Abstract
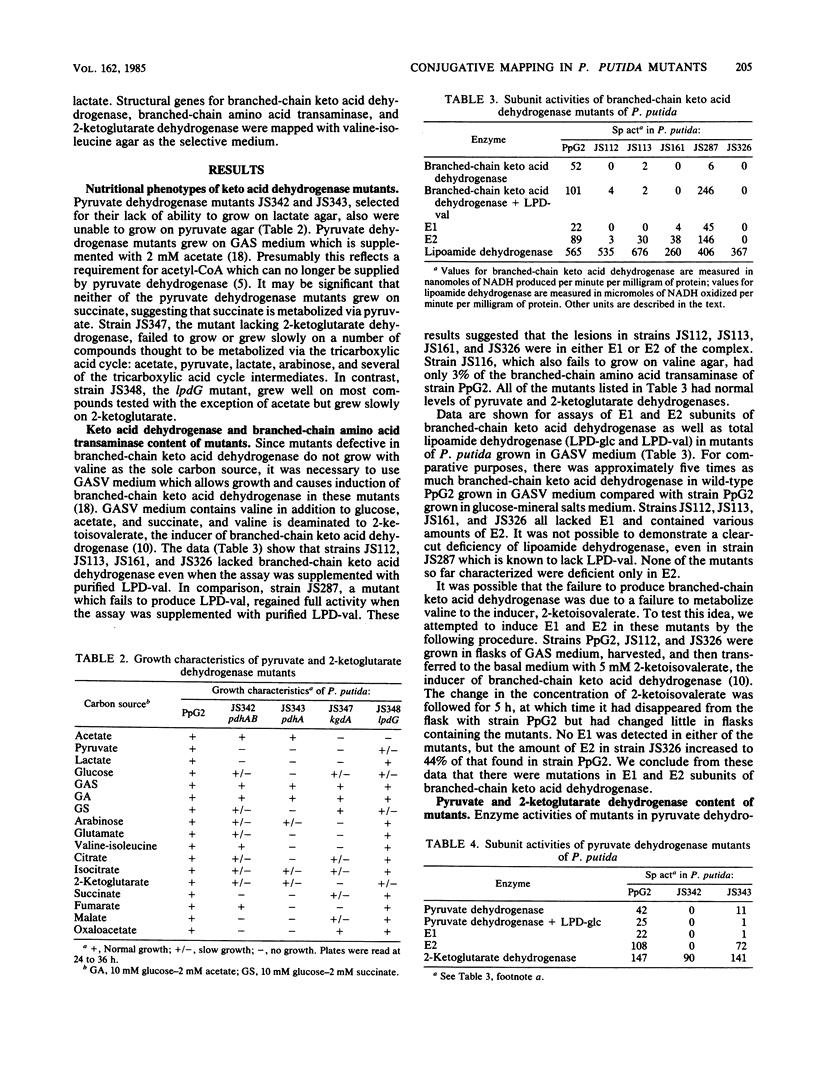
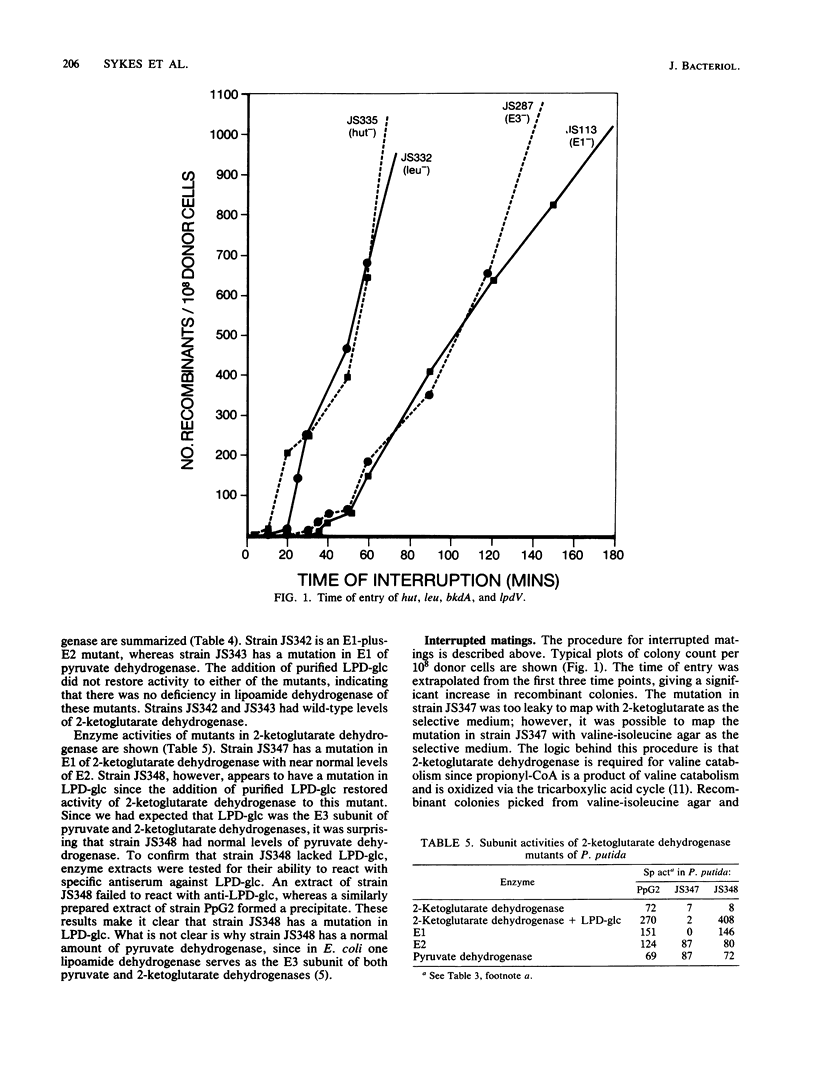
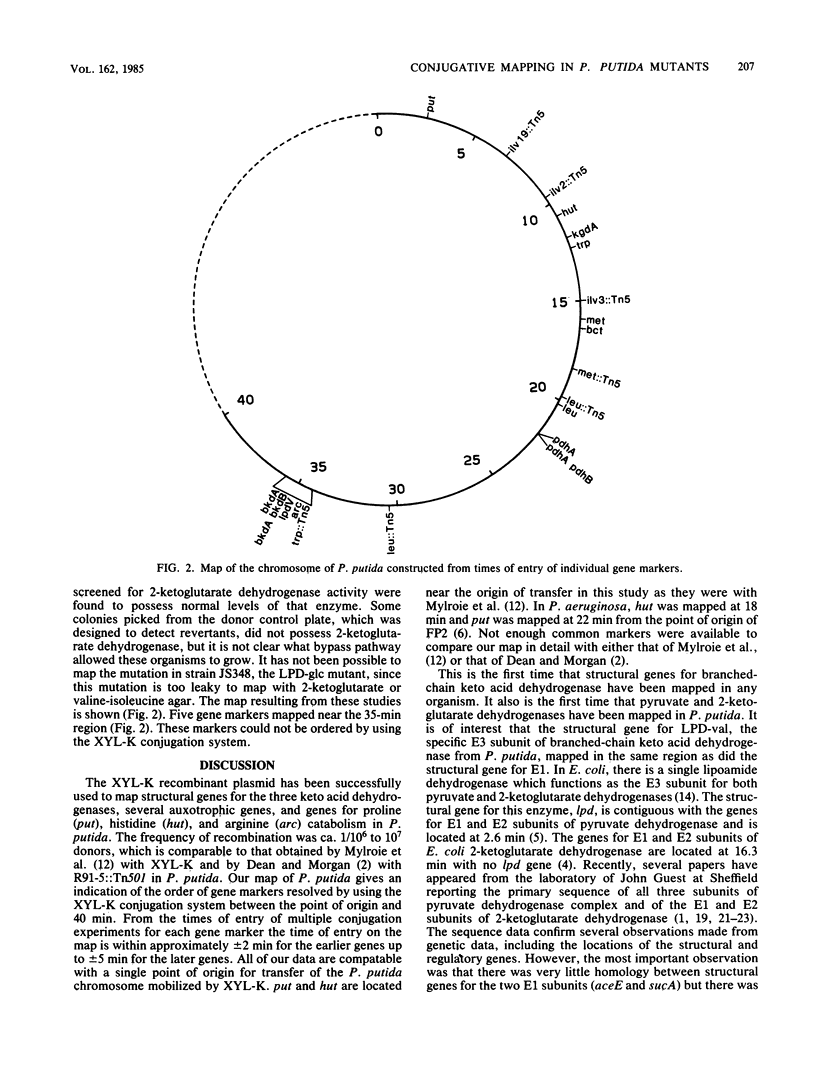
Branched-chain keto acid dehydrogenase, an enzyme in the common pathway of branched-chain amino acid catabolism of Pseudomonas putida, is a multienzyme complex which catalyzes the oxidative decarboxylation of branched-chain keto acids. The objective of the present study was to isolate strains with mutations of this and other keto acid dehydrogenases and to map the location of the mutations on the chromosome of P. putida. Several strains with mutations of branched-chain keto acid dehydrogenase, two pyruvate and two 2-ketoglutarate dehydrogenase, were isolated, and the defective subunits were identified by biochemical analysis. By using a recombinant XYL-K plasmid to mediate conjugation, these mutations were mapped in relation to a series of auxotrophic and other catabolic mutations. The last time of entry recorded was at approximately 35 min, and the data were consistent with a single point of entry. Branched-chain keto acid dehydrogenase mutations affecting E1, E1 plus E2, and E3 subunits mapped at approximately 35 min. One other strain affected in the common pathway was deficient in branched-chain amino acid transaminase, and the mutation was mapped at 16 min. The mutations in the two pyruvate dehydrogenase mutants, one deficient in E1 and the other deficient in E1 plus E2, mapped at 22 minutes. The 2-ketoglutarate dehydrogenase mutation affecting the E1 subunit mapped at 12 minutes. A 2-ketoglutarate dehydrogenase mutant deficient in E3 was isolated, but the mutation proved too leaky to map.