both.
All Hpl constructs were able to complement, to a greater or a lesser extent, the nhx1 null mutant, raising the possibility that, although the insertion of the HA tag did not affect the membrane topology of the antiporter, it modified AtNHX1 function.
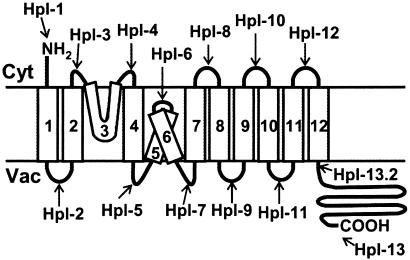
On the basis of the results of the protease protection assays, we propose a topological model of AtNHX1 (Fig. 6). The overall structure of AtNHX1 is similar to, but distinct from, NHE1 or any known Na+/H+ antiporter. It is comprised of nine transmembrane domains, with a hydrophilic C-terminal domain and three hydrophobic regions that do not appear to span the tonoplast membrane but yet appear to be membrane-associated. It should be noted that TM3 contains a putative amiloridebinding domain (FFIYLLPPI), which is hypothesized to span the membrane among vertebrate Na+/H+ antiporters (15). Also, TM5 and -6 of AtNHX1 correspond to TM6 and -7 of NHE1, are highly conserved among the NHE isoforms identified to date, and are considered critical for antiporter transport activity (2, 15, 35, 36). Because these hydrophobic regions are thought to form membrane-spanning helices and contain important residues for Na+ binding, it has been suggested that these regions are involved in the formation of a pore structure for cation transport in NHE1. It is interesting that these regions of AtNHX1 (i.e., TM5 and -6) do not appear to be transmembrane segments as assayed by protease protection. Because there is very little structure/function information for AtNHX1, it might be too early to assume that these hydrophobic regions of AtNHX1 play a similar role as those of NHE1. It is possible, however, that TM3 and TM5–6 comprise the cytosolic (upper) and vacuolar (lower) structure of the ionic pore, respectively.
Fig. 6.
Proposed topological model of AtNHX1. The boxes indicate putative transmembrane domains, and the arrows (Hpl-1 to -13.2) indicate the positions of 3xHA-tag insertion in pHpl-1 to -13.2, respectively.
A comparison of the topology of NHE1 (37) with our model for AtNHX1 raises several important points. Despite the high similarity of the transmembrane domains, the direction of ion transport for these antiporters is reversed: NHE1 mediates Na+ influx to cytosol and H+ efflux from cytosol, whereas AtNHX1 mediates Na+ influx from cytosol and H+ efflux to cytosol. The orientation of TM1, -2, and -10–12 of AtNHX1 (corresponding to TM2, -3, -11, and -13 of NHE1, respectively) appears to be similar to that of NHE1, whereas TM4, -7, and -8 of AtNHX1 (TM5, -8, and -9 of NHE1) are embedded in the opposite direction into the membrane. Also, as noted above, TM3 and -5–6 do not span the membrane, suggesting that the orientation of these regions may determine the direction of ion movement. In NHE1, it has been shown that the C-terminal hydrophilic region is cytosolic, and that it participates in the regulation of Na+/H+ antiporter activity (2). However, our results indicate that almost the entire C-terminal hydrophilic region of AtNHX1 resides in the vacuolar lumen. It has been suggested by others (28) that the yeast NHX1 C-terminal domain is at least partially exposed to the vacuolar lumen, because the Asp residues at 515 and 550 of the C-terminal hydrophobic region had been shown to be glycosylated. Although there is no homologous sequence in the corresponding region of AtNHX1, it is possible that AtNHX1 has a similar conformation, because AtNHX1 is able to complement NHX1 activity. In addition, AtNHX1 also runs as multiple bands (similar to NHX1) on Western blots, suggesting that AtNHX1 is also glycosylated in yeast cells (data not shown; ref. 28). Notably, the protease protection assay with the strains harboring C terminus HA tags showed a larger band in SDS/PAGE than that predicted by hydropathy analysis (Fig. 4, 13.2 and 13; Table 1), indicating that the C-terminal portion of AtNHX1 may be glycosylated or otherwise modified.
