Abstract
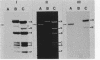
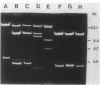
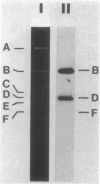
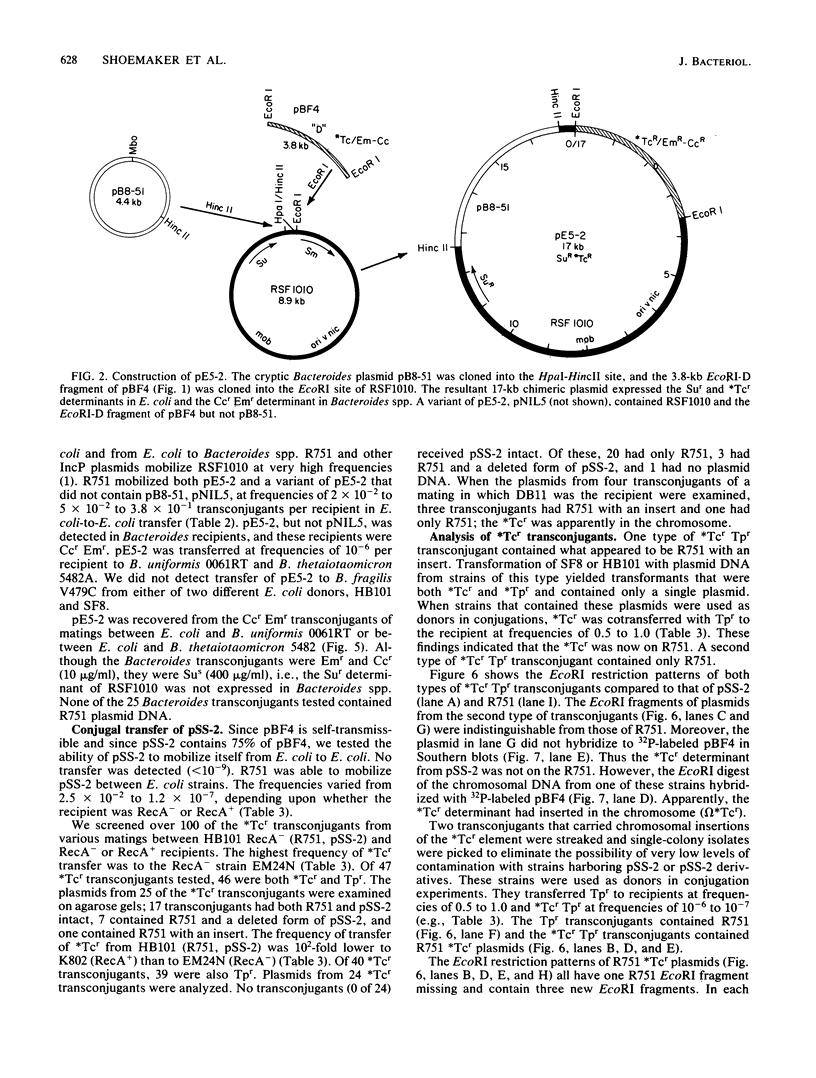
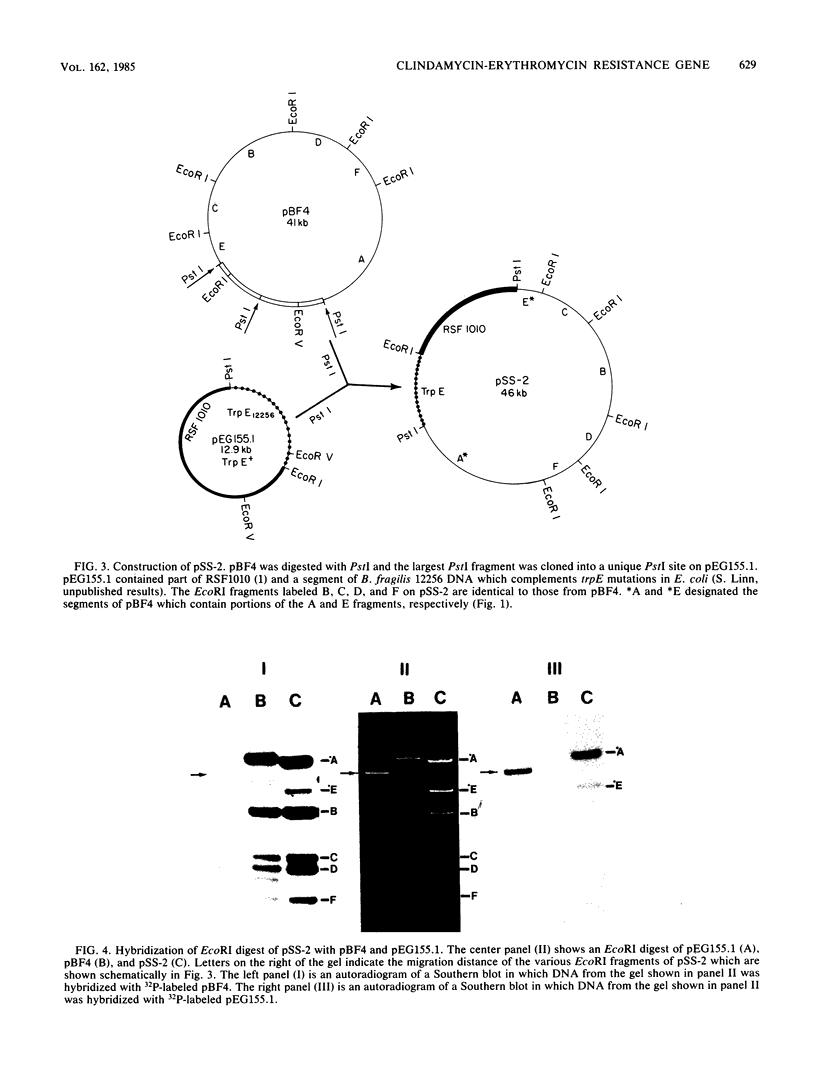
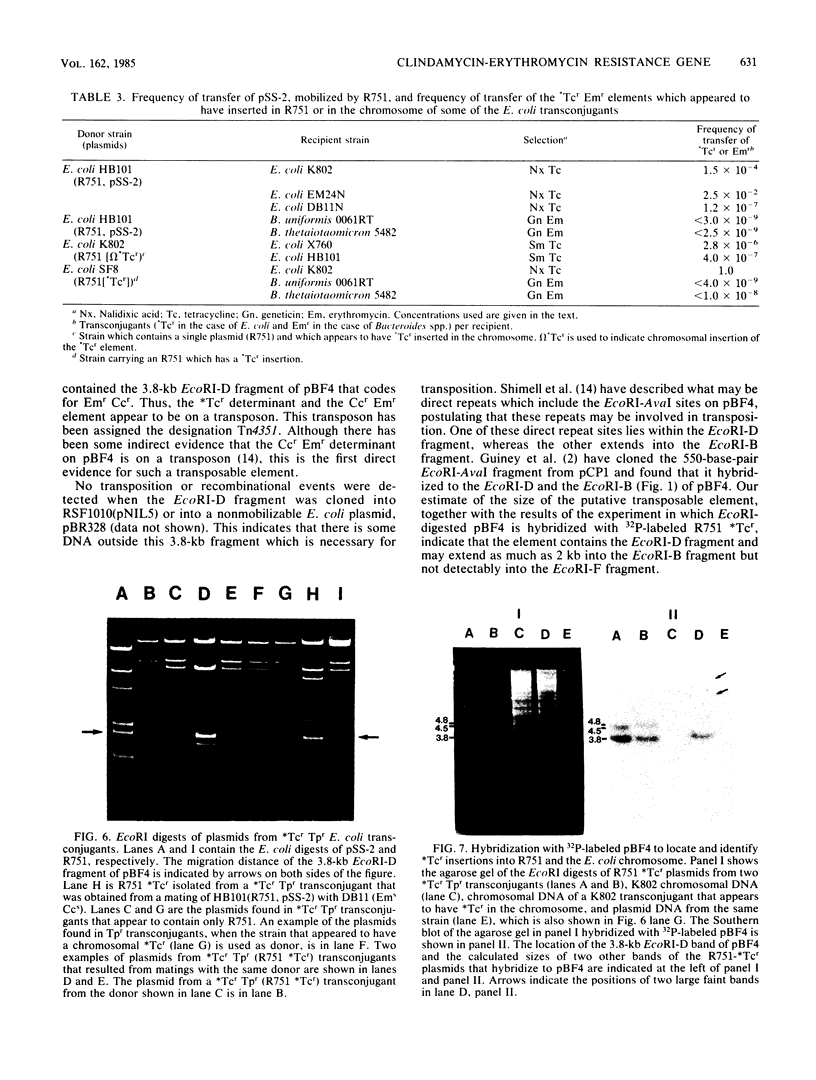
We constructed a shuttle vector, pE5-2, which can replicate in both Bacteroides spp. and Escherichia coli. pE5-2 contains a cryptic Bacteroides plasmid (pB8-51), a 3.8-kilobase (kb) EcoRI-D fragment from the 41-kb Bacteroides fragilis plasmid pBF4, and RSF1010, an IncQ E. coli plasmid. pE5-2 was mobilized by R751, an IncP E. coli plasmid, between E. coli strains with a frequency of 5 X 10(-2) to 3.8 X 10(-1) transconjugants per recipient. R751 also mobilized pE5-2 from E. coli donors to Bacteroides uniformis 0061RT and Bacteroides thetaiotaomicron 5482 with a frequency of 0.9 X 10(-6) to 2.5 X 10(-6). The Bacteroides transconjugants contained only pE5-2 and were resistant to clindamycin and erythromycin. Thus, the gene for clindamycin and erythromycin resistance must be located within the Eco RI-D fragment of BF4. A second recombinant plasmid, pSS-2, which contained 33 kb of pBF4 (including the EcoRI-D fragment and contiguous regions) could also be mobilized by R751 between E. coli strains. In some transconjugants, a 5.5-kb (+/- 0.3 kb) segment of the pBF4 portion of pSS2 was inserted into one of several sites on R751. In some other transconjugants this same 5.5-kb segment was integrated into the E. coli chromosome. This segment could transfer a second time onto R751. Transfer was RecA independent. The transferred segment included the entire EcoRI-D fragment, and thus the clindamycin-erythromycin resistance determinant, from pBF4.