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Darlison M. G., Spencer M. E., Guest J. R. Nucleotide sequence of the sucA gene encoding the 2-oxoglutarate dehydrogenase of Escherichia coli K12. Eur J Biochem. 1984 Jun 1;141(2):351–359. doi: 10.1111/j.1432-1033.1984.tb08199.x. [DOI] [PubMed] [Google Scholar]
- Dean H. F., Morgan A. F. Integration of R91-5::Tn501 into the Pseudomonas putida PPN chromosome and genetic circularity of the chromosomal map. J Bacteriol. 1983 Jan;153(1):485–497. doi: 10.1128/jb.153.1.485-497.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney R., Burns G., Sokatch J. R. Relationship of lipoamide dehydrogenases from Pseudomonas putida to other FAD-linked dehydrogenases. FEBS Lett. 1984 Mar 26;168(2):265–270. doi: 10.1016/0014-5793(84)80259-8. [DOI] [PubMed] [Google Scholar]
- Duggan D. E., Wechsler J. A. An assay for transaminase B enzyme activity in Escherichia coli K-12. Anal Biochem. 1973 Jan;51(1):67–79. doi: 10.1016/0003-2697(73)90453-3. [DOI] [PubMed] [Google Scholar]
- Guest J. R. Aspects of the molecular biology of lipoamide dehydrogenase. Adv Neurol. 1978;21:219–244. [PubMed] [Google Scholar]
- Holloway B. W., Krishnapillai V., Morgan A. F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979 Mar;43(1):73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaseelan K., Guest J. R. Isolation and properties of pyruvate dehydrogenase complex mutants of Pseudomonas aeruginosa PAO. J Gen Microbiol. 1980 Oct;120(2):385–392. doi: 10.1099/00221287-120-2-385. [DOI] [PubMed] [Google Scholar]