It is noteworthy that some HA-tagged polypeptides appeared protected from protease K even in the presence of 1% Triton X-100. This resistance suggests that these regions are protected, presumably via interaction(s) with other protein(s) or through interaction with other region(s) of AtNHX1. The occurrences of these protease-resistant bands are not likely the artifacts of HA-tag insertion, because we did not see any protease-resistant bands for Hpl-1, -5, -7, -8, -11, and -13. It has been reported that vacuoles from budding yeast are covered by actin, which mediates vacuolar fusion (38). It might be possible that actins cover the vacuole and prevent access to the extravacuolar loops (Hpl-3, -4, -10, and -12) of AtNHX1 by proteinase K. Interestingly, yeast NHX1 (VPS44) is required for endosomal protein trafficking, and substitution of acidic amino acids in TM5 and -6 (TM6 and -7 in NHE1), which are presumably important for Na+/H+ antiporter activity, resulted in an impaired protein trafficking phenotype (18). It has also been suggested that in mammalian cells, NHE1 interacts with the cytoskeleton (39), and that the interaction with both actin and ion transport activity is needed for cell migration (40). It might be possible that protein trafficking in yeast, mediated by NHX1, undergoes a similar process to that seen in human cell migration, and that a similar interaction could occur with AtNHX1 expressed in yeast cells. There are a few precedents for the alteration of transport characteristics for cation/H+ exchange due to modification of the transporter N or C terminus. In cyanobacteria, the deletion of parts of the C terminus of the plasma membrane Na+/H+ antiporter from Synechocystis inhibited the antiporter activity (29), and replacement of the Synechocystis Na+/H+ antiporter C terminus with that from Aphanothece halophytica resulted in altered ion specificity. In the human Na+/H+ antiporter NHE1, the deletion of the C terminus altered the sensitivity of the antiporter to cytosolic pH (31). Plant vacuolar Ca2+/H+ antiporters (CAX) appear to be regulated by an autoinhibitory domain at their N terminus (32). Although in these cases alteration of transport could be interpreted with respect to the role of particular residues in intra- and intermolecular interactions, here we can bring to the interpretation of transport activity the context of AtNHX1 topology. Among a set of site-directed and deletion mutations, the two mutant forms of AtNHX1 selected for investigating transport characteristics in this paper were the N and C terminus truncated forms. In this heterologous expression and assay system, the deletion of the N terminus of AtNHX1 resulted in a small reduction of Na+/H+ transport and a small increase in K+/H+ transport; these differences, however, were not statistically significant. The deletion of the hydrophilic C terminus, which appears to reside in the vacuolar lumen, resulted in a dramatic increase in the relative rate of Na+/H+ transport. The ratio of Vmax (Na+)/Vmax (K+) was double that of the unmodified AtNHX1. The altered ratio resulted from a relatively small decrease in K+/H+ transport with a large increase in Na+/H+ transport.
In summary, on the basis of our protease protection studies, we propose a topological model of the vacuolar Na+/H+ antiporter with the N and C termini of AtNHX1 located in the cytosol and the vacuolar lumen, respectively. This model provides the basis for the study of the structure/function relationship of AtNHX1. Our transport studies demonstrate the modulation of antiporter cation selectivity by the C terminus of the protein. The vacuolar localization of the C terminus of the AtNHX1, the regulation of the antiporter selectivity by its C terminus, together with the regulation of the antiporter cation affinity by vacuolar pH (41), suggest the existence of vacuolar regulatory mechanisms of the antiporter activity.
Supplementary Material
Acknowledgments
We thank Helena Garcês for technical support. This work was supported by National Science Foundation Grant IBN-0110622 and the Will W. Lester Endowment from the University of California.
Abbreviations: HA, hemagglutinin; CPY, carboxypeptidase Y.
References
- 1.Orlowski, J. & Grinstein, S. (1997) J. Biol. Chem. 272, 22373-22376. [DOI] [PubMed] [Google Scholar]
- 2.Putney, L. K., Denker, S. P. & Barber, D. L. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 527-552. [DOI] [PubMed] [Google Scholar]
- 3.Padan, E., Venturi, M., Gerchman, Y. & Dover, N. (2001) Biochem. Biophys. Acta 1505, 144-157. [DOI] [PubMed] [Google Scholar]
- 4.Ivey, D. M., Guffanti, A. A., Zemsky, J., Pinner, E., Karpel, R., Padan, E., Schuldiner, S. & Krulwich, T. A. (1993) J. Biol. Chem. 268, 11296-11303. [PubMed] [Google Scholar]