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Hasegawa P., Davis C. E. Plasmid transfer from Escherichia coli to Bacteroides fragilis: differential expression of antibiotic resistance phenotypes. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7203–7206. doi: 10.1073/pnas.81.22.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Jr, Hasegawa P., Davis C. E. Expression in Escherichia coli of cryptic tetracycline resistance genes from bacteroides R plasmids. Plasmid. 1984 May;11(3):248–252. doi: 10.1016/0147-619x(84)90031-3. [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Jr, Hasegawa P., Davis C. E. Homology between clindamycin resistance plasmids in Bacteroides. Plasmid. 1984 May;11(3):268–271. doi: 10.1016/0147-619x(84)90035-0. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy M. H., Tally F. P. Mechanisms of drug-resistance transfer in Bacteroides fragilis. J Antimicrob Chemother. 1981 Dec;8 (Suppl 500):59–75. doi: 10.1093/jac/8.suppl_d.59. [DOI] [PubMed] [Google Scholar]
- Marsh P. K., Malamy M. H., Shimell M. J., Tally F. P. Sequence homology of clindamycin resistance determinants in clinical isolates of Bacteroides spp. Antimicrob Agents Chemother. 1983 May;23(5):726–730. doi: 10.1128/aac.23.5.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays T. D., Smith C. J., Welch R. A., Delfini C., Macrina F. L. Novel antibiotic resistance transfer in Bacteroides. Antimicrob Agents Chemother. 1982 Jan;21(1):110–118. doi: 10.1128/aac.21.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. J., Shapiro J. A. Genetic organization of the broad-host-range IncP-1 plasmid R751. J Bacteriol. 1980 Sep;143(3):1362–1373. doi: 10.1128/jb.143.3.1362-1373.