- Lowe P. N., Hodgson J. A., Perham R. N. Dual role of a single multienzyme complex in the oxidative decarboxylation of pyruvate and branched-chain 2-oxo acids in Bacillus subtilis. Biochem J. 1983 Oct 1;215(1):133–140. doi: 10.1042/bj2150133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall V. D., Sokatch J. R. Regulation of valine catabolism in Pseudomonas putida. J Bacteriol. 1972 Jun;110(3):1073–1081. doi: 10.1128/jb.110.3.1073-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. R., Marshall V. D., Sokatch J. R., Unger L. Common enzymes of branched-chain amino acid catabolism in Pseudomonas putida. J Bacteriol. 1973 Jul;115(1):198–204. doi: 10.1128/jb.115.1.198-204.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey L. K., Sokatch J. R., Conrad R. S. Branched-chain amino acid catabolism in bacteria. Bacteriol Rev. 1976 Mar;40(1):42–54. doi: 10.1128/br.40.1.42-54.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. IV. Regulation. J Biol Chem. 1966 Aug 25;241(16):3800–3810. [PubMed] [Google Scholar]
- Pettit F. H., Reed L. J. Alpha-keto acid dehydrogenase complexes. 8. Comparison of dihydrolipoyl dehydrogenases from pyruvate and alpha-ketoglutarate dehydrogenase complexes of Escherichia coli. Proc Natl Acad Sci U S A. 1967 Sep;58(3):1126–1130. doi: 10.1073/pnas.58.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REED L. J., KOIKE M., LEVITCH M. E., LEACH F. R. Studies on the nature and reactions of protein-bound lipoic acid. J Biol Chem. 1958 May;232(1):143–158. [PubMed] [Google Scholar]