- 5.Apse, M. P., Aharon, G. S., Snedden, W. A. & Blumwald, E. (1999) Science 285, 1256-1258. [DOI] [PubMed] [Google Scholar]
- 6.Blumwald, E. (1987) Physiol. Plant. 69, 731-734. [Google Scholar]
- 7.Ward, J. M. (2001) Bioinformatics 17, 560-563. [DOI] [PubMed] [Google Scholar]
- 8.Mäser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K., Sze, H., Talke, I. N., Amtmann, A. & Maathuis, F. J. M., et al. (2001) Plant Physiol. 126, 1646-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokoi, S., Quintero, F. J., Cubero, B., Ruiz, M. T., Bressan, R. A., Hasegawa, P. M. & Pardo, J. M. (2002) Plant J. 30, 529-539. [DOI] [PubMed] [Google Scholar]
- 10.Aharon, G. S., Apse, M. P., Duan, S., Hua, X., Zhang, H.-X. & Blumwald, E. (2003) Plant Soil 253, 245-256. [Google Scholar]
- 11.Bañuelos, M. A., Garciadeblas, B., Cubero, B. & Rodoríguez-Navarro, A. (2002) Plant Physiol. 130, 784-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, H.-X. & Blumwald, E. (2001) Nat. Biotechnol. 19, 765-768. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi, T., Fukada-Tanaka, S., Inagaki, Y., Saito, N., Yonekura-Sakakibara, K., Tanaka, Y., Kusumi, T. & Iida, S. (2001) Plant Cell Physiol. 42, 451-461. [DOI] [PubMed] [Google Scholar]
- 14.Dibrov, P., Young, P. G. & Fliegel, L. (1998) Biochemistry 36, 8682-8288. [DOI] [PubMed] [Google Scholar]
- 15.Wakabayashi, S., Shigekawa, M. & Pouyssegur, J. (1997) Physiol. Rev. 77, 51-74. [DOI] [PubMed] [Google Scholar]
- 16.The Arabidopsis Genome Initiative (2000) Nature 408, 796-815. [DOI] [PubMed] [Google Scholar]
- 17.Longtine, M. S., Mckenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P. & Pringle, J. R. (1998) Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- 18.Bowers, K., Levi, B. P., Patel, F. I. & Stevens, T. H. (2000) Mol. Biol. Cell 11, 4277-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mumberg, M., Müller, R. & Fink, M. (1995) Gene 156, 119-122. [DOI] [PubMed] [Google Scholar]
- 20.Brunelli, J. P. & Pall, M. L. (1993) Yeast 9, 1309-1318. [DOI] [PubMed] [Google Scholar]
- 21.Ohsumi, Y. & Anraku, Y. (1981) J. Biol. Chem. 256, 2079-2082. [PubMed] [Google Scholar]
- 22.Landolt-Marticorena, C., Kahr, W. H., Zawarinski, P., Correa, J. & Manolson, M. F. (1999) J. Biol. Chem. 274, 26057-26064. [DOI] [PubMed] [Google Scholar]
- 23.Kyte, J. & Doolittle, R. F. (1982) J. Mol. Biol. 157, 105-132. [DOI] [PubMed] [Google Scholar]
- 24.Nagashima, K., Kasai, M., Nagata, S. & Kaziro, Y. (1986) Gene 45, 265-273. [DOI] [PubMed] [Google Scholar]
- 25.Schimaier, F. & Philippsen, P. (1984) EMBO J. 3, 3311-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaxiola, R. A., Rao, R., Sherman, A., Grisafi, P., Alper, S. L. & Fink, G. R. (1999) Proc. Natl. Acad. Sci. USA 96, 1480-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venema, K., Quintero, F. J., Pardo, J. M. & Donaire, J. P. (2002) J. Biol. Chem. 277, 2413-2418. [DOI] [PubMed] [Google Scholar]
- 28.Wells, K. M. & Rao, R. (2001) J. Biol. Chem. 276, 3401-3407. [DOI] [PubMed] [Google Scholar]
- 29.Hamada, A., Hibino, T., Nakamura, T. & Takabe, T. (2001) Plant Physiol. 125, 437-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waditee, R., Hibino, T., Tanaka, Y., Nakamura, T., Incharoensakdi, A. & Takabe, T. (2001) J. Biol. Chem. 276, 36931-36938. [DOI] [PubMed] [Google Scholar]
- 31.Wakabayashi, S., Fafournoux, P., Sardet, C. & Pouyssegur, J. (1992) Proc. Natl. Acad. Sci. USA 89, 2424-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pittman, J. K. & Hirschi, K. D. (2001) Plant Physiol. 127, 1020-1029. [PMC free article] [PubMed] [Google Scholar]
- 33.Nass, R. & Rao, R. (1998) J. Biol. Chem. 273, 21054-21060. [DOI] [PubMed] [Google Scholar]
- 34.Bryant, N. J. & Stevens, T. H. (1997) J. Cell Biol. 136, 287-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slepkov, E. & Fliegel, L. (2002) Biochem. Cell Biol. 80, 499-508. [DOI] [PubMed] [Google Scholar]
- 36.Murtazina, R., Booth, B. J., Bullis, B. L., Singh, D. N. & Fliegel, L. (2001) Eur. J. Biochem. 268, 4674-4685. [DOI] [PubMed] [Google Scholar]
- 37.Wakabayashi, S., Pang, T., Su, X. & Shigekawa, M. (2000) J. Biol. Chem. 275, 7942-7949. [DOI] [PubMed] [Google Scholar]
- 38.Eitzen, G., Wang, L., Thorngren, N. & Wickner W. (2002) J. Cell Biol. 158, 669-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denker, S. P., Huang, D. C., Orlowski, J., Furthmayr, H. & Barber, D. L. (2000) Mol. Cell 6, 1425-1436. [DOI] [PubMed] [Google Scholar]
- 40.Denker, S. P. & Barber, D. L. (2002) J. Cell Biol. 159, 1087-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blumwald, E. & Poole, R. J. (1985) Plant Physiol. 78, 163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.