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privitera G., Dublanchet A., Sebald M. Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J Infect Dis. 1979 Jan;139(1):97–101. doi: 10.1093/infdis/139.1.97. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Shimell M. J., Smith C. J., Tally F. P., Macrina F. L., Malamy M. H. Hybridization studies reveal homologies between pBF4 and pBFTM10, Two clindamycin-erythromycin resistance transfer plasmids of Bacteroides fragilis. J Bacteriol. 1982 Nov;152(2):950–953. doi: 10.1128/jb.152.2.950-953.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Macrina F. L. Large transmissible clindamycin resistance plasmid in Bacteroides ovatus. J Bacteriol. 1984 May;158(2):739–741. doi: 10.1128/jb.158.2.739-741.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Welch R. A., Macrina F. L. Two independent conjugal transfer systems operating in Bacteroides fragilis V479-1. J Bacteriol. 1982 Jul;151(1):281–287. doi: 10.1128/jb.151.1.281-287.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tally F. P., Snydman D. R., Shimell M. J., Malamy M. H. Characterization of pBFTM10, a clindamycin-erythromycin resistance transfer factor from Bacteroides fragilis. J Bacteriol. 1982 Aug;151(2):686–691. doi: 10.1128/jb.151.2.686-691.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch R. A., Jones K. R., Macrina F. L. Transferable lincosamide-macrolide resistance in Bacteroides. Plasmid. 1979 Apr;2(2):261–268. doi: 10.1016/0147-619x(79)90044-1. [DOI] [PubMed] [Google Scholar]
- Welch R. A., Macrina F. L. Physical characterization of Bacteroides fragilis R plasmid pBF4. J Bacteriol. 1981 Feb;145(2):867–872. doi: 10.1128/jb.145.2.867-872.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]