- Sokatch J. R., Burns G. Oxidation of glycine by Pseudomonas putida requires a specific lipoamide dehydrogenase. Arch Biochem Biophys. 1984 Feb 1;228(2):660–666. doi: 10.1016/0003-9861(84)90036-5. [DOI] [PubMed] [Google Scholar]
- Sokatch J. R., McCully V., Gebrosky J., Sokatch D. J. Isolation of a specific lipoamide dehydrogenase for a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981 Nov;148(2):639–646. doi: 10.1128/jb.148.2.639-646.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokatch J. R., McCully V., Sahm J. G., Reyes-Maguire M. Mutations affecting lipoamide dehydrogenases of Pseudomonas putida. J Bacteriol. 1983 Feb;153(2):969–975. doi: 10.1128/jb.153.2.969-975.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer M. E., Darlison M. G., Stephens P. E., Duckenfield I. K., Guest J. R. Nucleotide sequence of the sucB gene encoding the dihydrolipoamide succinyltransferase of Escherichia coli K12 and homology with the corresponding acetyltransferase. Eur J Biochem. 1984 Jun 1;141(2):361–374. doi: 10.1111/j.1432-1033.1984.tb08200.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the dihydrolipoamide acetyltransferase component. Eur J Biochem. 1983 Jul 1;133(3):481–489. doi: 10.1111/j.1432-1033.1983.tb07490.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the pyruvate dehydrogenase component. Eur J Biochem. 1983 Jun 1;133(1):155–162. doi: 10.1111/j.1432-1033.1983.tb07441.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Lewis H. M., Darlison M. G., Guest J. R. Nucleotide sequence of the lipoamide dehydrogenase gene of Escherichia coli K12. Eur J Biochem. 1983 Oct 3;135(3):519–527. doi: 10.1111/j.1432-1033.1983.tb07683.x. [DOI] [PubMed] [Google Scholar]


